Abstract
Selective movement of proteins between the nucleus and the cytoplasm is a regulatory mechanism exploited extensively by the eukaryotic cell. We have identified the evolutionarily conserved Sac3 protein, which was implicated previously in the regulation of mitosis [Bauer, A. & Kölling, R. (1996) J. Cell Sci. 109, 1575–1583] as a novel mediator of nuclear protein export. We show that Sac3p is localized to the nuclear pore, where it interacts with nucleoporins. Loss of SAC3 function results in a block in nuclear export of a nuclear export signal-containing reporter protein. Our results also demonstrate that SAC3 interacts genetically with the nuclear protein export factors Crm1p/Xpo1p and Yrb2p. Taken together, these data indicate a link between nuclear protein export and transition through the cell cycle.
The nuclear envelope provides a physical mechanism for regulation of numerous events in the eukaryotic cell. The segregation of essential cellular processes such as transcription and translation conferred by the separation of the nucleus and the cytoplasm allows cells to use compartmentalization as a method for controlling critical cellular processes. Efficient use of this regulatory mechanism requires rapid but highly regulated pathways for exchange of information between the nucleus and the cytoplasm. The route for this exchange in all cells is via nuclear pore complexes that form large proteinaceous channels in the double membrane that surrounds the nucleus (1). In addition to the nuclear pore proteins, which are called nucleoporins, a growing number of soluble factors are required to escort substrates through the nuclear pore complex (2). These factors can be divided into two general classes: those that target substrates to the nuclear pore, including the soluble receptors of the importin family (3), and those that are required for the actual translocation through the pore, primarily the small, GTP-binding protein Ran and its binding partner NTF2 (4, 5).
For many years, studies have focused on how entry of proteins into the nucleus can regulate cellular responses to external stimuli. Several recent developments have led researchers to consider the potential impact of selective export of proteins from the nucleus. Approximately 5 years ago, a nuclear export signal (NES) that targets substrate proteins for export from the nucleus was defined (6). This discovery was followed rapidly by the identification of an NES receptor called Xpo1p/Crm1p in Saccharomyces cerevisiae (7, 8) and CRM1 in other eukaryotes (9). The nuclear export receptor is a member of a large family of proteins related to the importin β-subunit of the classical nuclear localization signal (NLS) receptor. Despite these recent advances, the mechanism of nuclear protein export is not understood in detail. Thus far, only one additional soluble factor has been implicated in the process, the Ran-binding protein Yrb2p (10, 11).
Interest in the detailed mechanism of nuclear protein export has been sparked further by several recent studies that demonstrate that nuclear protein export serves to regulate critical cellular transitions. A recent study demonstrated that cyclin B must be exported from the nucleus to be degraded when cells exit mitosis (12, 13). Another study showed that the DNA-repair checkpoint protein Rad25 undergoes a regulated nuclear transport step when cells arrest in response to DNA damage (14, 15). The theme of dynamic localization emerging for a large number of regulatory proteins suggests that compartmentalization is a general mechanism for regulating numerous cellular events. If this is indeed the case, then a number of different pathways for both nuclear import and export should exist to direct this intracellular traffic.
We report here the identification of a nuclear export factor, Sac3p, a protein that previously has been implicated in control of mitosis (16). We identified the S. cerevisiae SAC3 gene in two complementary genetic screens for regulators of mitosis. Surprisingly, our subsequent experiments demonstrate that Sac3p is localized to the nuclear pore, where it interacts with several nucleoporins. Deletion of the SAC3 gene results in cells that are growth-compromised and defective in the export of an NES-containing reporter protein but not in protein import or poly(A)+ RNA export. Our findings support a model in which Sac3p impacts mitosis by controlling nuclear export of a factor that regulates cell cycle progression.
Materials and Methods
Strains, Plasmids, and Chemicals.
DNA manipulations were performed according to standard methods (17), and all media and yeast strains (see Table 1) were prepared by standard procedures (22). Plasmids used for the study are described in Table 2. Chemicals were obtained from Sigma or Fisher Scientific unless otherwise noted.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source/ reference |
|---|---|---|
| ACY193 (FY86) | MATα ura3-52 leu2Δ1 his3Δ200 | (18) |
| ACY137 | MATα cdc23-100 ade2 ade3 his3Δ200 leu2Δ1 lys2 trp1Δ63 ura3-52 | This study |
| ACY147 | ACY137 transformed with pAC39 | This study |
| ACY244 | MATa cdc28-1N ura3-52 leu2Δ1 ade2 | This study |
| ACY247 | MATa ura3 leu2ΔI his3Δ200 MATα ura3 leu2Δ1 his3Δ200 | This study |
| ACY159 | MATα ΔSAC3∷HIS3 ura3-52 leu2Δ1 lys2 his3Δ200 | This study |
| ACY160 | MATα ura3-52 leu2Δ1 lys2 his3Δ200 | This study |
| ACY276 | MATa SAC3∷SAC3-GFP-HIS3 his3Δ200 leu2Δ1 trp1Δ63 ura3-52 | This study |
| ACY311 | MATa ΔRAT2∷HIS3 ura3 leu2 his3Δ200 | (19) |
| ACY374 | MATα ΔYRB2∷HIS3 ura3 leu2Δ1 trp1 his3Δ200 | (20) |
| ACY371 | MATa crm1-2 ura3 leu2 his3 trp1 ade2 | (21) |
| ACY372 | MATa crm1-3 ura3 leu2 his3 trp1 ade2 | (21) |
Table 2.
