Abstract
We demonstrate here that the Tet repressor (TetR), a dimeric allosterical regulatory protein, can be converted to a fully functional monomer when connected by a 29 amino acid linker. TetR-based transregulators are widely used to regulate gene expression in eukaryotes. They can be fused to form single-chain (sc) Tet transregulators with two TetR moieties and one eukaryotic regulatory domain. Sc variants of transactivator and transsilencer exhibit the same regulatory properties as their respective dimeric counterparts in human cell lines. In particular, the reverse ‘tet-on’ phenotype of rtTA variants is also present in the sc variants. Coexpression of a reverse transactivator and sc transsilencer leads to reduced background expression and shows full activation upon induction. The data demonstrate that sc Tet transregulators exhibit the phenotype of their respective dimers and lack functional interference when coexpressed in the same cell.
INTRODUCTION
Many DNA-binding proteins recognize palindromic sequences as homodimers. Such proteins have previously been monomerized by the fusion of two genes to encode a polypeptide with two identical domains. Protein folding, dimerization and DNA binding are often coupled reactions and, hence, connecting the two monomers can enhance DNA affinity. This was shown for monomerized versions of lambda Cro, the N-terminal domain of 434 cI repressor, P22 Arc and the basic helix–loop–helix domain of MASH-1 (1–4). Furthermore, single-chain (sc) proteins may contain different functions in each domain. For example, sc DNA-binding domains of 434 cI repressor with different recognition specificities resulted in binding to asymmetric operators (2). Also, fusions of wild-type (wt) estrogen receptor α with a transactivation mutant have been used as a model for the functional analysis of naturally occurring heterodimeric receptor species (5).
We present here a sc version of the bacterial repressor of the tetracycline (tc) resistance operon (TetR). Each monomer contains a DNA reading head and a core mediating dimerization and inducer binding. Upon binding of tetracyclines, TetR undergoes complex conformational changes (6–8). To our knowledge such an allosteric repressor has not been monomerized to date.
TetR is the basis of widely used doxycycline (dox) controlled expression systems in eukaryotes (9). Several different TetR-based transregulators have been designed and used either alone or in combinations with each other. Fusing TetR to the activation domain of virion protein 16 from Herpes simplex (VP16) resulted in the tc-controlled transactivator (tTA) (10) which stimulates transcription in the absence of dox. The reverse transactivator rtTA obtained by mutagenesis of tTA activates transcription in the presence of dox (11,12). TetR fusions to modified activation domains circumvent possible adverse effects of VP16 (13–15). The dox-inducible tc-controlled transsilencer (tTS) contains the KRAB repressor domain fused to TetR (16).
Active repression of uninduced gene expression can be obtained when combining tTS with rtTA yielding even more stringent control (17–19). When several transregulator variants are coexpressed in the same cell, heterodimers will arise (20). To prevent this, naturally occurring sequence variants of TetR (21) have been used as platforms to attach mutated DNA reading heads (20) or the KRAB silencing domain (17,19). We demonstrate here that the mutations conferring rtTA properties do not show the same phenotype when transferred to other naturally occurring sequence variants of TetR. To provide a general basis for multiple use of TetR-based transregulators in the same cell, we report fully active sc transregulators and their use in human cell lines.
MATERIALS AND METHODS
Plamids constructions
Mutations conferring a reverse phenotype were introduced to transactivators based on TetR(E) as follows. A tetR(E) fragment containing the mutations DN95, LS101 and GD102 was PCR amplified with the primers TetR(E)-ApaI and TetR(E)-NgoMIV from the template pWH853Erev (P. Schubert and W. Hillen, unpublished results). This fragment was digested with ApaI/NgoMIV and ligated into equally restricted pWHE120(B), resulting in pWHE120(rE) encoding rtTA2E. To clone rtTA2E-M1, the TetR(B) DNA reading head containing the mutations SG12 and EG19 was isolated from pUHrT16-1 (12) with the primers LDsynNin and B-ApaI-rev. The fragment was cut with XbaI/ApaI and ligated into equally restricted pWHE120(B) yielding pWHE120(B)-1219. Mutation SP56 was introduced to tetR(E) by PCR with the primers TetR(E)-ApaI/SP56 and TetR(E)-NgoMIV using as template pCMV-TetR(B/E)-KRAB (18). The ApaI/NgoMIV restricted fragment was then ligated into pWHE120(B)-1219 resulting in pWHE120(E)-M1 which encodes rtTA2E-M1. To clone a vector encoding rtTA2-M1, pUHrT15-1 (12) was cut with XbaI/NruI. The tetR-M1 containing fragment was ligated into equally restricted pWHE120(B).
Sc tetR(B+B) was constructed by overlap extension PCR. Both first-step reactions were carried out using pWH1919SauI (22) with the primer pairs SC-1 and SC-3 or SC-4 and SC-2 to introduce the linker sequence. The first-step products were used as template in a second PCR with the outer primers SC-1 and SC-2. The PCR fragment was cut with StyI and cloned into the unique StyI site of tetR(B) in pWH1919SauI. The resulting plasmid pWH520(B+B) was digested with XbaI/BstEII and the fragment containing sc tetR(B+B) ligated into pWH853 (23).
