Abstract
Positive transcription elongation factor b (P-TEFb) comprises a cyclin (T1 or T2) and a kinase, cyclin-dependent kinase 9 (CDK9), which phosphorylates the carboxyl-terminal domain of RNA polymerase II. P-TEFb is essential for transcriptional elongation in human cells. A highly specific interaction among cyclin T1, the viral protein Tat, and the transactivation response (TAR) element RNA determines the productive transcription of the human immunodeficiency virus genome. In growing HeLa cells, half of P-TEFb is kinase inactive and binds to the 7SK small nuclear RNA. We now report on a novel protein termed MAQ1 (for ménage à quatre) that is also present in this complex. Since 7SK RNA is required for MAQ1 to associate with P-TEFb, a structural role for 7SK RNA is proposed. Inhibition of transcription results in the release of both MAQ1 and 7SK RNA from P-TEFb. Thus, MAQ1 cooperates with 7SK RNA to form a novel type of CDK inhibitor. According to yeast two-hybrid analysis and immunoprecipitations from extracts of transfected cells, MAQ1 binds directly to the N-terminal cyclin homology region of cyclins T1 and T2. Since Tat also binds to this cyclin T1 N-terminal domain and since the association between 7SK RNA/MAQ1 and P-TEFb competes with the binding of Tat to cyclin T1, we speculate that the TAR RNA/Tat lentivirus system has evolved to subvert the cellular 7SK RNA/MAQ1 system.
Phosphorylation of the RNA polymerase II (RNAP II) carboxyl-terminal domain (CTD) is a critical step required for transcription elongation (7) and for recruitment of the machinery involved in pre-mRNA maturation (3, 26, 46). The CTD is unphosphorylated when RNAP II assembles onto promoters (RNAP IIA). A class of negative transcription factors including the 5,6-dichlorozo-1-β-d-ribofuranosylbenzimidazole (DRB) sensitivity-inducing factor and the negative elongation factor causes transcriptional arrest shortly after initiation, during which the polymerase may fall off (60). To release this block, the CTD must be phosphorylated (RNAP IIO) by positive transcription elongation factor b (P-TEFb), a protein complex that comprises cyclin-dependent kinase 9 (CDK9) and a cyclin (T1 or T2) (45). P-TEFb kinase activity is required for transcription of most class II genes (6).
The human immunodeficiency virus (HIV) long terminal repeat (LTR) promoter uses a unique mechanism: the level of proviral DNA transcription is determined by recruitment of P-TEFb to the TAR (transactivation response) element, an RNA stem-loop structure that forms at the 5′ end of the viral transcript (4, 38, 59, 66). The viral genome encodes a very potent transactivator of its own transcription, the Tat protein. The formation of a quaternary complex among CDK9, cyclin T1, Tat, and TAR RNA determines the recruitment of human P-TEFb to the transcription elongation complex and the efficient synthesis of long productive viral transcripts (15, 18, 30, 33, 44, 65).
Binding of the 7SK small nuclear RNA (snRNA) to P-TEFb has recently been shown to be associated with the inhibition of CDK9 kinase activity (41, 62). Core P-TEFb is active, whereas the P-TEFb/7SK RNA complex is inactive. P-TEFb and 7SK associate in a reversible manner. Inhibition of cellular transcription by chemical agents or UV irradiation triggers the complete disruption of the P-TEFb/7SK complex and enhances CDK9 activity. In this study, we searched for additional cellular proteins that may be present in the P-TEFb/7SK RNA complex. A single novel P-TEFb subunit was found and termed MAQ1 (for ménage à quatre), alluding to MAT1 (for ménage à trois), which associates with CDK9-related CDK7 and cyclin H (10). The transcription-dependent interaction of P-TEFb with 7SK and MAQ1 may contribute to a feedback loop that modulates the activity of RNAP II.
MATERIALS AND METHODS
Plasmids.
pGST-Tat72, pGST-Tat72K41, and pGST-Tat48 (25) were provided by Monsef Benkirane (Montpellier, France); expression vectors for hemagglutinin (HA)-tagged P-TEFb subunits, pCMV-PITALRE-HA and pCMV-PITALRED167N-HA (19), were from Xavier Graña. pCDNA3-HA-CycT2a and pCDNA3-HA-CycT2b (44) were provided by David Price (Iowa City, Iowa). Full-length and C-terminal deletion-carrying cyclin T1 mutant pCDNA3-HA-CycT1FL (amino acids [aa] 1 to 726) and pCDNA3-HA-CycT1(1-333) were provided by Qiang Zhou (Berkeley, Calif.). pCDNA3-HA-CycT1(1-254) was a PCR-generated mutant consisting of the first N-terminal 254 aa. Tamás Kiss (Toulouse, France) provided human 7SK and U4 cDNAs cloned in pSP65. The pAdRSV-Sp-Flag3 vector was provided by François Giudicelli (Paris, France) (21). HEXIM1 cDNA clone 2-2 was provided by Masatoshi Kusuhara (32). A PCR-amplified fragment was inserted into the NheI and ClaI sites of pAdRSV-Sp-Flag3, thus fusing the Flag tag to the N terminus of MAQ1 and generating pAdRSV-FlagMAQ1. N-terminally Flag-tagged MAQ1 deletion mutants pAdRSV-FlagMAQ1(120-359), pAdRSV-FlagMAQ1(181-359), pAdRSV-FlagMAQ1(1-180), and pAdRSV-FlagMAQ1(1-240) were constructed by insertion of PCR-generated fragments of the corresponding MAQ1 sequences into the NheI and ClaI sites of pAdRSV-Sp-Flag3.
Yeast two-hybrid analysis.
The LexA DNA-binding domain in plasmid pBTM116 (2) was fused N terminally to CDK9 or cyclin T1 to provide plasmids pBTM-CycT1 and pBTM-CDK9 (14). MAQ1 from HEXIM1 cDNA clone 2-2 was PCR amplified and inserted into the EcoRI and SalI sites of pBTM116 to generate pBTM-MAQ1. Deletion mutants pBTM-MAQ1(120-359), pBTM-MAQ1(181-359), pBTM-MAQ1(1-180), and pBTM-MAQ1(1-240) were constructed by insertion of PCR-generated fragments of the corresponding MAQ1 sequences into the EcoRI and SalI sites of pBTM116. pBTM-MAQ1(1-274) was derived from pBTM-MAQ1 after KpnI digestion, followed by ligation. The GAL4 activation domain in plasmid pACTII (Clontech) was fused to CDK9 or cyclin T cDNA to provide pACTII-CycT1, pACTII-CycT1(1-290), pACTII-CycT1(290-726), pACTII-CycT2(1-285), and pACTII-CDK9 (14). pACTII-CycT1/2mut contains mutations K93L and E96K in the full-length cyclin T1 sequence (14). L40 yeast cells were cotransformed with LexA DNA-binding domain and GAL4 activation domain fusion constructs. Transformants were double selected on medium lacking both tryptophan and leucine. Quantitative β-galactosidase activities were determined from pools of more than 10 transformed yeast colonies.
Cells, drugs, and transfections.
HeLa (strain MRL2) or G3H (42) cells were cultured in Dulbecco modified Eagle medium supplemented with 10% (vol/vol) fetal calf serum. Log-phase cells were used for all experiments. Actinomycin D (1 μg ml−1) or DRB (10 μM) was added, and the mixture was incubated for 1 h. UV irradiation (40 J m−2) was performed at 254 nm. Cells were transfected by the standard calcium phosphate method with a coding plasmid supplemented with a noncoding carrier plasmid added (up to 10 μg per 25-cm2 dish).
Cell labeling and fractionation.
Cells were labeled for 20 h by addition of 50 μCi of Redivue Pro-mix L-[35S] (Amersham) ml−1. Actinomycin D was present or not present during the last hour of labeling. Cells were vortexed at 4°C in ice-chilled lysis buffer. Lysis buffer L was derived from buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 200 mM NaCl, 0.2 mM EDTA) supplemented with 1 mM dithiothreitol, 40 U of RNasin (Amersham) ml−1, protease inhibitor cocktail (P-8340; Sigma), and 0.5% Nonidet P-40. Cell lysates clarified by centrifugation for 5 min at 500 × g and for 5 min at 9,000 × g at 4°C were loaded on top of 5 to 45% glycerol gradients in buffer A supplemented with 1 mM dithiothreitol and 10 U of RNasin (Amersham) ml−1. The gradients were spun at 4°C for 16 h at 40,000 rpm in a Kontron TST41 rotor. Ten fractions were collected from the top of the gradients. Core P-TEFb complexes were recovered from fractions 3 and 4, whereas inactive P-TEFb/7SK complexes were in fraction 6 and 7 (41). This distribution was checked by Western blotting.
Antibodies.