Plasmids used in this study
| Plasmid | Source |
|---|---|
| pAC24 | URA3 ADE3 plasmid (2μ) (23) |
| pAC39 | SacI–BamHI genomic Cdc23p fragment (4.7 kb) cloned into pAC24 (URA3 ADE3, 2μ) |
| pAC173 | Original YCp50 isolate encoding Sac3p (URA3 CEN) |
| pAC174 | Genomic PstI–PstI fragment (5.8 kb) of pAC173 encoding Sac3p cloned into pRS315 (LEU2 CEN) |
| pAC175 | pAC174 was digested with XhoI, and oligonucleotides encoding the myc epitope were inserted (LEU2 CEN) |
| pAC45 | Nuf2p-GFP fusion plasmid with a unique XhoI site at the ATG of sGFP (S65T, V163A), used for making C-terminal GFP fusion proteins (24) |
| pAC60 | SAC3 was amplified by PCR by using an oligo 5′ of the SAC3 stop codon with a SalI site. The PCR product was cut with SnaBI (329 bp upstream of SAC3 ATG) and SalI and ligated into pAC45 cut with SmaI and XhoI to create SAC3-GFP (URA3, 2μ). |
| pAC176 | pAC60 cut with XhoI and ApaI to liberate a 1.8-kb fragment encoding the C-terminal 321 bp of Sac3p fused to GFP. This fragment was cloned into pAC174 cut with XhoI and ApaI (SAC3-GFP LEU2 CEN). |
| pAC61 | pAC60 was cut with HpaI and StuI to liberate the Sac3p-GFP fusion lacking the first 96 bp of SAC3 (including the ATG). This fragment was cloned into pRS303 (25) to generate a truncated integrating Sac3-GFP construct (HIS3). |
| pRS202 | URA3 plasmid backbone (2μ) for the high-copy library (gift from P. Hieter, Univ. of British Columbia) used to screen for suppressors |
| pAC17 (pPS311) | LEU2 CEN plasmid with a GAL1-10 promoter followed by several unique sites including BamHI and SalI |
| pAC188 | Oligonucleotides with a 5′ BamHI and a 3′ SalI site were used to amplify the SAC3 gene. The PCR product was cloned into pAC17 (LEU2 CEN GAL1-10) |
| PAC342 | Nup49p fused in-frame to GFP (26) |
| pAC403 (pPS892) | PGEX.2TB inserted into pPS293 (GAL1-10 URA3 CEN) (23) |
| PAC404 | Oligonucleotides with a 5′ BamHI and a 3′ SalI site were used to amplify the SAC3 gene. The PCR product was cloned into pAC403 to create an N-terminal GST fusion. |
| pAC485 | Oligonucleotides with a 5′ BamHI and a 3′ SalI site were used to amplify the NTF2 gene. The PCR product was cloned into pAC403 to create an N-terminal GST fusion. |
| pAC361 | Oligonucleotides with a 5′ BamHI and a 3′ SalI site were used to amplify the PRP20 gene. The PCR product was cloned into pAC403 to create an N-terminal GST fusion. |
| pAC213 | NES-NLS-containing GFP reporter protein (7, 10) |
| PAC520 pLDB419 | YAP1∷GFP LEU2 2μ plasmid with sGFP inserted at the NdeI site of YAP1 gene (21) |
Genetic Screens.
The synthetic lethal screen was performed as described (23) by taking advantage of a colony-sectoring assay (27). The parental strain (ACY147) for the screen was a temperature-sensitive allele of CDC23, cdc23–100 (R455P), which was identified in our laboratory. Ten mutants were identified that met all the criteria of the screen. These mutants were crossed to one another to define five distinct complementation groups. Three of these mutants were cloned by complementation. Two were identified by complementation of the sect− phenotype (SDS23 and SAC3), and one was identified by complementation of a temperature-sensitive phenotype (CDC28).
High-copy suppressors of the temperature-sensitive cdc28–1N allele were identified by transforming ACY244 with a pRS202-based high-copy S. cerevisiae genomic library (see Table 2). Transformants were grown on synthetic medium lacking uracil at 37°C to select for those plasmids able to complement the temperature-sensitive phenotype. Suppression by each plasmid (plasmid linkage) was confirmed by rescuing each suppressor plasmid and then retransforming them into the original temperature-sensitive strain.
Deletion of the SAC3 Gene.
A complete deletion of the SAC3 open reading was created by using a previously described PCR-based strategy (23) with the starting strain ACY247. The heterozygous diploid then was sporulated and subjected to tetrad dissection to generate the haploid deletion strain ACY159 (SAC3Δ) and the corresponding wild-type strain ACY160. The deletion was confirmed by PCR and Southern blotting.
Construction/Localization of Sac3-GFP (Green Fluorescent Protein).