For the construction of pWH853(B+sB), tetRs(B) was amplified by PCR from pUHT61-1 (12) with the primers synTetR-BglII and synTetR-BstNco. The fragment was restricted with BglII and BstEII and ligated to equally restricted pWH853(B+B). The corresponding high expression vector was contructed by excising the XbaI/NcoI fragment of pWH853(B+sB) and cloning it into pWH620 (22) to form pWH620(B+sB).
To construct tetR(sB+B), tetRs(B) was amplified by PCR from plasmid pUHT61-1 with the primers LDsynNin and BamHI-Linker-CsynB. The product was cut with XbaI/BamHI and ligated to restricted pWH520(B+B)oB (N. Conrad and W. Hillen, unpublished results) which gave rise to pWH520(sB+B).
Sc tetR was introduced into eukaryotic expression vectors by excising the XbaI/ApaI fragment of pWH853(B+sB) and ligating it to restricted pWHE120(sB) (pUHT61-1 with singular NgoMIV site; L. Drüppel and W. Hillen, unpub lished results) to form pWHE120(B+sB). The plasmid pWHE120(sB+B) was constructed by ligating the XbaI/MluI fragment of pWH520(sB+B) to equally cut pWHE120(B). These expression plasmids contain a CMV promoter which confers high level constitutive expression of the transactivator. The plasmids pWHE120(sS2+S2) and pWHE120(sM2+M2) were similarly constructed by isolating tetRs-S2 and tetRs-M2 from the respective template pUHrT61-1 and pUHrT62-1 (12) by PCR using the primers LDsynNin and BamHI-Linker-CsynB. The fragments were digested with XbaI/BamHI and introduced into pWH520(B+B)oB. The resulting plasmids pWH520(sS2+B) and pWH520(sM2+B) were restricted with XbaI/MluI. The fragment containing the reverse tetR allele was cloned into pWHE120(B) and generated pWHE120(sS2+B) and pWHE120(sM2+B). tetR(B) was exchanged by tetR-S2 and tetR-M2 which were obtained by BglII/NgoMIV restriction of the PCR product from pUHrT10-1 and pUHrT16-1 (12) with the primers B-BglII and B-NgoMIV. The resulting eukaryotic expression vectors encode the reverse transactivators sc rtTA2-S2 and sc rtTA2-M2.
To construct a sc transsilencer, the KRAB repression domain was isolated from pCMV-TetR(B/E)-KRAB (18) by PCR with the primers NgoM-KRAB and KRAB-XmaI. The product was cut with NgoMIV/SmaI and ligated into equally restricted pWHE120(sB+B) which resulted in pWHE122(sB+B). All primer sequences are available upon request.
β-Galactosidase assays in Escherichia coli
Repression, inducibility and negative transdominance of the tetR variants were assayed in E.coli WH207(λtet50) (23) carrying a tetPO-lacZ transcriptional fusion on a λ phage. Bacteria were grown at 37°C in LB supplemented with the appropriate antibiotics. Quantification of induction efficiencies was done with 0.2 µg/ml tetracycline in overnight and log-phase cultures. β-Galactosidase activities were determined as described (24). Four independent cultures were assayed for each strain and measurements were repeated at least twice.
Transient transfections
Transfections of HeLa and HEK293T cells were performed at 50–60% confluency with 1–1.5 µg of DNA and 3–5 µl of Lipofectamine (Gibco Life Technologies) in 35-mm dishes, according to the instructions of the producer. The respective DNA mixtures consisted of 0.1 µg of transregulator plasmid, 0.1 µg of reporter plasmid pUHC13-3 (10), 0.4 µg of lacZ expression vector pUHD16-1 (13), and the unspecific DNA pWHE121 (S. Grimm and W. Hillen, unpublished results). Dox (Sigma) was added at 5 µg/ml for induction. Cells were harvested 24 h after induction.
Luciferase assay
The transfected cells were lysed by incubation with 100 µl of 25 mM Tris-phosphate (pH 7.8), 2 mM EDTA (pH 8.0), 5% glycerol, 1% Triton X-100, 20 mM DTT and harvested from the 35-mm dishes. Luciferase activity of 5–30 µl aliquots from HeLa cell lysates was determined with 100 mM potassium phosphate (pH 7.8), 15 mM MgSO4, 5 mM ATP and 0.2 mM d-luciferin (Boehringer Mannheim). Protein concentrations were measured with the Bio-Rad protein assay kit. Relative light units were normalized for β-galactosidase activity (24) to correct for different transfection efficiencies.
Western blot analysis of transactivators in HeLa cells
Aliquots containing 10 µg total protein from cell extracts prepared for the luciferase assays were subjected to western blot analysis. A polyclonal TetR serum from rabbit was used to detect transregulators (laboratory stock). Signal intensities were normalized to the β-actin signals (Sigma). Bound antibodies were visualized using the ECL plus kit (Amersham).
RESULTS
Reverse properties are not readily transferable to TetR class E
Tetracycline resistance determinants isolated from different bacteria have been categorized into classes according to their amino acid sequence identity. They are designated by a Roman letter or a number (25). The original tTA (10) and rtTA (26) as well as rtTA-S2/M2 (12) are based on TetR class B. In current dual setups class B derived transregulators are often combined with regulators originating from class E because they will not form heterodimers (19,20). A future, versatile Tet controlled multi-gene regulation system should involve reverse transactivators. We thus tested whether different sets of mutations that introduce a reverse phenotype to class B derived regulators would also have this effect in class E based proteins. The first rtTA (11) contained the essential amino acid substitutions DN95, LS101, GD102 (U. Wellmann, V. Helbl and W. Hillen, unpublished results), while the improved rtTA-M1 (12) has the mutations SG12, EG19 and AP56. These two sets of mutations were used to generate the class E based rtTA2E and rtTA2E-M1 which contain TetR(B) DNA-reading heads to retain operator recognition and the VP16 derived minimal activation domains ‘FFF’ (13).