Anti-cyclin T1 and anti-CDK9 rabbit antisera were generously provided by David Price. The other antibodies used were rabbit anti-cyclin T1 antibody (H-245; Santa Cruz), goat anti-cyclin T1 antibody (T-18; Santa Cruz), anti-CDK9 mouse monoclonal antibody (D-7; Santa Cruz), rabbit anti-HA antibody (Y11; Santa Cruz), anti-Flag mouse monoclonal antibody (M2, Sigma), and anti-HA mouse monoclonal antibody (12CA5; BAbCo). Primary antibodies were detected with horseradish peroxidase-labeled (Promega) or Cy3-conjugated (Amersham) antibodies or horseradish peroxidase-labeled protein A (Zymed). A rabbit was immunized with the MAQ1 C-terminal peptide LHRQQERAPLSKFGD to provide the MAQ1 antiserum (C4; Eurogentec). For immunoprecipitations, antibodies were cross-linked or not cross-linked to protein A-Sepharose beads with dimethylpimelidiminate (Amersham). The beads were incubated with clarified lysates or gradient fractions at 4°C. The beads were washed four times with buffer A supplemented with 0.5% Nonidet P-40.
Northern blot assays.
RNAs were electrotransferred onto Hybond N+ (Amersham). The membranes were cross-linked by UV irradiation, hybridized overnight in Church buffer at 65°C, and washed at 65°C first for 30 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer and twice for 30 min each time in 0.2× SSC buffer supplemented with 0.5% sodium dodecyl sulfate (SDS) (48). 7SK and U4 cDNA probes were labeled by random priming (Megaprime DNA labeling system; Amersham).
Immunofluorescence assays.
HeLa cells were grown on glass coverslips, washed in phosphate-buffered saline (PBS), and fixed for 15 min at room temperature with 3.7% formaldehyde in PBS. The cells were permeabilized for 15 min in 0.2% Triton X-100 in PBS and then incubated for 10 min in 10 mM glycine-PBS and for 30 min in 3% bovine serum albumin-PBS. Antibodies were diluted in 5% bovine serum albumin-PBS-0.1% Tween 20. Coverslips were mounted in Vectashield (Vector Laboratories).
Mass spectrometric peptide identification.
Five grams of G3H cells that constitutively express HA-tagged cyclin T1 (42) was lysed in 15 ml of the above-described lysis buffer with a Dounce homogenizer. The lysate was loaded on top of a glycerol gradient. Fractions 6 and 7 were pooled and incubated overnight with 200 μl of protein A beads that had been cross-linked to the 12CA5 anti-HA monoclonal antibody. The beads were washed in buffer A supplemented with 0.5% Nonidet P-40 and extracted with Laemmli buffer. Samples were electrophoresed in a polyacrylamide gel. A Coomassie blue-stained band was cut out and digested with trypsin (EC 3.421.4; Roche) (51). Digests were suspended in 20 μl of 1% formic acid, desalted on Zip Tips C18 (Millipore), and eluted with 50 and 80% acetonitrile. Eluates were dried and dissolved in 3 μl of 1% formic acid. A saturated solution of 2,5-dihydroxybenzoic acid in 0.1% trifluoroacetic acid was used as a matrix. Matrix-assisted laser desorption ionization-time-of-flight (mass spectrometry) spectra were obtained with a Voyager-DE STR Biospectrometry Workstation mass spectrometer (PE Biosystems Inc., Framingham, Mass.). Spectra were calibrated externally with the [M+H]+ ion from Des-Arg bradykinin peptide (molecular weight [MW], 904.4681) and ACTH peptide (MW, 2465.1989). Trypsin autoproteolytic fragments (132-to-142 fragment [MW, 1,153.57] and 56-to-75 fragment [MW, 2,163.06]) were used as second calibrators. Data mining was performed with the ProFound software allowing a 0.1-Da mass deviation in the database search. Alternatively, dried in-gel digests were dissolved in 3 μl of water-formic acid-methanol (49:1:50) and analyzed by tandem mass spectrometry on a nanoESI quadrupole time-of-flight tandem mass spectrometer (Micromass, Manchester, United Kingdom). The identification was performed with MS-Tag and Mascot software.
GST-Tat-binding assays.
Escherichia coli BL21 cells transformed with the appropriate plasmids were induced with isopropyl-β-d-thiogalactopyranoside (IPTG) at 30°C. Bacteria were washed and sonicated in EBC buffer (50 mM Tris-HCl [pH 8.0], 120 mM NaCl, 0.5% Nonidet P-40, 5 mM dithiothreitol) supplemented with a protease inhibitor cocktail. The lysate was spun for 15 min at 9,000 × g. Glutathione S-transferase (GST)-Tat proteins in the supernatants were retained on glutathione beads (Amersham) in EBC buffer supplemented with 0.075% SDS and stored at 4°C (25, 61). Beads were equilibrated at room temperature for 30 min in buffer A supplemented with 0.5% Nonidet P-40, 50 μM ZnCl2, and 50 U of E. coli tRNA (Sigma) ml−1. The Tat-binding assay was performed with the same buffer for 40 min at room temperature. Beads were washed four times in buffer A supplemented with 0.5% Nonidet P-40 and 50 μM ZnCl2 and suspended in 1× Laemmli buffer.
RESULTS
A 65-kDa protein copurifies with the P-TEFb/7SK RNA complex.
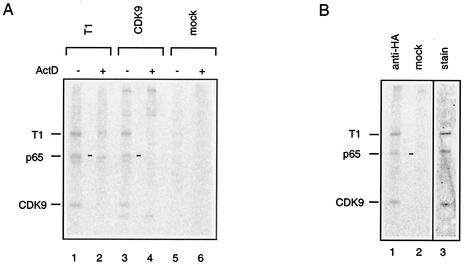
P-TEFb sediments in two sets of fractions when a HeLa cell lysate is ultracentrifuged on a glycerol gradient (41). Upper fractions contain active core P-TEFb, whereas the large kinase-inactive P-TEFb/7SK RNA complex is found in the lower fractions. To identify proteins associated with P-TEFb/7SK RNA complexes, immunoprecipitations were performed from the appropriate gradient fractions. To avoid interference with antiserum proteins, HeLa cells were labeled with [35S]methionine. Anti-CDK9 and anti-cyclin T1 antisera immunoprecipitated several proteins (Fig. 1A, lanes 1 and 3). As P-TEFb/7SK RNA complexes are disrupted upon transcription arrest (41, 62), we looked for proteins that would not immunoprecipitate from the same gradient fractions when cells had been treated with actinomycin D prior to lysis (lanes 2 and 4). Three protein bands met this criterion. A 40-kDa band comigrated with CDK9, and an 85-kDa band comigrated with cyclin T1. Extracts from G3H cells expressing HA-tagged cyclin T1 were next loaded onto the gradients. An anti-HA monoclonal antibody also coimmunoprecipitated a 65-kDa protein along with CDK9 and cyclin T1 from the fractions containing the P-TEFb/7SK RNA complex (Fig. 1B, lane 1). Despite numerous attempts with different detection procedures, no other actinomycin D-sensitive protein bands were detected.
FIG. 1.
Selective coimmunoprecipitation of a 65-kDa protein with CDK9 and cyclin T1. (A) Lysates from 35S-labeled HeLa cells were fractionated by ultracentrifugation. The large P-TEFb/7SK complex was found in gradient fraction 7 (−) unless transcription had been inhibited with actinomycin D (ActD) prior to lysis (+) (41). Gradient fraction 7 was immunoprecipitated with anti-cyclin T1 (lanes 1 and 2) or anti-CDK9 (lanes 3 and 4) antibodies or a control mock serum (lanes 5 and 6). (B) Lysates from G3H cells were fractionated by ultracentrifugation. P-TEFb was immunoprecipitated with protein A beads cross-linked to 12CA5 (anti-HA) antibodies (lanes 1 and 3) or control mock serum (lane 2) from gradient fraction 7. Lanes 1 and 2 correspond to an autoradiogram of the 35S-labeled immunoprecipitates. Lane 3 is Coomassie blue staining of a preparative 12CA5 immunoprecipitate. The positions of cyclin T1, CDK9, and a coimmunoprecipitating 65-kDa protein (p65) are indicated.
The 65-kDa P-TEFb/7SK-binding protein is encoded by the HEXIM1 RNA.