The SAC3 reading frame was fused in-frame to GFP (28) to generate pAC60. To examine the localization of Sac3p when expressed at endogenous levels, the SAC3-GFP construct was integrated into the genome such that the GFP fusion protein was the only functional copy of Sac3p. This was accomplished by cloning an N-terminally truncated Sac3-GFP fragment into an integrating vector to create pAC61. The integrating plasmid was linearized within the SAC3 coding region and transformed into the diploid strain ACY247. This diploid was sporulated to generate the haploid ACY276. This integration method results in a duplication at the SAC3 locus, where one copy of Sac3p is N-terminally truncated (lacking the ATG) and the second functional copy is expressed from the endogenous SAC3 promoter and fused in-frame to GFP.
The Sac3-GFP fusion protein was localized either by directly viewing the GFP signal in living cells through a GFP-optimized filter (Chroma Technology, Brattleboro, VT) using an Olympus BX60 epifluorescence microscope equipped with a Photometrics Quantix digital camera or by indirect immunofluorescence using a polyclonal anti-GFP antibody (29).
Indirect Immunofluorescence.
Samples were prepared for indirect immunofluorescence as described (30), with the exception that fixation was in methanol for 6 min once samples were adhered to the polylysine-coated slides. The GFP antibody was used at 1:5,000 dilution, the anti-80-kDa spindle pole body antigen antibody (gift of J. Kilmartin, Medical Research Council, Cambridge, U.K.) was used at 1:1,000, the mAb414 (Berkeley Antibody, Richmond, CA) was used at 1:2,000, and the antitubulin antibody (gift of M. Fuller, Stanford University) was used at 1:100. Secondary antibodies (Jackson ImmunoResearch) (1:1,000 dilution) were anti-rabbit FITC (GFP) or anti-mouse Texas red (80-kDa antigen, mAb414, tubulin).
Protein Interaction Assay.
To test for biochemical interactions between Sac3p and the nucleoporin Nsp1p, a galactose-inducible N-terminal glutathione S-transferase (GST)-Sac3p fusion protein was generated (pAC404). Expression of the GST-Sac3p in wild-type cells (ACY193) was induced by the addition of galactose (2% final concentration), and cells were lysed by glass-bead lysis in PBS containing 5 mM MgCl2 and 0.5% Triton X-100 (PBSMT). Lysates (≈2 mg total protein) were incubated with glutathione-Sepharose beads for 1 hr at 4°C. Beads then were washed two times in PBSMT and two times in PBSM. Bound and unbound samples were analyzed by immunoblotting with anti-Nsp1p (1:5,000 dilution) (gift of M. Stewart, Medical Research Council). Samples also were analyzed with anti-GST antibodies (Santa Cruz Biotechnologies) to demonstrate that the GST-fusion protein was bound to the beads. Control GST fusion proteins include vector that expresses GST alone (pAC403), GST-Ntf2p (pAC485), and GST-Prp20p (pAC361).
Nuclear Export Assay.
The nuclear protein export assay takes advantage of a GFP–reporter construct that contains both an NLS and an NES (7, 10). Wild-type (ACY160) and SAC3Δ (ACY159) cells were transformed with this reporter construct (pAC213, gift of H. Krebber and P. Siwer, Harvard Medical School). Cells expressing the reporter protein were examined by direct fluorescence microscopy.
Nuclear Import Assay and Poly(A)+ RNA Localization.
Indirect immunofluorescence microscopy was used to examine the localization of either an endogenous nuclear protein, Npl3p, or an artificial reporter protein, simian virus 40 (SV40)-NLS invertase. Assays were carried out as described (31). Protein localization was examined in wild-type (ACY160) and SAC3Δ (ACY159) cells grown at 25°C or shifted to 37°C for 3 hr.
Poly(A)+ RNA export was examined as described (31) through the use of an oligo(dT) probe for fluorescence in situ hybridization. Poly(A)+ RNA localization was examined in wild-type (ACY160) and SAC3Δ (ACY159) cells grown at 25°C or shifted to 37°C for 3 hr.
Accession Numbers.
Sac3-related proteins are found in Schizosaccharomyces pombe (SPCC576, GenBank accession no. AL031798), Caenorhabditis elegans [cDNA cm16h1 (32), GenBank accession no. U40933], and humans [KIAA0572 (33), GenBank accession no. AB011144].
Results
Identification of SAC3.
To identify critical regulators of mitotic progression, we carried out two complementary genetic screens. The first screen sought to identify mutants that were lethal in combination with a novel temperature-sensitive allele (cdc23–100) of the essential anaphase-promoting complex (APC)/cyclosome component, CDC23 (34, 35). This screen identified five mutant alleles that are lethal in combination with cdc23–100. Complementation of the synthetic lethal phenotype in conjunction with genetic linkage analysis revealed that three of the mutations were in the SAC3, SDS23, and CDC28 genes. The function of SAC3 has not been delineated, but it was identified first as a suppressor of actin (36) and subsequently implicated in mitotic regulation (16). The SDS23 gene encodes one of two S. cerevisiae homologues (Ssd23p and Sds24p) of the fission yeast Sds23 protein, which genetic studies have implicated in APC/cyclosome regulation (37). CDC28 encodes the cell cycle regulatory, cyclin-dependent kinase in S. cerevisiae (38). The allele of CDC28 identified in the screen is temperature-sensitive and contains a mutation (P250L) identical to the mutation in the previously characterized cdc28–1N allele (39). This allele of CDC28 is unique in that it arrests in mitosis with an elongated spindle and substantial histone H1 kinase activity (38, 40). Most other temperature-sensitive alleles of CDC28 arrest at the G1/S transition with reduced H1 kinase activity (38). The unique phenotype of the cdc28–1N allele along with our reisolation of this previously identified mutation in the synthetic lethal screen led us to carry out a second screen. This screen sought high-copy suppressors of the cdc28–1N mutant allele (ACY244). A number of suppressors were identified, some of which have been implicated directly in the regulation of mitosis [CDC20 (41–43); ESP1 (44, 45); and ZDS1/ZDS2 (46, 47)]. The most striking finding was that the SAC3 gene was obtained as a high-copy suppressor of cdc28–1N. This dual identification of the SAC3 gene in two independent but complementary screens led us to examine the in vivo function of the Sac3 protein.