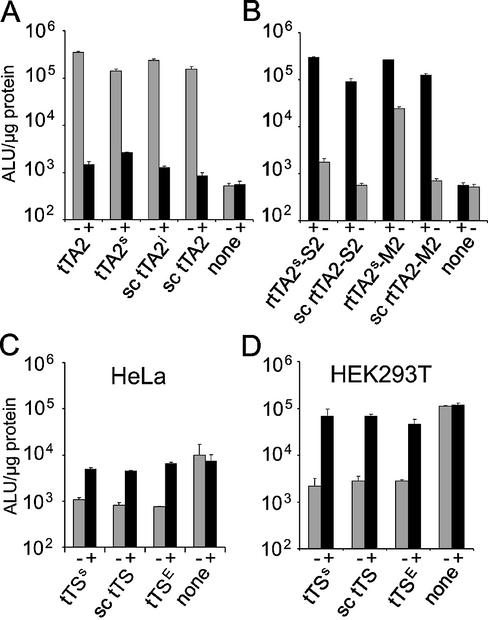
The regulatory properties of the reverse transactivators rtTA2, rtTA2E, rtTA2-M1 and rtTA2E-M1 were compared in transiently transfected HeLa cells. Plasmids encoding the transactivators were cotransfected with the reporter plasmid pUHC13-3 containing the luciferase gene under Ptet-1 control (10). Activation of expression as quantified by the luciferase activity is shown in Figure 1. The maximum levels of activation are ∼1200-fold above background and vary by <2.5-fold. Differences in regulation are seen in the absence of dox. Uninduced activity is not observed for rtTA2-M1, but for rtTA2E-M1, which, therefore, shows only 12-fold induction. rtTA2E exhibits a tTA phenotype with very low inducibility. Thus, the mutations leading to a reverse phenotype in the class B platform do not display the same properties in the class E background. As an alternative, we explore monomerization of Tet transregulators for the prevention of heterodimer formation.
Figure 1.
Transfer of the reverse phenotype to class E transregulators. HeLa cells were transiently cotransfected with plasmid pUHC13-3 carrying the luciferase gene under Ptet-1 control and plasmids encoding the indicated transactivators. Cells were grown in the absence (–) or presence (+) of 5 µg/ml dox. Luciferase activity was determined from cell extracts after 24 h. Values represent the means of triplicate samples with standard deviations given in arbitrary light units (ALU) per microgram of total cell protein.
Design of single-chain regulators
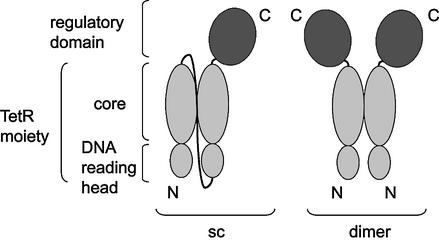
Two TetR (class B) proteins were fused genetically head to tail by a sequence encoding an (SG4)5 linker to yield a gene encoding a monomeric, sc version of the natural TetR dimer (Fig. 2). The 29 amino acid linker should be long and flexible enough to allow intramolecular assembly of the two TetR subunits according to the X-ray structure of TetR (6,7). TetR-(SG4)5-TetR is called sc TetR, and all other regulators containing this artificial monomer are designated by the prefix sc to distinguish them from their respective dimeric forms.
Figure 2.
Schematic outline of an sc and a dimeric Tet-transregulator. Sc: the C-terminus of the first TetR monomer (gray) is fused to the N-terminus of the second monomer by a (SG4)5 linker (black line). Sc TetR is converted into an eukaryotic transcription factor by fusing a regulatory domain (dark grey) to the C-terminus of the second subunit. Dimer: the C-terminus of each TetR monomer is fused to a regulatory domain. The DNA reading heads and core domains of TetR are indicated.
To avoid genetic instability due to identical sequence repeats, we have used the sequence variations wt and synthetic tetR, which encode the same amino acid sequence but differ in codon usage. Synthetic tetR was designed for expression in mammalian cells (12). All sc regulators contain one wt and one synthetic tetR gene. This allows two different arrangements having either the wt or synthetic tetR in front. All tests in E.coli were done with wt in the 5′ position.
Eukaryotic sc Tet transregulators contain an sc TetR moiety and one eukaryotic regulatory domain. To generate sc tTA2, sc TetR was fused to ‘FFF’ minimal activation domains derived from VP16 (13). Sc transactivators with a reverse phenotype were constructed using rtTA alleles S2 and M2 and their synthetic counterparts sS2 and sM2 (12), yielding sc rtTA2-S2 and sc rtTA2-M2, respectively. An sc Tet trans-silencer (sc tTS) was constructed by fusing a KRAB repression domain to sc TetR (16).