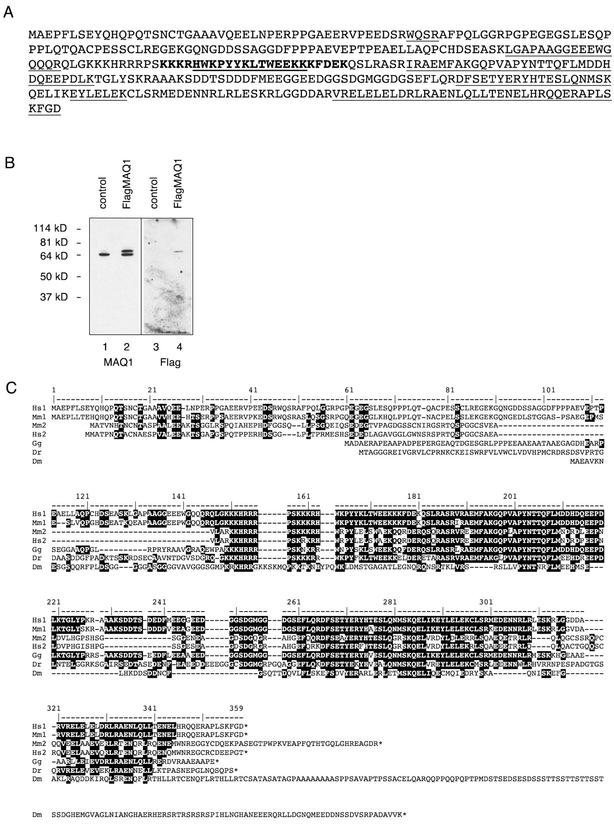
The P-TEFb/7SK complex was isolated from a large number of G3H cells after ultracentrifugation, immunoprecipitation with an HA monoclonal antibody, and SDS-polyacrylamide gel electrophoresis. Three bands were stained with Coomassie blue (Fig. 1 B, lane 3). The tryptic peptides were identified by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. According to their mass, the tryptic peptides of the highest and lowest bands matched those of human cyclin T1 and CDK9, respectively. Tryptic peptides from the intermediate 65-kDa protein matched 26% of a protein sequence potentially coded by the HEXIM1 RNA (32) (Fig. 2A). Sequencing of three peptides by nanoESI quadrupole time-of-flight tandem mass spectrometry confirmed this identification. The protein encoded by the HEXIM1 RNA was termed MAQ1.
FIG. 2.
Identification of MAQ1, a protein coded by the HEXIM1 RNA. (A) Amino acid sequence predicted from the HEXIM1 RNA sequence (32). Tryptic peptides identified in the MALDI/TOF spectrum are underlined. A putative nuclear localization sequence (bold) was predicted by the PredictNLS server (http://cubic.bioc.columbia.edu/predictNLS/). (B) HeLa cells transiently transfected (FlagMAQ1, lanes 2 and 4) or not transfected (control, lanes 1 and 3) with pAdRSV-FlagMAQ1 were lysed and analyzed by Western blotting for endogenous MAQ1 and Flag-MAQ1 by anti-MAQ1 (lanes 1 and 2) or anti-Flag antibodies (lanes 3 and 4). (C) Alignment of human, mouse, chicken, zebra fish, and drosophila MAQ protein sequences deduced from cDNAs in the GenBank database. Accession numbers are given in parentheses. The Homo sapiens MAQ1 (Hs1; AB021179), H. sapiens MAQ1 paralogue (Hs2; AK056946), Mus musculus MAQ1 (Mm1; AY090614), M. musculus MAQ1 orthologue to Hs2 (Mm2; BC026458), G. gallus MAQ1 orthologue (Gg; BU488401 and BU374772), D. renio MAQ1 orthologue (Dr; BG307670), and Drosophila melanogaster MAQ1 orthologue (Dm; AY051786) sequences are shown. The human MAQ1 amino acid numbering is indicated above the sequences. The dark boxes enclose residues that are conserved in other species.
The HEXIM1 RNA codes for a protein of 359 aa; it was not expected to migrate as a protein of 65 kDa. Indeed, CLP-1 (cardiac lineage protein 1), a murine MAQ1 orthologue of 356 aa, had been described as a 41-kDa protein (27). Rabbit antibodies were raised against a carboxyl-terminal peptide of MAQ1 and reacted with a single 65-kDa protein from HeLa cell extracts (Fig. 2B, lane 1). Furthermore, when cells were transfected with an expression vector fusing the Flag epitope to MAQ1, the Flag protein was detected by the MAQ1 antiserum as an additional band migrating slightly above the endogenous 65-kDa protein (lane 2). The difference between the apparent MW and the predicted MW is not likely to be due to posttranslational modifications as the electrophoretic mobility of the recombinant protein made in bacteria was the same (data not shown). Thus, MAQ1 indeed migrates as a 65-kDa protein.
MAQ1 homologues are present in higher eukaryotes.
A blast search through cDNA sequence databases indicated the existence of two sets of MAQ1-related human sequences (Fig. 2C). Sequences in one set were almost identical to the MAQ1 cDNA sequence. Sequences in the other set were almost identical to another cDNA that potentially codes for a 286-amino-acid protein distinct from MAQ1. These paralogues are found in a very close tandem at the same locus (10614) at position q21.32 in human chromosome 17, and they are separated by only 8,700 bp. It is noteworthy that the human MAQ1-encoding gene has no introns whereas the second gene contains four exons. cDNAs coding for orthologues of both proteins are found in expressed sequence tags (ESTs) from mouse, rat, cow, and pig cells (Fig. 2C and data not shown). In contrast, only one MAQ1 cDNA ortholog is found among ESTs or genomes from birds (Gallus gallus), frogs (Xenopus tropicalis and X. laevis), and fish (Danio renio and Fugu rubripes). Sequences sharing blocks of homologies including a bipartite nuclear localization sequence are found within insect genomic and EST databases (Apis mellifera, Bombyx mori, Anopheles gambiae, and Drosophila melanogaster) (Fig. 2C and data not shown). However, no MAQ1 orthologues were detected in worm (Caenorhabditis elegans) or yeast (Saccharomyces cerevisiae and Schizosaccharomyces pombe) genomes. Notably, the 7SK RNA is conserved among vertebrates, but no 7SK RNAs have yet been reported in insects, worms, plants, or yeast (22).
Transcription-dependent association of MAQ1 with P-TEFb complexes.
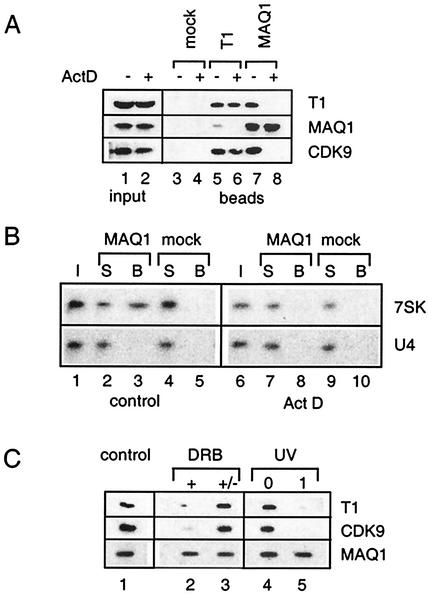
The anti-MAQ1 antiserum was next used to verify MAQ1 binding to P-TEFb. Indeed, MAQ1 coimmunoprecipitated with cyclin T1 and CDK9 from a HeLa cell extract (Fig. 3A, lanes 5 and 7). Anti-MAQ1 antibodies retained 7SK RNA and not nonspecific RNA such as U4 snRNA (Fig. 3B, lane 3). Thus, MAQ1 indeed associates with P-TEFb and 7SK RNA.
FIG. 3.
Transcription-dependent association of MAQ1 with P-TEFb/7SK RNA complexes. (A) Lysates, from cells treated (lane 2) or not treated (lane 1) with 1 μg of actinomycin D ml−1 for 1 h were immunoprecipitated with anti-cyclin T1 (lanes 5 and 6), anti-MAQ1 (lanes 7 and 8), or preimmune (mock) antiserum (lanes 3 and 4). Cyclin T1, MAQ1, and CDK9 in inputs and immunoprecipitates (beads) were analyzed by Western blotting. (B) Lysates from cells treated (Act D) or not treated (control) with actinomycin D were immunoprecipitated with anti-MAQ1 and analyzed for 7SK and U4 RNA by Northern blotting. I, inputs; S, supernatants; B, beads. Preimmune serum was used as a negative control (mock). (C) Cells were treated or not treated (control) with 10 μM DRB and lysed after 1 h (+), or DRB was washed out and the cells were allowed to recover for another hour in fresh medium before lysis (+/−). Alternatively, cells were irradiated at 254 nm (UV) and lysed immediately (lane 0) or allowed to recover for 1 h at 37°C (lane 1). Cyclin T1, MAQ1, and CDK9 retained on beads by anti-MAQ1 were detected by Western blotting.
P-TEFb/7SK complexes are disrupted when transcription is arrested (41, 62). When cells were exposed to actinomycin D prior to lysis, MAQ1 failed to coimmunoprecipitate with cyclin T1 and CDK9 (Fig. 3A, lanes 6 and 8) or 7SK RNA (Fig. 3B, lane 8). To generalize this observation, treatments that inhibit transcription through different mechanisms were investigated. Actinomycin D arrests transcription presumably through intercalation into DNA, DRB prevents phosphorylation of the CTD of RNAP II and of the Spt5 subunit of DRB sensitivity-inducing factor (36), whereas UV irradiation directly damages DNA. Coimmunoprecipitation of CDK9 and cyclin T1 with MAQ1 was strongly reduced when the cells had been treated with DRB at a concentration as low as 10 μM (Fig. 3C, lane 2). The transcriptional arrest by DRB is rapid and reversible. Consistently, removal of DRB from the culture medium restored the MAQ1-P-TEFb interaction within 1 h (lane 3). Transcriptional arrest following UV irradiation is not immediate (47). Consistently, anti-MAQ1 antibodies did immunoprecipitate CDK9 or cyclin T1 from extracts of cells lysed immediately after UV irradiation (lane 4), but the interaction was lost when the cells were allowed to recover for 1 h (lane 5). Thus, the release of both MAQ1 and 7SK RNA from P-TEFb coincides with an arrest in transcription.