The Sac3 Protein Is a Member of an Evolutionarily Conserved Family of Proteins.
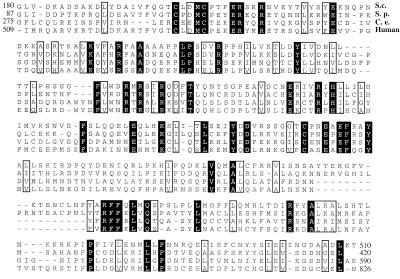
The S. cerevisiae SAC3 gene encodes a 1,293-aa protein of approximately 150-kDa molecular mass. Although the Sac3 protein does not contain any domains of known function, database searches did indicate that Sac3p is a member of an evolutionarily conserved family of proteins (Fig. 1). Proteins of comparable size to Sac3p (≈1,024–1,152 aa) that share a conserved central domain are found in the S. pombe, C. elegans, and human genomes. The presence of related proteins in a variety of higher eukaryotes suggests that Sac3p may serve an evolutionarily conserved function.
Figure 1.
Sequence comparison of Sac3-related proteins. Database searches revealed S. cerevisiae Sac3-related proteins in S. pombe, C. elegans, and humans. A conserved central region of the related proteins is shown. Amino acid residues that are identical in all four species are shaded in black, and those that are similar are contained in the unshaded boxes.
Alterations in the Level of Sac3p Expression Affect Mitotic Spindle Morphology.
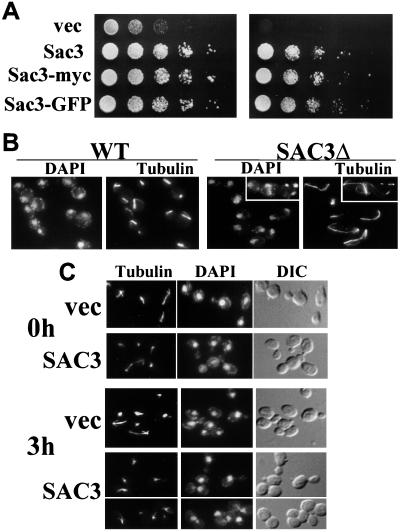
As a first step toward defining the in vivo function of Sac3p, we generated a complete deletion of the SAC3 gene. The SAC3 gene is not essential but cells grow slowly at room temperature (Fig. 2 A Left) and are unable to grow at 37°C (Fig. 2 A Right). This growth defect is rescued by a plasmid-borne copy of SAC3. To assess the function of Sac3p, we examined the morphology of SAC3Δ cells after a shift to 37°C. We found that cells accumulated in mitosis as large-budded cells with extended microtubules (Fig. 2B). This is consistent with a loss of function phenotype previously reported for SAC3 (16). As a complementary experiment, we examined the effect of overexpressing the Sac3 protein from a galactose-inducible promoter. Wild-type cells were transformed either with an inducible SAC3 plasmid (pAC188) or with vector alone (pAC17). Upon induction with galactose, cells expressing Sac3p arrested as large-budded cells (77 ± 7% of cells expressing SAC3 as compared with 24 ± 5% of cells expressing vector alone) with short mitotic spindles (Fig. 2C). Visualization of the spindle pole bodies with a Nuf2p-GFP fusion protein (pAC45) indicated that the spindle pole bodies had duplicated but not separated (data not shown). Thus, depletion of Sac3p and overexpression of Sac3p have opposite effects on the mitotic spindle.
Figure 2.
Levels of Sac3p affect cell cycle progression. (A) The growth of SAC3Δ cells transformed with vector alone (vec), wild-type SAC3 (Sac3), myc-tagged SAC3 (Sac3-myc), or GFP-tagged SAC3 (Sac3-GFP) is shown at 25°C (Left) and 37°C (Right). Growth was assessed by serial dilutions of logarithmically growing cultures. (B) Wild-type and SAC3Δ cells were grown to log phase in yeast extract/peptone/dextrose medium and then shifted to 37°C for 2 hr. Samples were stained with an antitubulin antibody to visualize microtubules and with 4′,6-diamidino-2-phenylindole to visualize DNA. (C) Wild-type cells were transformed with a galactose-inducible SAC3 plasmid (SAC3) or with the empty vector (vec). Cells were grown overnight in minimal medium containing raffinose. At time zero (0 h), expression of Sac3p was induced by the addition of galactose (2% final concentration). After a 3-hr induction (3 h), cells were prepared for indirect immunofluorescence and stained with antitubulin and 4′,6-diamidino-2-phenylindole.
Sac3p Is Associated with the Nuclear Pore.