Activity profile of sc TetR in E.coli
Repression, induction by tc, and heterodimerization of sc TetR was compared with that of wt TetR in E.coli. Regulation of a tetA-lacZ transcriptional fusion located in single-copy in the genome of E.coli WH207 by tc was monitored at 37°C with high level and low level constitutive sc TetR expression plasmids (Table 1). Lowly expressed sc TetR leads to slightly enhanced repression and full inducibility compared to the wt. Highly expressed sc TetR shows slightly impaired induction. This is most likely due to an increased intracellular protein level of sc TetR compared to wt TetR as was determined by western blotting (data not shown).
Table 1. DNA-binding, inducibility by tc, and transdominance of sc TetR.
| TetR allele | β-Galactosidase activity (%) | Derepression factor | ||
|---|---|---|---|---|
| –TetRΔ9-11 | +TetRΔ9-11 | |||
| –tc | +tc | |||
| High expression | ||||
| None | 100 ± 5.0 | 100 ± 1.8 | ||
| wt | 1.0 ± 0.1 | 87 ± 2.4 | ||
| sc | 1.1 ± 0.1 | 51 ± 3.9 | ||
| Low expression | ||||
| None | 100 ± 3.7 | 100 ± 0.9 | 100 ± 3.4 | |
| wt | 2.2 ± 0.1 | 101 ± 4.9 | 36 ± 4.1 | 16.4 |
| sc | 1.3 ± 0.1 | 96 ± 4.9 | 1.2 ± 0.1 | 1.0 |
β-Galactosidase activities were determined according to Miller (24) at 37°C. The strain used was WH207(λtet50) (23) and the plasmids are either derivatives of pWH520 (27) in the high expression system or derivatives of pWH806 (23) in the low expression system. 100% β-galactosidase activity corresponds to 6784 units (high expression; –tc) (24), to 5709 units (high expression; +tc), to 6380 units (low expression; –tc), to 6002 units (low expression; +tc) or to 7168 units (low expression; +TetRΔ9-11), respectively.
Sc TetR could either assemble intramolecularly to form a monomer or aggregate intermolecularly to form higher order oligomers. To check whether sc TetR can interact with a second TetR variant in vivo, negative transdominance by TetRΔ9-11 was determined. In N-terminally truncated TetRΔ9-11, DNA binding is abolished but dimerization with the wt is efficient. Heterodimers containing wt and truncated subunits are inactive (27) so that their formation leads to decreased repression. A low amount of sc TetR or wt TetR was coexpressed with a large amount of TetRΔ9-11. Coexpression of TetRΔ9-11 and wt TetR yields a 15-fold derepression of β-gal expression indicating heterodimer formation (Table 1, columns 4 and 5). In contrast, repression exerted by sc TetR is not affected by the presence of TetRΔ9-11 and thus not subject to negative transdominance of TetRΔ9-11. This result suggests intramolecular assembly of sc TetR. In summary, sc TetR is an efficient repressor with tc inducibility like the wt protein but is not affected by negative transdominance.
Sc transregulator variants are active in HeLa cells
Two genes encoding sc tTA2 were constructed to determine potential effects of codon usage on expression efficiency. In one construct called sc tTA2, synthetic tetR was placed in front. The other construct called sc tTA2i has the wt tetR sequence in the front. Both constructs were analyzed in transiently transfected HeLa cells and compared to the corresponding dimeric regulators tTA2 and tTA2s as described above. Their identical TetR moieties are encoded by tetR and synthetic tetR, respectively, and are fused to the ‘FFF’ minimal activation domain. All transactivators show similar activation potentials in the absence of dox (Fig. 3A). Addition of dox leads to expression levels which are only ∼3-fold above background for tTA2, sc tTA2 and sc tTA2i and 5-fold above background for tTA2s. Sc tTA2 and sc tTA2i resemble their dimeric counterparts in activation and response to dox and are, therefore, equally efficient regulators. As no effects in regulatory properties could be seen with respect to the position of the synthetic tetR sequence, all other sc transregulator genes contain synthetic tetR in the front.
Figure 3.
Dox-dependent gene regulation by transregulators. Cells were cotransfected transiently with plasmid pUHC13-3 carrying the luciferase gene under Ptet-1 control and plasmids encoding the indicated transactivators. Cells were grown in the absence (–) or presence (+) of 5 µg/ml dox. Luciferase activity was determined as described in Figure 1. (A) Characterization of the transactivator variants tTA2, tTA2s, sc tTA2i and sc tTA2 in HeLa cells. The dimeric and the monomeric transactivators are identical, respectively, but encoded by different DNA sequences. (B) Characterization of the reverse transactivator variants rtTA2s-S2, sc rtTA2-S2, rtTA2s-M2 and sc rtTA2-M2 in HeLa cells. (C) Dox-dependent repression by the transsilencers tTSs, sc tTS and tTSE in HeLa cells and (D) in HEK293 cells.
Two transactivator variants with a reverse phenotype, sc rtTA2-S2 and sc rtTA2-M2 were compared with the corresponding dimeric rtTA2s-S2 and rtTA2s-M2 after transient transfection in HeLa cells as described above. Their regulatory properties are shown in Figure 3B. Two differences are observed: (i) activation of expression by sc rtTA2 variants is ∼2-fold lower; (ii) both sc rtTA2 variants show a noticeably decreased uninduced activity in the absence of dox that is only barely above the background level. It is up to 30-fold reduced compared to rtTA2s-S2 and rtTA2s-M2. As a result, the efficiency of induction of sc rtTA2 variants is excellent.