MAQ1 is a subunit of P-TEFb/7SK complexes.
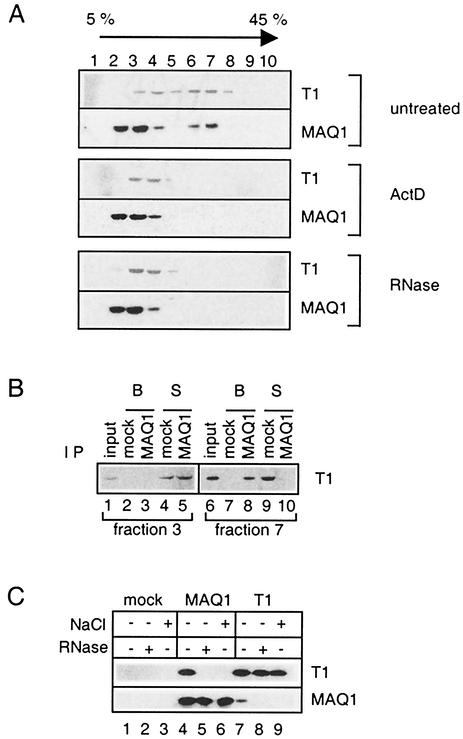
Following ultracentrifugation of HeLa cell extracts on a glycerol gradient, P-TEFb is distributed almost equally into two sets of fractions (41; Fig. 4A, top). Cyclin T1 in fractions 3 and 4 corresponded to active core P-TEFb, whereas fractions 6 and 7 contained the inactive P-TEFb/7SK complex. A major proportion of MAQ1 was present in gradient fractions 2 and 3, but a significant amount was found in fractions 6 and 7. The P-TEFb/7SK complex is disrupted following transcriptional arrest (41). Consistently, neither MAQ1 nor cyclin T1 was found in fractions 6 and 7 of gradients loaded with a lysate from actinomycin D-treated cells (Fig. 4A, middle). Although both MAQ1 and cyclin T1 were present in gradient fraction 3, which contained core P-TEFb complexes, they did not coimmunoprecipitate (Fig. 4B, lane 3). In contrast, MAQ1 antibodies did coimmunoprecipitate cyclin T1 from gradient fraction 7, which contained the P-TEFb/7SK complex (lane 8). These findings demonstrate that MAQ1 is associated with the inactive P-TEFb/7SK complex but not with the small core P-TEFb. It should be emphasized that cyclin T1 was immunodepleted from fraction 7 by the MAQ1 antibodies (lane 10). This observation demonstrates that all P-TEFb/7SK RNA complexes contain MAQ1 molecules. Addition of salt prevented the coimmunoprecipitation of cyclin T1 with MAQ1 (Fig. 4C, lanes 6 and 9). Furthermore, addition of RNase A to cell extracts prevented the coimmunoprecipitation of cyclin T1 with MAQ1 (Fig. 4C, lanes 5 and 8). Consistently, when a lysate from HeLa cells was treated with RNase A to destroy 7SK RNA and loaded onto a glycerol gradient, neither MAQ1 nor cyclin T1 remained in fractions 6 and 7 (Fig. 4A, bottom). These findings indicate that an RNA is required to maintain a stable association between MAQ1 and P-TEFb. Large complexes of MAQ1 and cyclin T1 all contain 7SK RNA. A structural role for 7SK RNA in stabilizing the P-TEFb/MAQ1 association is therefore suggested.
FIG. 4.
MAQ1 is a subunit of P-TEFb/7SK RNA complexes. (A) Fractions from glycerol gradients. Gradients were loaded with a lysate from HeLa cells treated (ActD) or not treated (untreated) with 1 μg of actinomycin D ml−1. Alternatively, RNase A was added to the lysate from untreated cells (RNase). (B) Supernatants (S) or protein A beads (B) after immunoprecipitation (IP) with either preimmune (mock) or MAQ1 antiserum from gradient fractions containing either core P-TEFb (fraction 3) or P-TEFb/7SK complexes (fraction 7). (C) Cell lysates treated (+) or not treated (−) with 500 mM NaCl or RNase A were immunoprecipitated with preimmune serum (mock), MAQ1 antiserum (MAQ1), or anti-cyclin T1 (T1). Samples were analyzed for cyclin T1 and MAQ1 by Western blotting.
MAQ1 associates with P-TEFb subunits in transfected HeLa cells.
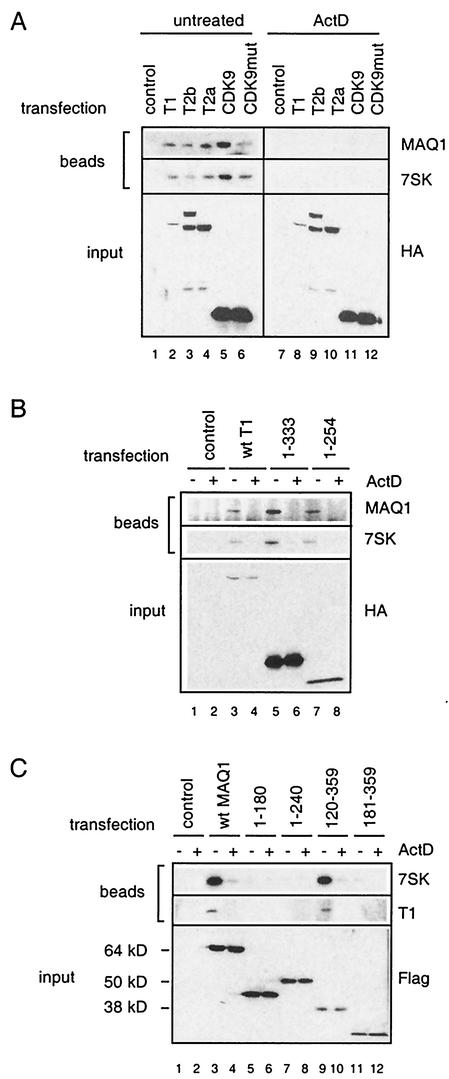
To investigate requirements for the association of MAQ1 with P-TEFb, HeLa cells were transfected with various HA-tagged P-TEFb subunits. MAQ1 and 7SK RNA were immunoprecipitated with tagged cyclin T1, cyclin T2, or CDK9 (Fig. 5A, lanes 2 to 6). However, MAQ1 and 7SK RNAs were not found in the immunoprecipitates when the cells had been exposed to actinomycin D prior to lysis (lanes 8 to 12). P-TEFb containing kinase-inactive mutant CDK9D167N bound MAQ1 and 7SK RNA with a lower efficiency than did tagged wild-type CDK9, although both proteins were expressed at the same level (lanes 5 and 6). Kinase-inactive CDK9D167N may have a decreased intrinsic capacity to associate with MAQ1. This mutant is incapable of forming a stable complex with Tat and TAR (13). Instead, as the CTD kinase activity of CDK9 is required for class II gene transcription (6), overexpression of a kinase-inactive protein may lead to an inhibition of transcription that accounts for the partial disruption of P-TEFb/7SK complexes. Importantly, MAQ1 binding to CDK9D167N was also disrupted by actinomycin treatment (lane 12). In summary, epitope-tagged CDK9, cyclin T1, and cyclin T2 interacted with 7SK RNA and MAQ1 in a transcription-dependent manner in transient transfection assays.
FIG. 5.
Requirements for MAQ1 association with P-TEFb. (A) HeLa cells were transfected with carrier plasmid (control) or expression vectors for HA-tagged full-length cyclin T1, cyclin T2b, cyclin T2a, CDK9, or CDK9D167N (CDK9mut). Cells were treated (ActD) or not treated (untreated) with actinomycin D. Lysates were immunoprecipitated with protein A beads and anti-HA antibodies. Notably, the cyclin T2b cDNA generated both cyclin T2a and T2b proteins because of alternative splicing (44). (B) HeLa cells were transfected with the carrier plasmid (control) or expression vectors for HA-tagged full-length cyclin T1 (wt T1) and C-terminal deletion mutants CycT1(1-333) and CycT1(1-254). Cells were treated (+) or not treated (−) with actinomycin D. Lysates were immunoprecipitated with protein A beads and anti-HA antibodies. (C) HeLa cells were transfected with the carrier plasmid (control) or expression vectors for Flag-tagged full-length MAQ1 (wt MAQ1), C-terminal deletion mutants MAQ1(1-180) and MAQ1(1-240), or N-terminal deletion mutants MAQ1(120-359) and MAQ1(181-359). Transfected cells were treated (+) or not treated (−) with actinomycin D and lysed. Lysates were immunoprecipitated with protein A beads and anti-Flag antibodies. Transfected epitope-tagged proteins (input), endogenous MAQ1 protein, and endogenous cyclin T1 were detected by Western blotting with appropriate antibodies. 7SK RNA was detected by Northern blotting. Positions of molecular size markers are indicated.