To gather information about the in vivo function of Sac3p, we determined the intracellular localization of the protein. For these experiments we used a C-terminal Sac3-GFP fusion. The Sac3-GFP fusion protein is functional because it can rescue the slow-growth phenotype of the SAC3Δ strain (see Fig. 2A). Immunoblotting with an anti-GFP antibody confirms the expression of Sac3-GFP as a single band corresponding to the expected size (≈180 kDa) of the fusion protein (data not shown). For all localization experiments the fusion protein was integrated into the genome as the only functional copy of Sac3p as described in Materials and Methods.
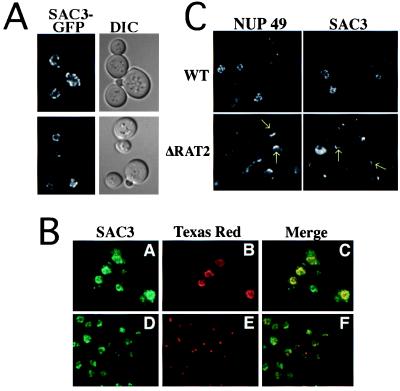
In living cells the Sac3-GFP protein localized to the rim of the nucleus in a punctate pattern (Fig. 3A). This perinuclear staining is suggestive of nuclear pore staining. To characterize further the intracellular localization of Sac3-GFP, cells expressing Sac3-GFP were costained either with mAb414, which recognizes nuclear pore proteins (48), or with an antibody directed against the 80-kDa spindle pole body antigen (49). In Fig. 3B, Sac3-GFP staining is shown in green whereas either the nuclear pore staining (Fig. 3B) or the spindle pole body (Fig. 3E) is shown in red. Colocalization is indicated by the yellow in the merged image. The data demonstrate that the Sac3-GFP protein is localized at the nuclear periphery and that it colocalizes with nuclear pore proteins. A similar pattern of localization was observed with a functional myc-tagged Sac3p (pAC175) (data not shown).
Figure 3.
Sac3-GFP is localized to the nuclear rim and associated with nuclear pore complexes. (A) Localization of Sac3-GFP in living cells (ACY276). (B) Cells (ACY276) expressing Sac3-GFP were prepared for indirect immunofluorescence. Sac3-GFP signal (A and D) is shown in green. Nuclear pores (B) and spindle pole bodies (E) are shown in red. Overlap between the signal for Sac3-GFP and either nuclear pore antigens (C) or spindle pole bodies (F) is indicated by the yellow in the merged images. (C) Wild-type and NUP120/RAT2Δ cells expressing either Sac3-GFP or Nup49-GFP were grown at 25°C and then shifted to the nonpermissive temperature (37°C) for 2 hr. Areas of nuclear pore clustering are indicated by arrows.
To determine whether Sac3p is associated with nuclear pores or simply localized in a punctate pattern within the nuclear envelope, we took advantage of a nuclear pore clustering phenomenon observed in cells lacking the nucleoporin Nup120p/Rat2p (19, 26). If Sac3p is associated with nuclear pores, then it should cluster with other nucleoporins in the NUP120/RAT2 deletion mutant. For this experiment plasmids encoding Sac3-GFP (pAC176) or Nup49-GFP (pAC342), a well-characterized nucleoporin (26), were transformed either into wild-type cells (ACY193) or NUP120/RAT2Δ cells (ACY311) (19). When NUP120/RAT2Δ cells are shifted to the nonpermissive temperature, Sac3-GFP clusters in a manner that is similar to that observed for Nup49-GFP, demonstrating that it is associated with nuclear pore structures (Fig. 3C).
Sac3p Interacts with the Nucleoporin Nsp1p.
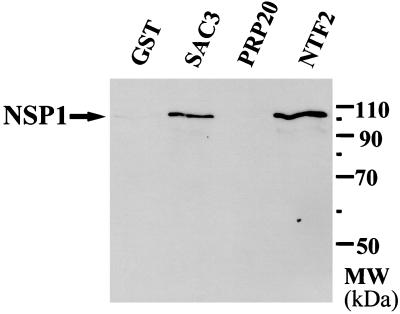
To determine whether Sac3p is physically associated with components of the nuclear pore complex, interaction with the yeast nucleoporin Nsp1p was examined. For these experiments, Sac3p was expressed in yeast as an N-terminal GST fusion protein (pAC404). This fusion protein is toxic when overexpressed, as is wild-type Sac3p. Cells expressing the fusion protein were lysed by glass-bead lysis, and GST-Sac3p was purified by binding to glutathione-Sepharose beads (23). Copurifying proteins were identified by immunoblotting. Results indicate that the nucleoporin Nsp1p copurifies with GST-Sac3p (Fig. 4) as well as with a known Nsp1p-interacting protein, Ntf2p (50), but not with GST-Prp20p, a nuclear protein that is not nuclear pore-associated (51). This biochemical data confirm that Sac3p physically interacts with the nuclear pore. Sac3p also was found to interact with the nucleoporin Nsp1p (data not shown), as well as Nup159p and Nup1p, in a standard two-hybrid assay (52).
Figure 4.
Sac3p interacts with the nuclear pore complex component, Nsp1p. Cells expressing N-terminal GST fusions to Sac3p, Prp20p, Ntf2p, or the GST protein alone were lysed as described in Materials and Methods and analyzed for copurification of the nucleoporin Nsp1p. GST alone and Prp20p serve as negative controls for this experiment. Ntf2p serves as a positive control.
