Abstract
Lactic acid-producing bacteria are associated with various plant and animal niches and play a key role in the production of fermented foods and beverages. We report nine genome sequences representing the phylogenetic and functional diversity of these bacteria. The small genomes of lactic acid bacteria encode a broad repertoire of transporters for efficient carbon and nitrogen acquisition from the nutritionally rich environments they inhabit and reflect a limited range of biosynthetic capabilities that indicate both prototrophic and auxotrophic strains. Phylogenetic analyses, comparison of gene content across the group, and reconstruction of ancestral gene sets indicate a combination of extensive gene loss and key gene acquisitions via horizontal gene transfer during the coevolution of lactic acid bacteria with their habitats.
Keywords: evolutionary genomics, fermentation
Lactic acid bacteria (LAB) are historically defined as a group of microaerophilic, Gram-positive organisms that ferment hexose sugars to produce primarily lactic acid. This functional classification includes a variety of industrially important genera, including Lactococcus, Enterococcus, Oenococcus, Pediococcus, Streptococcus, Leuconostoc, and Lactobacillus species. The seemingly simplistic metabolism of LAB has been exploited throughout history for the preservation of foods and beverages in nearly all societies dating back to the origins of agriculture (1). Domestication of LAB strains passed down through various culinary traditions and continuous passage on food stuffs has resulted in modern-day cultures able to carry out these fermentations. Today, LAB play a prominent role in the world food supply, performing the main bioconversions in fermented dairy products, meats, and vegetables. LAB also are critical for the production of wine, coffee, silage, cocoa, sourdough, and numerous indigenous food fermentations (2).
LAB species are indigenous to food-related habitats, including plant (fruits, vegetables, and cereal grains) and milk environments. In addition, LAB are naturally associated with the mucosal surfaces of animals, e.g., small intestine, colon, and vagina. Isolates of the same species often are obtained from plant, dairy, and animal habitats, implying wide distribution and specialized adaptation to these diverse environments. LAB species employ two pathways to metabolize hexose: a homofermentative pathway in which lactic acid is the primary product and a heterofermentative pathway in which lactic acid, CO2, acetic acid, and/or ethanol are produced (3).
Complete genome sequences have been published for eight fermentative and commensal LAB species: Lactococcus lactis, Lactobacillus plantarum, Lactobacillus johnsonii, Lactobacillus acidophilus, Lactobacillus sakei, Lactobacillus bulgaricus, Lactobacillus salivarius, and Streptococcus thermophilus (4–11). This study examines nine other LAB genomes representing the phylogenetic and functional diversity of lactic acid-producing microorganisms. The LAB have small genomes encoding a range of biosynthetic capabilities that reflect both prototrophic and auxotrophic characters. Phylogenetic analyses, comparison of genomic content across the group, and reconstruction of ancestral gene sets reveal a combination of gene loss and gain during the coevolution of LAB with animals and the foods they consumed.
Results and Discussion
General Features of the LAB Genomes.
The major features of the sequenced LAB genomes are summarized in Table 1, which is published as supporting information on the PNAS web site. The number of predicted protein-coding genes in the LAB differs from ≈1,700 to ≈2,800. Given the close phylogenetic relationship of these organisms, such a difference suggests substantial gene loss and/or gain in their evolution. In addition, all LAB genomes harbor pseudogenes. Strikingly, the number of pseudogenes differs by an order of magnitude, from <20 in Leuconostoc mesenteroides and Pediococcus pentosaceus to ≈200 in S. thermophilus (5) and Lactobacillus delbrueckii, indicating an active, ongoing process of genome degeneration. The LAB also differ in the number of rRNA operons, from two in Oenococcus oeni to nine in Lb. delbrueckii, which correlates with the number of tRNA genes (Table 1) and may reflect differences in the ecological competitiveness (e.g., capacity for rapid growth and production of lactic acid) between these bacteria (12, 13). All LAB genomes contained transposons, ranging from ≈0.2% of the genome in Lactobacillus gasseri to nearly 5% in Lc. lactis ssp. cremoris. Many LAB harbor plasmids, some of which are essential for growth in specific environments and carry genes for metabolic pathways, membrane transport, and bacteriocin production (14). The contribution of plasmid-encoded genes in the LAB ranges from 0% to 4.8% of total gene content (Table 1).
Phylogenetic Analysis and Impact of Horizontal Gene Transfer (HGT) on LAB Evolution.