Cyclin T1 and MAQ1 domains required for MAQ1 association with P-TEFb/7SK RNA in HeLa cells.
Transient transfection assays were used next to map the cyclin T1 and MAQ1 domains required to form the P-TEFb/7SK RNA/MAQ1 complex. HeLa cells were transfected with HA-tagged C-terminal deletion-carrying cyclin T1 mutants. Anti-HA antibodies coimmunoprecipitated MAQ1 and 7SK RNA with a wild-type construct or with deletion mutant CycT1(1-333) or CycT1(1-254) (Fig. 5B, lanes 3, 5, and 7). This immunoprecipitation did not occur following actinomycin D treatment (lanes 4, 6, and 8). Although the N-terminal 254 aa in CycT1(1-254) lacked a functional TAR RNA recognition motif (18), this mutant was still effective in associating both the 7SK RNA and MAQ1 in a transcription-dependent manner (lane 7).
HeLa cells were next transfected with Flag-tagged deletion mutants of MAQ1. Although they had roughly the same number of amino acids, the MAQ1(1-180) and MAQ1(1-240) proteins migrated much slower than MAQ1(181-359) and MAQ1(120-359), respectively (Fig. 5C, compare lanes 5 and 11 and lanes 7 and 9). This finding indicates that an element in the structure of the N-terminal half of MAQ1 confers its abnormal electrophoretic mobility. Removal of the N-terminal 119 aa did not significantly affect the interaction of MAQ1(120-359) with P-TEFb and 7SK RNA (Fig. 5C, compare lanes 3 and 9). In contrast, removal of the N-terminal 180 aa of MAQ1(181-359) abolished this interaction (lane 11). The C-terminal domain appears to be more critical, as removal of the C-terminal 119 aa abolished MAQ1(1-240) binding to P-TEFb (lane 7). In any case, the MAQ1-P-TEFb interaction was always suppressed after actinomycin D treatment. Thus, MAQ1 interaction with P-TEFb involves amino acid sequences located after residue 120 of MAQ1 and in the N-terminal third of cyclin T1, which corresponds to the cyclin homology region.
Nuclear localization of the MAQ1 protein.
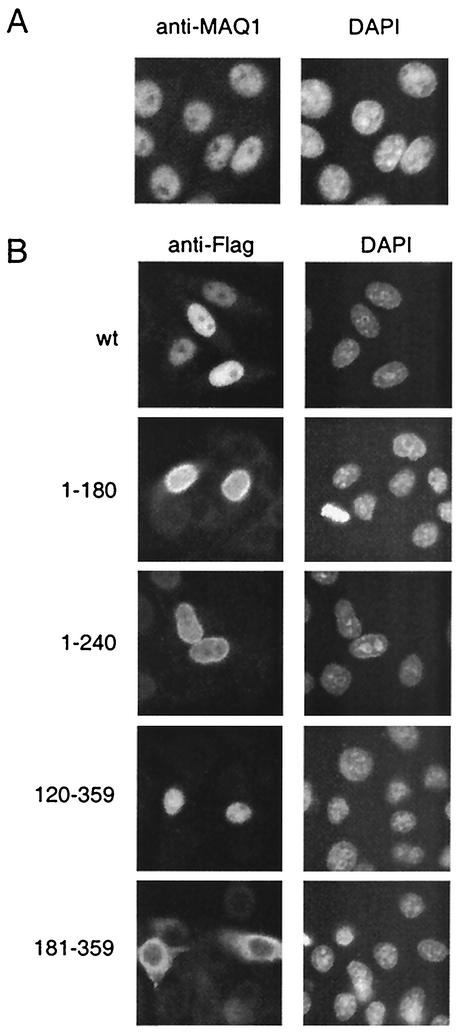
The localization of MAQ1 was investigated by immunofluorescence assay. The MAQ1 antiserum stained the nuclei of HeLa cells (Fig. 6A). MAQ1 is a nuclear protein, like CDK9 and cyclin T1, the other P-TEFb subunits (24). 7SK RNA is also located in the nucleus (39). Flag antibodies also stained the nuclei of cells transfected with a full-length Flag-MAQ1 expression vector (wt in Fig. 6B). Subsequently, the domain involved in nuclear localization was mapped by using transient transfections with Flag-MAQ1 deletion mutants. MAQ1(1-180), MAQ1(1-240), and MAQ1(120-359) were also localized in the nucleus. In contrast, MAQ1(181-359) was cytoplasmic. A bipartite nuclear localization signal may be recognized in the MAQ1 sequence between aa 159 and 181 (Fig. 2A). The immunofluorescence data indicated that nuclear localization required sequences between aa 120 and 180, which include this putative nuclear localization signal.
FIG. 6.
MAQ1 is a nuclear protein. (A) Immunofluorescence of HeLa cells stained with anti-MAQ1 antibodies (left) or 4′,6′-diamidino-2-phenylindole (DAPI; right). (B) HeLa cells transiently transfected with full-length Flag-MAQ1 (wt) or deletion mutants Flag-MAQ1(1-180), Flag-MAQ1(1-240) Flag-MAQ1(120-359), and Flag-MAQ1(181-359). Immunofluorescence assay with anti-Flag antibodies is shown on the left. Chromatin staining with DAPI is shown on the right.
MAQ1 interacts with cyclins T1 and T2 in a two-hybrid test.
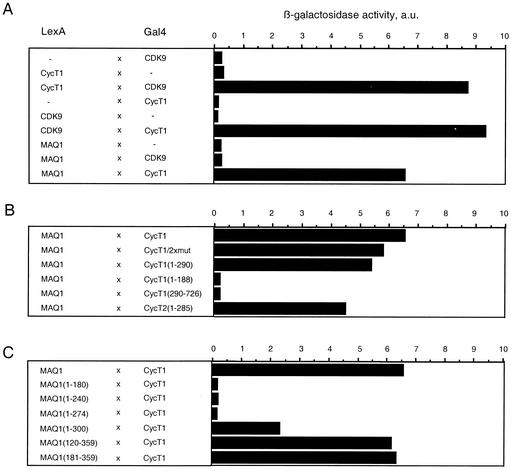
To identify which P-TEFb subunit directly interacted with MAQ1, a yeast two-hybrid study was undertaken. MAQ1 was fused to the LexA DNA-binding domain, while CDK9 and cyclin T1 were fused to the LexA or GAL4 activation domain with vectors pBTM116 and pACTII, respectively. Robust growth on medium lacking histidine and β-galactosidase activities was obtained with cells cotransformed with either LexA-CDK9 and GAL4-cyclin T1 or LexA-cyclin T1 and GAL4-CDK9 as positive controls (5, 14). Elevated β-galactosidase activities were obtained with cells cotransformed with LexA-MAQ1 and GAL4-cyclin T1 (Fig. 7A) or with LexA-CycT1 and GAL4-MAQ1 (data not shown). In contrast, cells cotransformed with LexA-MAQ1 and GAL4-CDK9 failed to induce β-galactosidase activity higher than that observed with GAL4-CDK9 and the empty control vector. Thus, MAQ1 efficiently interacts directly with cyclin T1 and not with CDK9. The MAQ1-cyclin T1 interaction did not require the presence of 7SK RNA in two-hybrid tests. This apparent discrepancy may reflect a lower stability of MAQ1-cyclin T1 association in HeLa cell extracts than in intact yeast cells. Alternatively, the absence of CDK9 in the two-hybrid analysis may increase the strength of MAQ1-cyclin T1 interactions. Overall, it is suggested that association of MAQ1 with P-TEFb is mediated by direct interactions between MAQ1 and cyclin T1.
FIG. 7.
The MAQ1 C-terminal domain directly interacts with the N-terminal domain of cyclins T1 and T2. The interactions of MAQ1 with CDK9 and cyclin T were analyzed with a yeast two-hybrid system coexpressing LexA DNA-binding domain fusion proteins with GAL4 activation domain fusion proteins. β-Galactosidase activities (in arbitrary units [a.u.]) were quantified in pools of more than 10 yeast double transformants. Fusion protein combinations are described on the left. A, full-length proteins; B, cyclin T deletion mutants; C, MAQ1 deletion mutants.
The MAQ1 C-terminal region interacts with the cyclin T N terminus in two-hybrid analysis.