SAC3 Functions in Nuclear Protein Export.
Previously, the growth defects observed in SAC3Δ cells have been correlated with defects in executing anaphase (16). Our results demonstrating that Sac3p is associated with nuclear pores led us to investigate whether Sac3p might play a role in trafficking of proteins between the cytoplasm and the nucleus. The SAC3 deletion and a wild-type control were used to examine different types of nucleocytoplasmic trafficking, including nuclear protein export, poly(A)+ RNA export, and nuclear protein import.
To examine nuclear protein export, either SAC3Δ cells (ACY159) or wild-type cells (ACY160) were transformed with a GFP reporter protein plasmid (pAC213) containing both a simian virus 40 T antigen NLS and a protein kinase inhibitor (PKI) NES sequence (7, 10). In wild-type cells this reporter protein is distributed throughout the cell as it rapidly cycles between the cytoplasm and the nucleus (Fig. 5A). In contrast, in SAC3Δ cells this reporter protein accumulates in the nucleus even in cells grown at room temperature (Fig. 5A). This suggests that Sac3p plays an important role in the export of NES-containing proteins from the nucleus. Consistent with this hypothesis, SAC3Δ cells are also defective in the nuclear export of the HIV Rev protein, which contains a classical, leucine-rich NES (data not shown).
Figure 5.
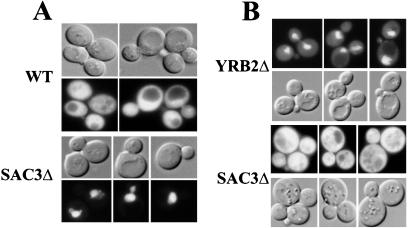
SAC3 functions in nuclear protein export. (A) Wild-type (ACY160) or SAC3Δ (ACY159) cells were transformed with an NES-NLS-GFP reporter plasmid (7, 10). Cells were grown at 25°C. The localization of the reporter protein (GFP) was examined in living cells by direct fluorescence microscopy. Corresponding, differential interference contrast microscopy images are shown. (B) SAC3Δ (ACY159) and YRB2Δ (ACY374) cells were transformed with a plasmid encoding Yap1-GFP (21). Localization of Yap1-GFP was examined in living cells incubated at 25°C.
To determine whether Sac3p is a general nuclear export factor or whether it might mediate selective nuclear export, we examined the export of Yap1-GFP (21) in cells lacking Sac3p. The stress-response factor, Yap1p, is an endogenous yeast export substrate (21). Although the NES in Yap1p (ID–VDG-LCS) is quite divergent from the leucine-rich NES found in the PKI (LALKLAGLDI) and Rev (LPP-LER-LTL) proteins, it is the only export substrate that has been shown to bind to the yeast export receptor, Crm1p/Xpo1p (21).
Yap1-GFP localization was examined in cells lacking either Sac3p or the soluble nuclear export factor Yrb2p (20). Control experiments demonstrated that, as reported previously (21), Yap1p-GFP is distributed throughout wild-type cells but is retained in the nucleus of crm1–3 cells (data not shown). Results shown in Fig. 5B reveal that Yap1-GFP (pAC520) is retained in the nucleus of YRB2Δ cells (ACY374) but is exported normally in SAC3Δ cells (ACY159). This finding is consistent with a model in which Sac3p is not a general nuclear export factor but, rather, plays a role in the selective nuclear export of specific substrates.
Two of the nuclear export substrates used for the experiments described above accumulated within the nucleus of SAC3Δ cells. This implies that loss of Sac3p does not severely impact nuclear protein import as the reporter proteins enter the nucleus and are retained there. No defect in protein import was observed in SAC3Δ cells (data not shown). Similarly, poly(A)+ RNA export from the nucleus was indistinguishable in wild-type and SAC3Δ cells (data not shown).
Genetic Interactions Between SAC3 and Other Nuclear Transport Factors.
Cells lacking Sac3p are defective in the export of a reporter protein from the nucleus. Furthermore, Sac3p is located at the nuclear rim and colocalizes with nuclear pores. These findings suggest that Sac3p should functionally interact with other proteins known to play a role in nuclear export.
There are two soluble transport factors that have been implicated in the export of NES-containing proteins from the nucleus. These two proteins are the NES receptor Crm1p/Xpo1p (7, 9) and the Ran-binding protein Yrb2p (10, 11). Previous work has demonstrated that YRB2 interacts both physically and genetically with yeast XPO1/CRM1 (10). To test whether Sac3p might have functional overlap with either Yrb2p or Xpo1p/Crm1p, yeast lacking the SAC3 gene were crossed to either YRB2Δ (ACY374) (20) or to several crm1/xpo1 mutants (ACY371, ACY372) (8) to determine whether any synthetic lethal interactions were observed. We found that the deletion of SAC3 was synthetically lethal in combination with the deletion of YRB2 or with two different mutant alleles, crm1–2 and crm1–3, of CRM1/XPO1 (data not shown). These results indicate that the function of Sac3p becomes essential in the absence of Yrb2p or when the function of Xpo1p/Crm1p is compromised.