The LAB analyzed here belong to the phylum Firmicutes, class Bacilli and order Lactobacillales, a sister taxon to Firmicutes, Bacilli, and Bacillales. Classification of Lactobacillales remains an unresolved issue in particular because phenotypic classification, which is traditionally based on the type of fermentation, does not match the rRNA-based phylogeny (15). Whole-genome DNA and DNA–RNA hybridization and GC content studies led to the delineation of three closely related lineages of Lactobacillales (16): the Leuconostoc group (Le. mesenteroides and O. oeni), the Lactobacillus casei–Pediococcus group (Lb. plantarum, Lb. casei, P. pentosaceus, and Lactobacillus brevis), and the Lb. delbrueckii group (Lb. delbrueckii, Lb. gasseri, and Lb. johnsonii) (15); streptococci (S. thermophilus) and lactococci (Lc. lactis ssp. lactis and Lc. lactis ssp. cremoris) formed a separate branch (16).
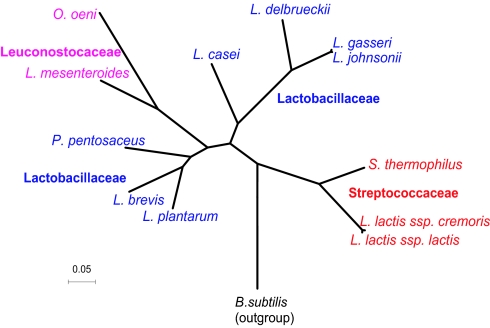
The availability of complete genomes for all major branches of Lactobacillales enables a more definitive analysis of their evolutionary relationships. We constructed phylogenetic trees from concatenated protein sequences, an approach shown to improve the resolution and increase robustness of phylogenetic analyses (17). We supplemented the ribosomal protein data set (Fig. 1) with concatenated RNA polymerase subunits (Fig. 4, which is published as supporting information on the PNAS web site), which also undergo little horizontal transfer. Both trees, constructed with a variety of methods, display the same topology with strongly supported internal branches. The streptococci–lactococci branch is basal in the Lactobacillales tree, and the Pediococcus group is a sister to the Leuconostoc group within the Lactobacillus clade. Thus, the Lactobacillus genus appears to be paraphyletic with respect to the Pediococcus–Leuconostoc group. Lactobacillus casei is confidently placed at the base of the Lb. delbrueckii group, which contradicts the previous classifications (16, 18).
Fig. 1.
Phylogenetic trees of Lactobacillales constructed on the basis of concatenated alignments of ribosomal proteins. All branches are supported at >75% bootstrap values. Species are colored according to the current taxonomy: Lactobacillaceae, blue; Leuconostocaceae, magenta; Streptococcaceae, red.
A molecular clock test (19) showed a high heterogeneity of evolutionary rates within Lactobacillales. Most of the root-to-tip distances are significantly unequal to the mean tree height; the previously reported (20) accelerated evolution of the Leuconostoc group (by a factor of 1.7–1.9 relative to the sister Pediococcus group) was especially prominent.
The strength of purifying selection acting on Lactobacillales species can be estimated by using two closely related pairs of genomes: Lb. gasseri/Lb. johnsonii and Lc. lactis/Lc. cremoris. Synonymous and nonsynonymous substitution rates were estimated from concatenated coding sequence alignments of 443 orthologous genes (142,031 codons). The dS/dN (distance at synonymous sites/distance at nonsynonymous sites) ratio was 38.5 ± 0.5 for the Lb. gasseri/Lb. johnsonii pair and 29.8 ± 0.4 for Lc. lactis/Lc. cremoris pair, showing unusually strong evolutionary pressure as compared with Proteobacteria, which has a characteristic dS/dN ratio of 5–10 (21). This is likely to reflect the large effective population size and/or high mutation rate of the Lactobacillales species because the intensity of purifying selection is known to be proportional to these quantities (22).
Clusters of Orthologous Genes in Lactobacillales.