The behavior of cyclin T deletion mutants was next investigated in the yeast two-hybrid assay. MAQ1 efficiently interacted with CycT1(1-290) (Fig. 7B). In contrast, MAQ1 failed to interact with CycT1(1-188) or with CycT1(290-726). Thus, a domain within the cyclin T1 N-terminal 290 aa is involved in MAQ1-cyclin T1 interaction. MAQ1 also interacted with CycT2(1-285), indicating that the interacting domain was located within the N-terminal 285 aa of cyclin T2. CDK9-cyclin T1 or T2 binding also involves the N-terminal domain of cyclin T1 or T2. In contrast to CDK9, MAQ1 does not bind to CycT1(1-188). Furthermore, the point mutations in cyclin T1/2xmut that prevent interaction with CDK9 (14) did not affect the MAQ1-cyclin T1 interaction (Fig. 7B).
To determine the MAQ1 domains involved in cyclin T1 binding in the two-hybrid experiments, MAQ1 deletion mutants were investigated. The interaction decreased upon deletion of the C-terminal domain in mutant MAQ1(1-300), and it was lost in the MAQ1(1-274) and MAQ1(1-240) mutants (Fig. 7C). In contrast, deletion of the N-terminal half of the protein in the MAQ1(181-359) mutant did not affect the binding. The latter finding was in apparent contradiction with data shown above for transfected HeLa cells, as the corresponding deletion abolished MAQ1 binding to P-TEFb and 7SK RNA. However, it should be remembered that the MAQ1(181-359) mutant is cytoplasmic in HeLa cells (Fig. 6B), whereas in two-hybrid experiments, it is addressed to the nucleus by the fused LexA DNA-binding domain.
Taken together, the two-hybrid data were consistent with immunoprecipitations with transfected HeLa cells and indicate that the MAQ1 C-terminal region interacts with the cyclin T N-terminal cyclin homology domain.
MAQ1/7SK RNA and Tat compete for binding to cyclin T1.
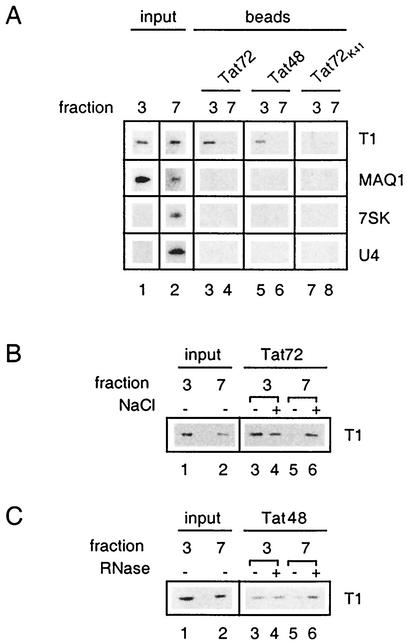
A highly specific interaction between the HIV protein Tat and the N-terminal domain of human cyclin T1 determines the formation of a P-TEFb/TAR RNA/Tat complex (15, 18, 33, 44, 65). The class II transactivator that binds to the N-terminal domain of cyclin T1 competes with Tat for P-TEFb binding (28). MAQ1 also binds the N-terminal domain of cyclin T1. Moreover, MAQ1 and Tat bind the same cyclin T1 mutants in the two-hybrid experiments (Fig. 7B; see reference 14). Hence, a possible interference of MAQ1 with P-TEFb/Tat complex formation was investigated with a GST-Tat pull-down assay (25, 61). Glycerol gradient fractions containing either core P-TEFb or inactive P-TEFb/7SK RNA/MAQ1 complexes were incubated with beads coated with either GST-Tat72 or GST-Tat48 protein. The GST-Tat72K41 protein, in which lysine 41 has been replaced with an arginine, was used as a negative control (59). Cyclin T1 was efficiently pulled down from gradient fractions containing core P-TEFb (Fig. 8A, lanes 3 and 5). In contrast, when gradient fractions containing P-TEFb/7SK/MAQ1 complexes were used, cyclin T1 binding to GST-Tat72 or GST-Tat48 was hardly detectable (lanes 4 and 6). Furthermore, neither MAQ1 nor 7SK RNA was retained on GST-Tat beads.
FIG. 8.
P-TEFb/7SK RNA/MAQ1 complexes are impaired for Tat binding. (A) The small core active P-TEFb complex (glycerol gradient fraction 3) and the large inactive P-TEFb complex (glycerol gradient fraction 7) were tested for the ability to associate with GST-Tat72, GST-Tat72K41, and GST-Tat48. Cyclin T1 and MAQ1 were probed by Western blotting, and 7SK and U4 RNAs were probed by Northern blotting. (B) GST-Tat72 was tested for the ability to bind cyclin T1 from gradient fraction 3 or 7 after addition (+) or no addition (−) of NaCl up to 500 mM. (C) GST-Tat48 was tested for the ability to bind cyclin T1 from gradient fraction 3 or 7 after addition (+) or no addition (−) of RNase A. Inputs corresponded to 25% of the fractions incubated with GST-Tat.
To demonstrate that no posttranslational modification prevented Tat from binding P-TEFb/7SK RNA/MAQ1 complexes, these were disrupted in vitro by RNase treatment or by an increased salt concentration. The GST-Tat pull-down of cyclin T1 from core P-TEFb was not affected by such treatments. In contrast, GST-Tat72 binding to cyclin T1 from P-TEFb/7SK RNA/MAQ1 markedly increased in the presence of salt and reached levels similar to that of core P-TEFb (Fig. 8B, compare lanes 5 and 6). GST-Tat48, which lacked the arginine-rich RNA-binding domain (16), was used with RNase-treated P-TEFb complexes to avoid the requirement of carrier RNA in the assay. Again, GST-Tat48 binding to cyclin T1 from P-TEFb/7SK RNA/MAQ1 increased and reached levels similar to that of core P-TEFb (Fig. 8C, compare lanes 5 and 6). Thus, the presence of 7SK RNA and/or MAQ1 in P-TEFb complexes prevents Tat binding to P-TEFb.
DISCUSSION
In this study, we identified MAQ1, a novel cellular protein that belongs to the inactive P-TEFb/7SK complex and prevents Tat binding to P-TEFb. A direct interaction between MAQ1 and cyclin T1 or T2 is suggested by two-hybrid studies. This interaction requires the C-terminal region of MAQ1 and the N-terminal cyclin homology domain of cyclin T1 or T2. The P-TEFb/7SK RNA/MAQ1 association is disrupted in vivo when transcription is arrested and in vitro when the 7SK RNA is degraded. The latter finding suggests a structural role for 7SK RNA.
MAQ1 is an evolutionarily conserved protein.
MAQ proteins are fairly conserved in vertebrates, and distant orthologues may be present in insects. Sequences in the C-terminal part of the MAQ proteins are the most conserved and contain those required to interact with cyclin T. The region of strong homology (aa 149 to 309) contains a putative bipartite nuclear localization sequence (aa 159 to 181). Indeed, sequences between aa 120 and 180 are required for nuclear localization, while sequences between aa 181 and 300 are involved in MAQ1-cyclin T1 interaction.
MAQ1 is encoded by the HEXIM1 mRNA that is induced when vascular smooth muscle cells are treated with hexamethylene bisacetamide (32). CLP-1, the chicken and mouse orthologues of MAQ1, has been suggested to accumulate in cardiac muscle cells early during heart formation (20, 27). Interestingly, P-TEFb/7SK complexes are disrupted during cardiac muscle cell hypertrophy, and the resulting activation of CDK9 induces cardiac myocyte hypertrophy in mice (49, 50).
MAQ1 is a major P-TEFb-associated protein.
Several proteins have been reported to associate with P-TEFb (reviewed in reference 53). The association of Tat is the most documented. Tat is essential for recruitment of P-TEFb to the HIV LTR and for elongation of transcription of the viral genome (15, 18, 33, 44, 65). However, P-TEFb is also required for efficient transcription of a large number of cellular class II genes (6). Thus, not surprisingly, P-TEFb interacts with and may be recruited by proteins involved in transcription. Examples are class II transactivators such as the class II major histocompatibility transactivator (28), B-Myb (9), Tat-SF1 and RNAP II (13), the androgen receptor (37), NF-κB (1), the Purα protein (8), GRIP1, the nuclear steroid receptor coactivator p160 protein (31), the MyoD transcription factor (54) and MCEF, a member of the AF4 family of transcription factors (12). Furthermore, the HSP90 and CDC37 chaperones coimmunoprecipitate with CDK9 in cells that overexpress epitope-tagged CDK9 (12, 42) and binding to the p45SKP2 E3 ubiquitin ligase component targets CDK9 for degradation (29). Although these associations may have important functions, little is known about their stoichiometry and their influence on CDK9 kinase activity. The MAQ1-P-TEFb interaction is unique in several aspects. First, more than half of all P-TEFb is associated with 7SK RNA/MAQ1 in HeLa cells. Second, this association is lost rapidly following changes in transcriptional activity. Lastly, binding of MAQ1 to P-TEFb requires 7SK RNA, thereby suggesting a structural role for this RNA in the regulation of P-TEFb. It may be a general finding that small RNAs contribute to the structure and activity of transcription factors. For example, SRA RNA acts as a steroid hormone receptor regulator (35, 52, 58), and U1 snRNA binds to and stimulates TFIIH, a transcription initiation factor that includes CDK7/cyclin H/MAT1 among its nine subunits (34). In the latter case, it should be noted that U1-containing TFIIH is devoid of proteins usually bound to U1 snRNA.