Discussion
We have identified the Sac3 protein as a nuclear pore-associated protein that impacts cell cycle progression. We provide genetic, biochemical, and cell biological evidence that Sac3p is a nuclear pore-associated protein that is required for efficient export of an NES-containing reporter protein from the nucleus. The genetic screens that led to the identification of SAC3 were designed to identify regulators of mitosis. Previous experiments indeed have provided evidence that Sac3p is required for efficient mitosis (16). Our findings now suggest that the critical function of Sac3p during the cell cycle may be linked to nuclear protein export.
In fact, nuclear protein export is emerging as a mechanism for controlling cell cycle transitions. Work from several laboratories has demonstrated that a number of critical regulators of the cell cycle must be exported from the nucleus in a precisely orchestrated manner. Recent studies have demonstrated that both cyclin B and the Cdc14p phosphatase, which acts at the end of mitosis (53), exit the nucleus in the course of the cell cycle (12, 13, 54, 55). Furthermore, cyclin B is modified with ubiquitin in an APC/cyclosome-dependent manner before being exported and degraded by the proteasome (56). Because cyclin B has to both exit the nucleus and be degraded, this could explain the genetic connection that we observe between SAC3 and CDC23, a component of the APC/cyclosome (34, 35).
Because CRM1/XPO1 is an essential gene (7), it is possible that most NES-dependent traffic requires the function of this protein. In contrast, SAC3 and YRB2 are not essential for viability; rather, they appear to have overlapping functions. Cells are viable, although growth-compromised (20), if they have a single copy of either SAC3 or YRB2, but loss of the two genes in combination is lethal. This suggests that both Sac3p and Yrb2p might be members of substrate-specific NES export pathways that converge at Xpo1p/Crm1p. Consistent with this model, our results indicate that Yrb2p is required for the export of Yap1p, but Sac3p is not. Thus, one of the major questions that remains is to identify relevant in vivo export substrates for Sac3p.
We find that Sac3p is required for the export of either a reporter protein containing the PKI NES or the Rev NES. These two NES sequences are very closely related to one another and contain the four leucine residues that were defined originally as the hallmark of an NES sequence (6). In contrast, the experimentally defined Yap1p NES (21) contains only a single leucine. This suggests that the endogenous export substrate for Sac3p may contain a nuclear export signal that is more closely related to the PKI and Rev NES sequences rather than to the Yap1p NES. Numerous candidates exist for this export substrate. One interesting candidate is tubulin, which may have to be exported rapidly from the nucleus upon dissolution of the mitotic spindle in cells that undergo a closed mitosis. Interestingly, the S. cerevisiae Tub1 protein contains a putative NES (LGDVLDRIRKL) that closely resembles the PKI NES (LALKLAGLDI). Furthermore, this sequence lies in an exposed helix that is predicted to form a structure similar to that of another described NES (57).
Thus, Sac3p represents a class of nuclear protein export factor that may be involved in the selective export of proteins from the nucleus. Proteins related to Sac3p exist in higher eukaryotes, and efforts currently are underway to test whether these proteins play a role in nuclear protein export in their cognate organisms. Further work will be required to fully understand how the cell cycle is modulated through the regulated transport of proteins between the cytoplasm and the nucleus, but the current work suggests that Sac3p plays a role in this process.
Acknowledgments
We thank L. Davis, C. Cole, H. Krebber, and T. Taura for strains and plasmids. We thank P. Fanara and N. Akhtar for reading the manuscript. This work was supported by grants from Burroughs Wellcome and the National Institutes of Health to A. Corbett, by grants from the National Institutes of Health and the Novartis/Dana Farber Cancer Institute Drug Discovery Program to P.A.S., and by a collaborative grant from the Human Frontiers in Science Program.
Abbreviations
- NLS
nuclear localization signal
- NES
nuclear export signal
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- PKI
protein kinase inhibitor
- APC
anaphase-promoting complex
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050432997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050432997
References
- 1.Doye V, Hurt E. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- 2.Mattaj I W, Englmeier L. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 3.Wozniak R W, Rout M P, Aitchison J D. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- 4.Görlich D. Curr Opin Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- 5.Cole C, Hammel C. Curr Biol. 1998;8:R368–R372. doi: 10.1016/s0960-9822(98)70239-8. [DOI] [PubMed] [Google Scholar]
- 6.Gerace L. Cell. 1995;82:341–344. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- 7.Stade K, Ford C, Guthrie C, Weis K. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 8.Neville M, Stutz F, Lee L, Davis L I, Rosbash M. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 9.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K, Fransen J, Grosveld G. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taura T, Krebber H, Silver P. Proc Natl Acad Sci USA. 1998;95:7427–7432. doi: 10.1073/pnas.95.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noguchi E, Saitoh Y H, Sazer S, Nishimoto T. J Biochem (Tokyo) 1999;125:574–585. doi: 10.1093/oxfordjournals.jbchem.a022323. [DOI] [PubMed] [Google Scholar]