Steady state expression levels of sc transregulators in HeLa cells
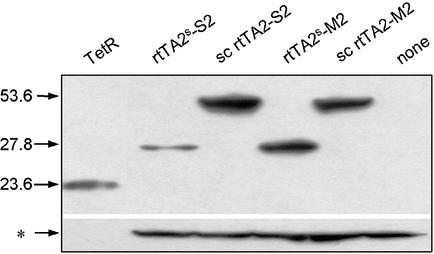
To investigate whether the lower activation exerted by the sc rtTA2 variants in the induced and the noninduced states is an effect of low protein concentration in the cell, the steady state amounts of the regulators were examined. Crude lysates of transiently transfected HeLa cells were subjected to SDS–PAGE followed by western blot analysis with polyclonal anti-TetR antibodies. As shown in Figure 4, the dimeric regulators rtTA2s-S2 and rtTA2s-M2 are stable during the incubation time (12). The protein amounts of sc rtTA2 variants are equal to or higher than those of the dimeric reverse transactivators. Furthermore, no degradation of the sc rtTA2 variants is detectable. Thus, the regulatory properties of the sc reverse transactivators are intrinsic features of these novel regulators.
Figure 4.
Western blot of reverse transactivator variants. Ten micrograms of total proteins from HeLa cell extracts were separated by SDS–PAGE. Western blot analysis was performed with anti-TetR polyclonal rabbit serum (laboratory stock) and detected with ECL+ (Amersham). Detection of purified TetR (15 ng) is shown in the first lane. The molecular weights of TetR, rtTA2s-S2, -M2 and those of their sc counterparts are indicated in kDa on the left side. The cellular protein marked with an asterisk was detected with antibody against β-actin and served as an internal loading standard for estimating the amounts of transactivators.
Sc and dimeric tTS show the same efficiency of regulation
The basal activity of the CMV minimal promoter or residual DNA-binding activity of a reverse transactivator can be repressed by the dox-inducible transsilencer tTS. We constructed a monomeric sc tTS by fusing sc TetR with the N-terminus of the KRAB repression domain of the mammalian Kox1 protein (16). The resulting sc tTS was compared with tTSE [referred to as transrepressor TetR(B/E)-KRAB in Forster et al. (18)] and its direct dimeric analog tTSs in transient transfections of HeLa cells (Fig. 3C). The HeLa cell line exhibits only weak but distinct basal activity of the Tet-responsive minimal promoter in the absence of a regulator. We also tested this construct in HEK 293T cells (Fig. 3D) because the CMV minimal promoter shows an up to 50-fold increased basal activity in this background (19). The three silencer constructs reduce luciferase activity in both cell lines. Repression was ∼8-fold in HeLa cells and 30-fold in HEK 293T cells irrespective of the transsilencer variant. Repression is completely relieved in the presence of dox. Thus, the regulatory properties of sc tTS and tTSE are indistinguishable.
Combinatorial regulation: reverse transactivator and transsilencer in the same cell
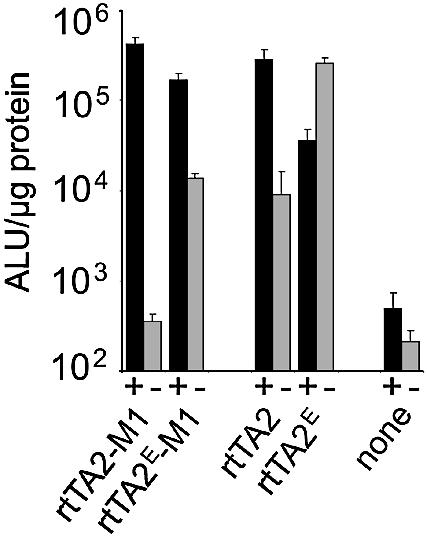
tTS type regulators bind to the target promoter in the absence of the effector while the rtTA type activators bind in its presence. Transsilencers are thus used to repress residual expression exerted by rtTA. Transcription is stimulated in the on-state of this dual system, whereas it is actively repressed in the off-state. We have constructed such a combinatorial set-up using monomeric regulators and compared it with one based on different TetR classes.
HeLa cells were transiently cotransfected with a plasmid encoding rtTA2s-M2 and a plasmid encoding either tTSE or the monomeric transsilencer sc tTS. Residual activity of rtTA2s-M2 in the absence of dox is efficiently reduced by both transsilencer constructs to the level reached when only the transsilencer is transfected (Fig. 5). Nevertheless, full activation by rtTA2s-M2 is achieved in the presence of dox. Thus, the dual system exhibits several hundred-fold induction. Sc tTS again shows the same repression characteristics as tTSE in this combinatorial approach.
Figure 5.