MAQ1 and 7SK RNA form a new type of protein kinase regulator.
CDKs comprise a kinase and a cyclin subunit. A third protein partner frequently regulates their kinase activity. A first group of CDK/cyclin regulators increases or stabilizes CDK kinase activity. The Suc1/Cks protein binds to the CDK1/cyclin B complex and stimulates phosphorylation of specific substrates (43). CDK7 forms a ternary complex with cyclin H and MAT1 (10). CTK1, a yeast CDK9 orthologue, forms a ternary complex with a cyclin, CTK2, and a third partner, CTK3 (55). In contrast to MAQ1, MAT1 and CTK3 are required for proper kinase activity of CDK7 and CTK1, respectively (10, 23). Another group of CDK regulators consists of CDK inhibitors such as p21/Cip1, p27/Kip1, p57Kip2, and Ink4, which bind to and inhibit CDK2-, CDK4-, and CDK6-cyclin complexes (11, 40, 57). P-TEFb is inactive when bound to MAQ1 and 7SK (41, 62). P-TEFb associates with 7SK and MAQ1 in a reversible manner. Inhibition of cellular transcription by chemical agents or UV irradiation enhances CDK9 activity. We now show that these treatments trigger the complete disruption of the P-TEFb/7SK RNA/MAQ1 complex. Thus, the 7SK RNA/MAQ1 system appears to be a new kind of CDK regulator that may contribute to a feedback loop modulating the activity of RNAP II.
The TAR RNA/Tat complex as a viral system that may subvert 7SK RNA/MAQ1.
The P-TEFb/7SK RNA/MAQ1 complex evokes the complex that forms among P-TEFb, the HIV Tat protein, and the TAR RNA structure. The latter is responsible for the strong activation of HIV LTR-driven transcription (17). However, there are major differences. (i) 7SK RNA is essential for MAQ1 binding to P-TEFb, whereas although TAR RNA enhances the interaction between Tat and cyclin T1 (63), Tat binding to P-TEFb can occur in the absence of TAR RNA (25, 61). (ii) Tat specifically associates with cyclin T1, whereas MAQ1 and 7SK RNA can associate with P-TEFb containing either cyclin T1 or T2. (iii) The TAR RNA recognition motif region in cyclin T1 is essential for TAR recognition (59) but is not required for 7SK RNA binding. (iv) P-TEFb bound to Tat may be more active than core P-TEFb (61, 64), whereas P-TEFb is inactive when bound to MAQ1 and 7SK (41, 62). Despite these differences, Tat and MAQ1 both bind the N terminus of cyclin T1 and 7SK RNA/MAQ1 association prevents Tat binding to P-TEFb in vitro. Conversely, Tat binding to free P-TEFb may prevent P-TEFb/7SK RNA/MAQ1 complex formation and the corresponding inhibition of P-TEFb activity. This competition model does not imply that Tat disrupts the P-TEFb/7SK RNA/MAQ1 complex in vitro. However, Tat may trap an active form of P-TEFb, as the P-TEFb/7SK RNA/MAQ1 complex appears to undergo continuous formation and disruption in vivo. It is rather common that viral proteins subvert cellular functions. For example, the herpesvirus genome codes for viral cyclins that allow CDKs to escape inhibition by CDK inhibitor proteins, thereby deregulating the cell cycle (56). It is tempting to speculate that the TAR RNA/Tat lentivirus system has evolved to subvert the 7SK RNA/MAQ1 cellular system.
Acknowledgments
This work was supported by grants from the Ligue Nationale Contre le Cancer, the Agence Nationale de Recherche sur le SIDA, the Association pour la Recherche sur le Cancer (O.B.), the Fondation pour la Recherche Médicale, the Federation of European Biochemical Societies, Sidaction (A.A.M.), and the National Research Program on AIDS, AIRC, and MURST (L.L.).
We thank Monsef Benkirane and Marie Françoise Dubois for extensive discussions and Xavier Graña, Masatoshi Kusuhara, David Price, and Qiang Zhou for providing plasmids and antibodies.
REFERENCES
- 1.Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327-337. [DOI] [PubMed]
- 2.Bartel, P. L., and S. Fields. 1995. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 254:241-263. [DOI] [PubMed] [Google Scholar]
- 3.Bentley, D. 2002. The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14:336-342. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout, B., R. H. Silverman, and K.-T. Jeang. 1989. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59:273-282. [DOI] [PubMed] [Google Scholar]
- 5.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1998. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 17:7056-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao, S. H., and D. H. Price. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 276:31793-31799. [DOI] [PubMed] [Google Scholar]
- 7.Dahmus, M. E. 1996. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 271:19009-19012. [DOI] [PubMed] [Google Scholar]
- 8.Darbinian, N., B. E. Sawaya, K. Khalili, N. Jaffe, B. Wortman, A. Giordano, and S. Amini. 2001. Functional interaction between cyclin T1/cdk9 and Purα determines the level of TNFα promoter activation by Tat in glial cells. J. Neuroimmunol. 121:3-11. [DOI] [PubMed] [Google Scholar]
- 9.De Falco, G., L. Bagella, P. P. Claudio, A. De Luca, Y. Fu, B. Calabretta, A. Sala, and A. Giordano. 2000. Physical interaction between CDK9 and B-Myb results in suppression of B-Myb gene autoregulation. Oncogene 19:373-379. [DOI] [PubMed] [Google Scholar]
- 10.Devault, A., A.-M. Martinez, D. Fesquet, J.-C. Labbé, N. Morin, J.-P. Tassan, E. A. Nigg, J.-C. Cavadore, and M. Dorée. 1995. MAT1 ('ménage à trois') a new RING finger protein subunit stabilizing cyclin H-cdk7 complexes in starfish and Xenopus CAK. EMBO J. 14:5027-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endicott, J. A., M. E. Noble, and J. A. Tucker. 1999. Cyclin-dependent kinases: inhibition and substrate recognition. Curr. Opin. Struct. Biol. 9:738-744. [DOI] [PubMed] [Google Scholar]
- 12.Estable, M. C., M. H. Naghavi, H. Kato, H. Xiao, J. Qin, A. Vahlne, and R. G. Roeder. 2002. MCEF, the newest member of the AF4 family of transcription factors involved in leukemia, is a positive transcription elongation factor-b-associated protein. J. Biomed. Sci. 9:234-245. [DOI] [PubMed] [Google Scholar]
- 13.Fong, Y. W., and Q. Zhou. 2000. Relief of two built-in autoinhibitory mechanisms in P-TEFb is required for assembly of a multicomponent transcription elongation complex at the human immunodeficiency virus type 1 promoter. Mol. Cell. Biol. 20:5897-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraldi, A., P. Licciardo, B. Majello, A. Giordano, and L. Lania. 2001. Distinct regions of cyclin T1 are required for binding to CDK9 and for recruitment to the HIV-1 Tat/TAR complex. J. Cell. Biochem. 36:247-253. [DOI] [PubMed] [Google Scholar]
- 15.Fujinaga, K., R. Taube, J. Wimmer, T. P. Cujec, and B. M. Peterlin. 1999. Interactions between human cyclin T, tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc. Natl. Acad. Sci. USA 96:1285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gait, M. J., and J. Karn. 1993. RNA recognition by the human immunodeficiency virus Tat and Rev proteins. Trends Biochem. Sci. 18:255-259. [DOI] [PubMed] [Google Scholar]
- 17.Garber, M. E., and K. A. Jones. 1999. HIV-1 Tat: coping with negative elongation factors. Curr. Opin. Immunol. 11:460-465. [DOI] [PubMed] [Google Scholar]
- 18.Garber, M. E., P. Wei, V. N. KewalRamani, T. P. Mayall, C. H. Herrmann, A. P. Rice, D. R. Littman, and K. A. Jones. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12:3512-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garriga, J., X. Mayol, and X. Graña. 1996. The CDC2-related kinase PITALRE is the catalytic subunit of active multimeric protein complexes. Biochem. J. 319:293-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghatpande, S., S. Goswami, S. Mathew, G. Rong, L. Cai, S. Shafiq, and M. A. Siddiqui. 1999. Identification of a novel cardiac lineage-associated protein (cCLP-1): a candidate regulator of cardiogenesis. Dev. Biol. 208:210-221. [DOI] [PubMed] [Google Scholar]
- 21.Giudicelli, F., E. Taillebourg, P. Charnay, and P. Gilardi-Hebenstreit. 2001. Krox-20 patterns the hindbrain through both cell-autonomous and non cell-autonomous mechanisms. Genes Dev. 15:567-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gürsoy, H. C., D. Koper, and B.-J. Benecke. 2000. The vertebrate 7S K RNA separates hagfish (Myxine glutinosa) and lamprey (Lampetra fluviatilis). J. Mol. Evol. 50:456-464. [DOI] [PubMed] [Google Scholar]