- 12.Yang J, Bardes E, Moore J, Brennan J, Powers M, Kornbluth S. Genes Dev. 1998;12:2131–2143. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toyoshima F, Moriguchi T, Wada A, Fukada M, Nishida E. EMBO J. 1998;17:2728–2735. doi: 10.1093/emboj/17.10.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Winkler K, Yoshida M, Kornbluth S. EMBO J. 1999;18:2174–2183. doi: 10.1093/emboj/18.8.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez-Girona A, Furnari B, Mondersert O, Russell P. Nature (London) 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- 16.Bauer A, Kölling R. J Cell Sci. 1996;109:1575–1583. doi: 10.1242/jcs.109.6.1575. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Winston F, Dollard C, Ricupero-Hovasse S L. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 19.Heath C, Copeland C, Amberg D C, DelPriore V, Snyder M, Cole C N. J Cell Biol. 1995;131:1677–1697. doi: 10.1083/jcb.131.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taura T, Schlenstedt G, Silver P. J Biol Chem. 1997;272:31877–31884. doi: 10.1074/jbc.272.50.31877. [DOI] [PubMed] [Google Scholar]
- 21.Yan C, Lee L, Davis L. EMBO J. 1998;17:7416–7429. doi: 10.1093/emboj/17.24.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams A, Gottschling D E, Kaiser C, Stearns T. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [Google Scholar]
- 23.Koepp D M, Wong D H, Corbett A H, Silver P A. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kahana J A, Schnapp B, Silver P A. Proc Natl Acad Sci USA. 1995;92:9707–9711. doi: 10.1073/pnas.92.21.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belgareh N, Doye V. J Cell Biol. 1997;136:747–759. doi: 10.1083/jcb.136.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kranz J, Holm C. Proc Natl Acad Sci USA. 1990;87:6629–6633. doi: 10.1073/pnas.87.17.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahana J A, Silver P A. In: GFP: Green Fluorescent Protein Strategies and Applications. Chalfie M, Kain S, editors. New York: Wiley; 1997. pp. 139–152. [Google Scholar]
- 29.Seedorf M, Damelin M, Kahana J, Taura T, Silver P A. Mol Cell Biol. 1999;19:1547–1557. doi: 10.1128/mcb.19.2.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spector D L, Goldman R D, Leinwand L A, editors. Cells: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1998. pp. 106.1–106.7. [Google Scholar]
- 31.Corbett A H, Silver P A. J Biol Chem. 1996;271:18477–18484. doi: 10.1074/jbc.271.31.18477. [DOI] [PubMed] [Google Scholar]
- 32.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 33.Nagase T, Ishikawa K, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. DNA Res. 1998;5:31–39. doi: 10.1093/dnares/5.1.31. [DOI] [PubMed] [Google Scholar]
- 34.Sikorski R, Michaud W, Hieter P. Mol Cell Biol. 1993;13:1212–1221. doi: 10.1128/mcb.13.2.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irniger S, Piatti S, Michaelis C, Nasmyth K. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 36.Novick P, Osmond B C, Botstein D. Genetics. 1989;121:659–674. doi: 10.1093/genetics/121.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishii K, Kumada K, Toda T, Yanagida M. EMBO J. 1996;15:6629–6640. [PMC free article] [PubMed] [Google Scholar]
- 38.Mendenhall M D, Hodge A E. Microbiol Mol Biol Rev. 1998;62:1191–1243. doi: 10.1128/mmbr.62.4.1191-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lörincz A T, Reed S I. Mol Cell Biol. 1986;6:4099–4103. doi: 10.1128/mcb.6.11.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher A B, Nasmyth K. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- 41.Sethi N, Monteagudo M, Koshland D, Hogan E, Burke D J. Mol Cell Biol. 1991;11:5592–5602. doi: 10.1128/mcb.11.11.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Visintin R, Prinz S, Amon A. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 43.Lim H H, Goh P Y, Surana U. Curr Biol. 1998;8:231–234. doi: 10.1016/s0960-9822(98)70088-0. [DOI] [PubMed] [Google Scholar]
- 44.McGrew J T, Goetsch L, Byers B, Baum P. Mol Biol Cell. 1992;3:1443–1454. doi: 10.1091/mbc.3.12.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- 46.Bi E, Pringle J R. Mol Cell Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu Y, Jiang Y, Wellinger R, Carlson K, Roberts J, Stillman D. Mol Cell Biol. 1996;16:5254–5263. doi: 10.1128/mcb.16.10.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davis L I, Blobel G. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- 49.Rout M P, Kilmartin J V. J Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paschal B M, Gerace L. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohtsubo M, Okazaki H, Nishimoto T. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finlay R L, Brent R. In: DNA Cloning 2, Expression Systems: A Practical Approach. Hames B D, Glover D M, editors. Oxford: Oxford Univ. Press; 1995. pp. 169–203. [Google Scholar]
- 53.Visintin R, Craig K, Hwang E, Prinz S, Tyers M, Amon A. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 54.Hagting A, Karlsson C, Clute P, Jackman M, Pines J. EMBO J. 1998;17:4127–4138. doi: 10.1093/emboj/17.14.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shou W, Seol J, Shevchenko A, Baskerville C, Moazed D, Chen Z W S, Jang J, Shevchenko A, Charbonneau H, Deshaies R J. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 56.Murray A. Cell. 1995;81:149–152. doi: 10.1016/0092-8674(95)90322-4. [DOI] [PubMed] [Google Scholar]
- 57.Rittinger K, Budman J, Xu J, Volinia S, Cantley L C, Smerdon S J, Gamblin S J, Yaffe M B. Mol Cell. 1999;4:153–166. doi: 10.1016/s1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]