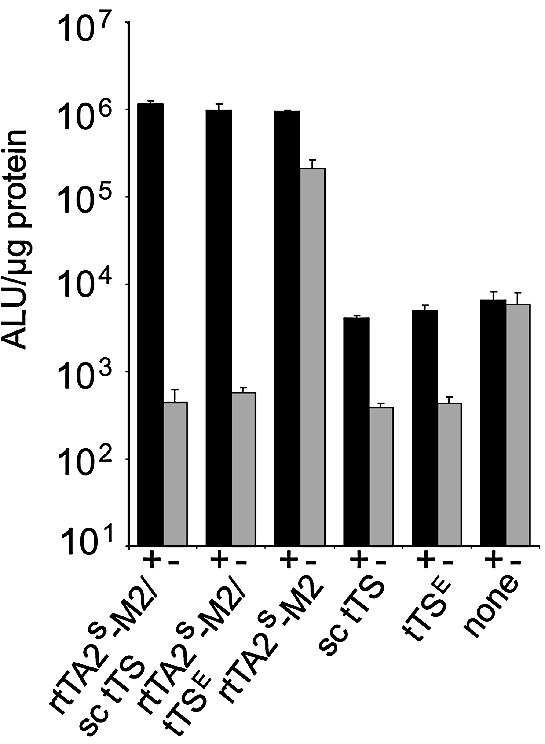
Combinatorial regulation by reverse transactivator and trans- silencer. HeLa cells were cotransfected with reporter plasmid pUHC13-3 and one or two regulator plasmids containing coding sequences for rtTA2s-M2, sc tTS or tTSE. Induction was performed by the addition of 5 µg/ml dox. Luciferase activity was determined as described in Figure 1.
DISCUSSION
sc TetR exhibits the same regulatory properties as the dimer but in contrast to the wt is not subject to negative transdominance. These findings indicate correct folding and intramolecular assembly of the monomerized repressor. In vitro experiments with other similarly designed sc DNA-binding proteins do not provide evidence for significant ‘cross-folding’ of the subunits (3,4).
When designing sc tetR, a wt and a synthetic tetR sequence optimized for mammalian codon usage were used to reduce potential recombination events. Low-usage codons are more likely to affect protein expression near the 5′ end of the mRNA (28). Sc tTA2 was constructed in both orientations to check for effects of codon usage. No differences in regulation were observed, suggesting that either wt and synthetic tetR sequences are suitable for good expression in HeLa or that translation efficiency is not limiting for regulation. These conclusions are in agreement with previous results indicating similar regulation efficiencies for synthetic and wt transregulators stably integrated into HeLa cell lines (29).
Regulation exerted by sc tTA2 and sc tTS variants in human cell lines is indistinguishable from regulation by the corresponding dimers. Sc rtTA2-S2/M2 show slightly reduced activation but also exhibit lower uninduced activity, which is not an effect of low protein amount. It should be emphasized that the uninduced activity of the dimeric rtTA2s-M2 is due to high regulator concentrations in transient assays and is abolished upon stable integration into the genome (12). Most applications feature stably transfected transregulators. However, the use of primary cells which can require transient assays may profit from a low activity in the uninduced state by transiently produced sc rtTA-M2.
Though the mechanisms underlying the reverse phenotype remain unknown, the reduction of activation should not be caused by structural constraints brought about by the linker sequence. Sufficient linker length and flexibility is required to ensure stability and activity of an sc protein (30). The sc TetR linker was designed to bridge the 61–66 Å between the C-terminus of the first subunit and the N-terminus of the second subunit as was estimated from the crystal structures of TetR(D) complexed with DNA or tetracycline (6,7). The distance covered by one amino acid residue in a fully extended polypeptide chain is 3.6 Å, thus, the minimum number of residues to cover this distance would be 19, assuming a fully extended conformation. However, linkers spanning ∼69 Å were modeled to pass around the protein surface, requiring a slightly longer linker. The linker sequence was based on earlier successful designs (3,31) and consists of glycine and serine residues which were chosen to maximize both flexibility and solubility. Thus, it seems reasonable to assume that the linker does not interfere with the three-dimensional structure of the transactivators. Nevertheless, we cannot exclude interactions of linker residues with the core protein. Alternatively, reduced activation could result from the smaller number of activation domains in the promoter region, although we consider this unlikely because this phenomenon is not observed with sc tTA2. Considering the complex structural changes of TetR upon induction, it is surprising that the conversion into an sc protein interferes so little with its regulatory properties. This makes it likely that altered phenotypes can be transferred to sc regulators in contrast to regulators based on different TetR classes.
Combinations of rtTA2s-M2 with tTSE or alternatively with sc tTS result in identically efficient regulation with high activation in the presence of the effector and active repression in its absence. Thus, both strategies are well suited to circumvent heterodimer formation, which is a prerequisite to set up multi-gene regulation in the same cell or organism using the Tet system. Though well functioning in present applications, exploiting different TetR classes may be a path of trial and tribulation for novel systems. Lack of heterodimerization has up to now only been demonstrated for regulators from the classes E and G with class B whereby the class G based trans-silencer exhibits a sensitivity for dox which is even lower than that of rtTA (32). More importantly, the different sets of mutations that introduce a reverse phenotype to first and second generation regulators based on TetR class B (11,12) failed to confer an efficient reverse phenotype to a TetR class E based regulator, which is routinely used in dual regulation setups (33–35). Though it may be possible to isolate reverse mutants in different TetR classes, the screening and characterization of new regulators is tedious. An elegant alternative is provided by sc transregulators which conserve the features of TetR-based regulators well. Taken together, the remodeling of Tet transregulators into monomers is not only an interesting paradigm for redesigning allosteric proteins but also adds promising new regulators to the Tet system.