- 23.Hautbergue, G., and V. Goguel. 2001. Activation of the cyclin-dependent kinase CTDK-I requires the heterodimerization of two unstable subunits. J. Biol. Chem. 276:8005-8013. [DOI] [PubMed] [Google Scholar]
- 24.Herrmann, C. H., and M. A. Mancini. 2001. The Cdk9 and cyclin T subunits of TAK/P-TEFb localize to splicing factor-rich nuclear speckle regions. J. Cell Sci. 114:1491-1503. [DOI] [PubMed] [Google Scholar]
- 25.Herrmann, C. H., and A. P. Rice. 1995. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 69:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14:1415-1429. [PubMed] [Google Scholar]
- 27.Huang, F., M. Wagner, and M. A. Siddiqui. 2002. Structure, expression, and functional characterization of the mouse CLP-1 gene. Gene 292:245-259. [DOI] [PubMed] [Google Scholar]
- 28.Kanazawa, S., T. Okamoto, and B. M. Peterlin. 2000. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity 12:61-70. [DOI] [PubMed] [Google Scholar]
- 29.Kiernan, R. E., S. Emiliani, K. Nakayama, A. Castro, J. C. Labbé, T. Lorca, K. Nakayama Ki, and M. Benkirane. 2001. Interaction between cyclin T1 and SCF (SKP2) targets CDK9 for ubiquitination and degradation by the proteasome. Mol. Cell. Biol. 21:7956-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, Y. K., C. F. Bourgeois, C. Isel, M. J. Churcher, and J. Karn. 2002. Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol. Cell. Biol. 22:4622-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kino, T., O. Slobodskaya, G. N. Pavlakis, and G. P. Chrousos. 2002. Nuclear receptor coactivator p160 proteins enhance the HIV-1 long terminal repeat promoter by bridging promoter-bound factors and the Tat-P-TEFb complex. J. Biol. Chem. 277:2396-2405. [DOI] [PubMed] [Google Scholar]
- 32.Kusuhara, M., K. Nagasaki, K. Kimura, N. Maass, T. Manabe, S. Ishikawa, M. Aikawa, K. Miyazaki, and K. Yamaguchi. 1999. Cloning of hexamethylene-bis-acetamide-inducible transcript, HEXIM1, in human vascular smooth muscle cells. Biomed. Res. 20:273-279. [Google Scholar]
- 33.Kwak, Y. T., D. Ivanov, J. Guo, E. Nee, and R. B. Gaynor. 1999. Role of the human and murine cyclin T proteins in regulating HIV-1 tat-activation. J. Mol. Biol. 288:57-69. [DOI] [PubMed] [Google Scholar]
- 34.Kwek, K. Y., S. Murphy, A. Furger, B. Thomas, W. O'Gorman, H. Kimura, N. J. Proudfoot, and A. Akoulitchev. 2002. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat. Struct. Biol. 9:800-805. [DOI] [PubMed] [Google Scholar]
- 35.Lanz, R. B., N. J. McKenna, S. A. Onate, U. Albrecht, J. Wong, S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 1999. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 97:17-27. [DOI] [PubMed] [Google Scholar]
- 36.Lavoie, S. B., A. L. Albert, H. Handa, M. Vincent, and O. Bensaude. 2001. The peptidyl-prolyl isomerase Pin1 interacts with hSpt5 phosphorylated by Cdk9. J. Mol. Biol. 312:675-685. [DOI] [PubMed] [Google Scholar]
- 37.Lee, D. K., H. O. Duan, and C. Chang. 2001. Androgen receptor interacts with the positive elongation factor P-TEFb and enhances the efficiency of transcriptional elongation. J. Biol. Chem. 276:9978-9984. [DOI] [PubMed] [Google Scholar]
- 38.Mancebo, H. S., G. Lee, J. Flygare, J. Tomassini, P. Luu, Y. Zhu, J. Peng, C. Blau, D. Hazuda, D. Price, and O. Flores. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matera, A. G., and D. C. Ward. 1993. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J. Cell Biol. 121:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakayama, K., and K. Nakayama. 1998. Cip/Kip cyclin-dependent kinase inhibitors: brakes of the cell cycle engine during development. Bioessays 20:1020-1029. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK snRNA binds to and inhibits the activity of Cdk9/cyclin T complexes. Nature 414:322-325. [DOI] [PubMed] [Google Scholar]
- 42.O'Keeffe, B., Y. Fong, D. Chen, S. Zhou, and Q. Zhou. 2000. Requirement for a kinase-specific chaperone pathway in the production of a Cdk9/cyclin T1 heterodimer responsible for P-TEFb-mediated Tat stimulation of HIV-1 transcription. J. Biol. Chem. 275:279-287. [DOI] [PubMed] [Google Scholar]
- 43.Patra, D., S. X. Wang, A. Kumagai, and W. G. Dunphy. 1999. The xenopus Suc1/Cks protein promotes the phosphorylation of G2/M regulators. J. Biol. Chem. 274:36839-36842. [DOI] [PubMed] [Google Scholar]
- 44.Peng, J., Y. Zhu, J. T. Milton, and D. H. Price. 1998. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 12:755-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 47.Rockx, D. A., R. Mason, A. van Hoffen, M. C. Barton, E. Citterio, D. B. Bregman, A. A. van Zeeland, H. Vrieling, and L. H. Mullenders. 2000. UV-induced inhibition of transcription involves repression of transcription initiation and phosphorylation of RNA polymerase II. Proc. Natl. Acad. Sci. USA 97:10503-10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Sano, M., M. Abdellatif, H. Oh, M. Xie, L. Bagella, A. Giordano, L. H. Michael, F. J. DeMayo, and M. D. Schneider. 2002. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 1310-1317. 8: [DOI] [PubMed]
- 50.Sano, M., and M. D. Schneider. 2003. Cyclins that don't cycle: cyclin t/cyclin-dependent kinase-9 determines cardiac muscle cell size. Cell Cycle 2:99-104. [PubMed] [Google Scholar]
- 51.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 52.Shi, Y., M. Downes, W. Xie, H. Y. Kao, P. Ordentlich, C.-C. Tsai, M. Hon, and R. M. Evans. 2001. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 15:1140-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simone, C., and A. Giordano. 2001. New insight in cdk9 function: from Tat to MyoD. Front Biosci. 6:D1073-D1082. [DOI] [PubMed] [Google Scholar]
- 54.Simone, C., P. Stiegler, L. Bagella, B. Pucci, C. Bellan, G. De Falco, A. De Luca, G. Guanti, P. L. Puri, and A. Giordano. 2002. Activation of MyoD-dependent transcription by cdk9/cyclin T2. Oncogene 21:4137-4148. [DOI] [PubMed] [Google Scholar]
- 55.Sterner, D. E., J. M. Lee, S. E. Hardin, and A. L. Greenleaf. 1995. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol. Cell. Biol. 15:5716-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swanton, C., G. L. Card, D. Mann, N. McDonald, and N. Jones. 1999. Overcoming inhibitions: subversion of CKI function by viral cyclins. Trends Biochem. Sci. 24:116-120. [DOI] [PubMed] [Google Scholar]
- 57.Vidal, A., and A. Koff. 2000. Cell-cycle inhibitors: three families united by a common cause. Gene 247:1-15. [DOI] [PubMed] [Google Scholar]
- 58.Watanabe, M., J. Yanagisawa, H. Kitagawa, K. Takeyama, S. Ogawa, Y. Arao, M. Suzawa, Y. Kobayashi, T. Yano, H. Yoshikawa, Y. Masuhiro, and S. Kato. 2001. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J. 15:1341-1352. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Wei, P., M. E. Garber, S.-M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 60.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 61.Yang, X., M. O. Gold, D. N. Tang, D. E. Lewis, E. Aguilar-Cordova, A. P. Rice, and C. H. Herrmann. 1997. TAK, an HIV Tat-associated kinase, is a member of the cyclin-dependent family of protein kinases and is induced by activation of peripheral blood lymphocytes and differentiation of promonocytic cell lines. Proc. Natl. Acad. Sci. USA 94:12331-12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317-322. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, J., N. Tamilarasu, S. Hwang, M. E. Garber, I. Huq, K. A. Jones, and T. M. Rana. 2000. HIV-1 TAR RNA enhances the interaction between Tat and cyclin T1. J. Biol. Chem. 275:34314-34319. [DOI] [PubMed] [Google Scholar]
- 64.Zhou, M., M. A. Halanski, M. F. Radonovich, F. Kashanchi, J. Peng, D. H. Price, and J. N. Brady. 2000. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 20:5077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou, Q., D. Chen, E. Pierstorff, and K. Luo. 1998. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 17:3681-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu, Y., T. Pe'ery, J. Peng, Y. Ramanathan, N. Marshall, T. Marshall, B. Amendt, M. B. Mathews, and D. H. Price. 1997. Transcription elongation factor P-TEFb is required for HIV-1 Tat transactivation in vitro. Genes Dev. 11:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]