Besides not needing a dimerization partner, sc transregulators exhibit a second difference to the known dimeric regulators: they contain only one regulatory domain per DNA binding unit. Thus, these sc regulators may avoid negative pleiotropic effects like squelching (36–38). Furthermore, the C-termini of TetR are only 23–26 Å apart in dimeric transregulators (6,7). Steric problems of two spacious complexes which have to assemble on the fused regulatory domain could arise in vivo and compromise regulation. Effects of the regulatory domain on transregulator properties have been reported for reverse transactivators: replacing the VP16 domain by ‘FFF’ minimal activation domains in the original rtTA resulted in drastically increased uninduced activity, while tTA and the second generation rtTA alleles allow this exchange without affecting specificity and inducibility (14). As sc transactivators harbor only one regulatory domain, they provide a possibility to minimize a potential influence of the fused domain on the properties of the transregulator.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr P. Schubert, L. Drüppel and S. Grimm for providing plasmids and C. Lang for technical assistance. This work was supported by the Bayerische Forschungsstiftung through the FORGEN initiative, by the Deutsche Forschungsgemeinschaft through SFB 473 and the Fonds der Chemischen Industrie Deutschlands.
REFERENCES
- 1.Jana R., Hazbun,T.R., Fields,J.D. and Mossing,M.C. (1998) Single-chain lambda Cro repressors confirm high intrinsic dimer-DNA affinity. Biochemistry, 37, 6446–6455. [DOI] [PubMed] [Google Scholar]
- 2.Simoncsits A., Chen,J., Percipalle,P., Wang,S., Toro,I. and Pongor,S. (1997) Single-chain repressors containing engineered DNA-binding domains of the phage 434 repressor recognize symmetric or asymmetric DNA operators. J. Mol. Biol., 267, 118–131. [DOI] [PubMed] [Google Scholar]
- 3.Robinson C.R. and Sauer,R.T. (1996) Covalent attachment of Arc repressor subunits by a peptide linker enhances affinity for operator DNA. Biochemistry, 35, 109–116. [DOI] [PubMed] [Google Scholar]
- 4.Sieber M. and Allemann,R.K. (1998) Single chain dimers of MASH-1 bind DNA with enhanced affinity. Nucleic Acids Res., 26, 1408–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muyan M., Yi,P., Sathya,G., Willmert,L.J., Driscoll,M.D., Hilf,R. and Bambara,R.A. (2001) Fusion estrogen receptor proteins: toward the development of receptor-based agonists and antagonists. Mol. Cell. Endocrinol., 182, 249–263. [DOI] [PubMed] [Google Scholar]
- 6.Kisker C., Hinrichs,W., Tovar,K., Hillen,W. and Saenger,W. (1995) The complex formed between Tet repressor and tetracycline-Mg2+ reveals mechanism of antibiotic resistance. J. Mol. Biol., 247, 260–280. [DOI] [PubMed] [Google Scholar]
- 7.Orth P., Schnappinger,D., Hillen,W., Saenger,W. and Hinrichs,W. (2000) Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nature Struct. Biol., 7, 215–219. [DOI] [PubMed] [Google Scholar]
- 8.Tiebel B., Garke,K. and Hillen,W. (2000) Observing conformational and activity changes of Tet repressor in vivo. Nature Struct. Biol., 7, 479–481. [DOI] [PubMed] [Google Scholar]
- 9.Bujard H. and Gossen,M. (2001) Tetracyclines in the control of gene expression in eukaryotes. In Nelson,M., Hillen,W. and Greenwald,R.A. (eds), Tetracyclines in Biology, Chemistry and Medicine. Birkhäuser Verlag, Berlin, pp. 139–157.
- 10.Gossen M. and Bujard,H. (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA, 89, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gossen M., Freundlieb,S., Bender,G., Müller,G., Hillen,W. and Bujard,H. (1995) Transcriptional activation by tetracyclines in mammalian cells. Science, 268, 1766–1769. [DOI] [PubMed] [Google Scholar]
- 12.Urlinger S., Baron,U., Thellmann,M., Hasan,M.T., Bujard,H. and Hillen,W. (2000) Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl Acad. Sci. USA, 97, 7963–7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baron U., Gossen,M. and Bujard,H. (1997) Tetracycline-controlled transcription in eukaryotes: novel transactivators with graded transactivation potential. Nucleic Acids Res., 25, 2723–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urlinger S., Helbl,V., Guthmann,J., Pook,E., Grimm,S. and Hillen,W. (2000) The p65 domain from NF-κB is an efficient human activator in the tetracycline-regulatable gene expression system. Gene, 247, 103–110. [DOI] [PubMed] [Google Scholar]
- 15.Akagi K., Kanai,M., Saya,H., Kozu,T. and Berns,A. (2001) A novel tetracycline-dependent transactivator with E2F4 transcriptional activation domain. Nucleic Acids Res., 29, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deuschle U., Meyer,W.K. and Thiesen,H.J. (1995) Tetracycline-reversible silencing of eukaryotic promoters. Mol. Cell. Biol., 15, 1907–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossi F.M., Guicherit,O.M., Spicher,A., Kringstein,A.M., Fatyol,K., Blakely,B.T. and Blau,H.M. (1998) Tetracycline-regulatable factors with distinct dimerization domains allow reversible growth inhibition by p16. Nature Genet., 20, 389–393. [DOI] [PubMed] [Google Scholar]
- 18.Forster K., Helbl,V., Lederer,T., Urlinger,S., Wittenburg,N. and Hillen,W. (1999) Tetracycline-inducible expression systems with reduced basal activity in mammalian cells. Nucleic Acids Res., 27, 708–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freundlieb S., Schirra-Muller,C. and Bujard,H. (1999) A tetracycline controlled activation/repression system with increased potential for gene transfer into mammalian cells. J. Gene Med., 1, 4–12. [DOI] [PubMed] [Google Scholar]
- 20.Baron U., Schnappinger,D., Helbl,V., Gossen,M., Hillen,W. and Bujard,H. (1999) Generation of conditional mutants in higher eukaryotes by switching between the expression of two genes. Proc. Natl Acad. Sci. USA, 96, 1013–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hillen W. and Berens,C. (1994) Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol., 48, 345–369. [DOI] [PubMed] [Google Scholar]
- 22.Berens C., Schnappinger,D. and Hillen,W. (1997) The role of the variable region in Tet repressor for inducibility by tetracycline. J. Biol. Chem., 272, 6936–6942. [DOI] [PubMed] [Google Scholar]
- 23.Wissmann A., Wray,L.V.,Jr, Somaggio,U., Baumeister,R., Geissendörfer,M. and Hillen,W. (1991) Selection for Tn10 tet repressor binding to tet operator in Escherichia coli: isolation of temperature-sensitive mutants and combinatorial mutagenesis in the DNA binding motif. Genetics, 128, 225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbour Laboratory Press, Cold Spring Harbour, NY.
- 25.Levy S.B., McMurry,L.M., Burdett,V., Courvalin,P., Hillen,W., Roberts,M.C. and Taylor,D.E. (1989) Nomenclature for tetracycline resistance determinants. Antimicrob. Agents Chemother., 33, 1373–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gossen M., Freundlieb,S., Bender,G., Muller,G., Hillen,W. and Bujard,H. (1995) Transcriptional activation by tetracyclines in mammalian cells. Science, 268, 1766–1769. [DOI] [PubMed] [Google Scholar]
- 27.Berens C., Altschmied,L. and Hillen,W. (1992) The role of the N terminus in Tet repressor for tet operator binding determined by a mutational analysis. J. Biol. Chem., 267, 1945–1952. [PubMed] [Google Scholar]
- 28.Goldman E., Rosenberg,A.H., Zubay,G. and Studier,F.W. (1995) Consecutive low-usage leucine codons block translation only when near the 5′ end of a message in Escherichia coli. J. Mol. Biol., 245, 467–473. [DOI] [PubMed] [Google Scholar]
- 29.Knott A., Garke,K., Urlinger,S., Guthmann,J., Müller,Y., Thellmann,M. and Hillen,W. (2002) Tetracycline-dependent gene regulation: combinations of transregulators yield a variety of expression windows. Biotechniques, 32, 796–807. [DOI] [PubMed] [Google Scholar]
- 30.Robinson C.R. and Sauer,R.T. (1998) Optimizing the stability of single-chain proteins by linker length and composition mutagenesis. Proc. Natl Acad. Sci. USA, 95, 5929–5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huston J.S., Levinson,D., Mudgett-Hunter,M., Tai,M.S., Novotny,J., Margolies,M.N., Ridge,R.J., Bruccoleri,R.E., Haber,E., Crea,R. et al. (1988) Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc. Natl Acad. Sci. USA, 85, 5879–5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossi F.M., Kringstein,A.M., Spicher,A., Guicherit,O.M. and Blau,H.M. (2000) Transcriptional control: rheostat converted to on/off switch. Mol. Cell, 6, 723–728. [DOI] [PubMed] [Google Scholar]
- 33.Mizuguchi H. and Hayakawa,T. (2001) Characteristics of adenovirus-mediated tetracycline-controllable expression system. Biochim. Biophys Acta, 1568, 21–29. [DOI] [PubMed] [Google Scholar]
- 34.Zhu Z., Ma,B., Homer,R.J., Zheng,T. and Elias,J.A. (2001) Use of the tetracycline controlled transcriptional silencer (tTS) to eliminate transgene leak in inducible overexpression transgenic mice. J. Biol. Chem., 30, 30. [DOI] [PubMed] [Google Scholar]
- 35.Lamartina S., Roscilli,G., Rinaudo,C.D., Sporeno,E., Silvi,L., Hillen,W., Bujard,H., Cortese,R., Ciliberto,G. and Toniatti,C. (2002) Stringent control of gene expression in vivo by using novel doxycycline-dependent trans-activators. Hum. Gene Ther., 13, 199–210. [DOI] [PubMed] [Google Scholar]
- 36.Gill G. and Ptashne,M. (1988) Negative effect of the transcriptional activator GAL4. Nature, 334, 721–724. [DOI] [PubMed] [Google Scholar]
- 37.Gilbert D.M., Heery,D.M., Losson,R., Chambon,P. and Lemoine,Y. (1993) Estradiol-inducible squelching and cell growth arrest by a chimeric VP16-estrogen receptor expressed in Saccharomyces cerevisiae: suppression by an allele of PDR1. Mol. Cell. Biol., 13, 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yueh Y.G., Yaworsky,P.J. and Kappen,C. (2000) Herpes simplex virus transcriptional activator VP16 is detrimental to preimplantation development in mice. Mol. Reprod. Dev., 55, 37–46. [DOI] [PubMed] [Google Scholar]