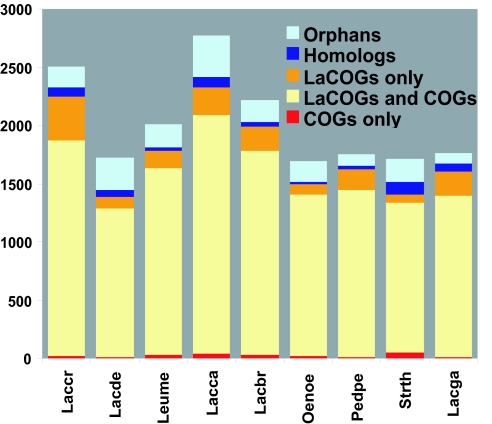
Robust identification of sets of orthologs (genes derived from the same ancestral gene) is a prerequisite for informative evolutionary–genomic analysis of any group of organisms. By using the computational procedure described previously (23), we constructed Lactobacillales-specific clusters of orthologous genes [LaCOGs (abbreviated COGs if clusters of orthologous genes are not Lactobacillales-specific)] for the Lactobacillales-specific set (Table 2, which is published as supporting information on the PNAS web site) from proteins encoded in 12 sequenced Lactobacillales genomes that were available at the time of this analysis. Many COGs include paralogous genes that evolved via duplications at different stages of evolution. The construction of orthologous clusters for a compact taxon, such as the Lactobacillales, results in much finer granularity, with a greater fraction of clusters containing a single member from all or most of the analyzed species. Altogether, 3,199 LaCOGs, which included from 2 species to all 12 species, were identified. On average, LaCOGs covered 86% of the genome (Fig. 2); 1,133 (35%) LaCOGs showed a one-to-one correspondence with the general COG set; 1,359 (43%) LaCOGs corresponded to 390 COGs that have been split into two or more paralogous groups. The remaining 707 (22%) LaCOGs have no counterparts in the general COG set (24); of these, 338 (11%) were shared with one or more non-Lactobacillales bacterial genomes among those reported recently and not yet included in the COGs, and 369 (11%) appeared to be specific to the Lactobacillales. Thus, the LaCOGs are a powerful resource for genome annotation and evolutionary analysis of Lactobacillales (for details, see Table 2).
Fig. 2.
Conserved and unique genes in the genomes of Lactobacillales. “Homolog” indicates genes with detectable homologs in organisms other than those analyzed here but could not be included in any of the COG sets. “Orphans” are predicted genes without detectable homologs. Lacga, Lb. gasseri; Lacbr, Lb. brevis; Pedpe, P. pentosaceus; Laccr, Lc. lactis ssp. cremoris; Strth, S. thermophilus; Oenoe, O. oeni; Leume, Le. mesenteroides; Lacca, Lb. casei; Lacde, Lb. delbrueckii; Lacla, Lc. lactis; Lacpl, Lb. plantarum; Lacjo, Lb. johnsonii. The vertical axis shows the number of genes per genome.
The conserved core of genes present in all 12 species analyzed in the Lactobacillales genomes (Table 2) consists of 567 LaCOGs (18%). Functional distribution of the LaCOGs in this core shows that the majority encode components of the information-processing systems (translation, transcription, and replication). However, the core also includes 41 uncharacterized genes and 50 genes with only a general prediction of biochemical activity. Because these genes are conserved throughout Lactobacillales, it is likely that they have essential functions, at least within this group. Furthermore, two core genes have no detectable orthologs outside lactobacilli. One of these unique genomic markers of Lactobacillales contains a LysM (peptidoglycan-binding) domain (LaCOG01826). In several lactobacilli, this gene is located next to the genes for ribosomal proteins and cytidylate kinase and might be coregulated with these housekeeping genes. The second genomic marker, the highly conserved LaCOG01237, contains no characterized domains. However, this gene is located in a conserved genomic neighborhood encoding two enzymes implicated in 4-thiouridine modification of tRNA [(5-methylaminomethyl-2-thiouridylate) methyltransferase and a predicted sulfurase] (LaCOG00578 and LaCOG01188; see Table 2), suggesting a role of LaCOG01237 proteins in specific modulation of this essential modification (25).
Local Molecular Clock and HGT.
We tested the consistency of a local molecular clock in individual LaCOGs with a technique developed recently (26). A matrix of interspecies distances for each LaCOG was compared with the matrix of baseline distances obtained from the concatenated alignment of ribosomal proteins. If a COG evolves in a clock-like manner relative to the evolution of ribosomal proteins, a linear dependence between the two matrices is observed. At least ≈25% of the LaCOGs showed strong deviations from the linear dependence, suggesting a high level of HGT and/or major local accelerations of evolution (Fig. 5, which is published as supporting information on the PNAS web site). Several functional groups of genes show statistically significant differences in their propensity to violate the local molecular clock (Table 3, which is published as supporting information on the PNAS web site). Of particular interest is the substantially elevated level of such violations among genes involved in sugar metabolism, including key enzymes, such as phosphoketolase, transketolase, and various components of phosphotransferase systems (Table 4, which is published as supporting information on the PNAS web site). HGT via bacteriophage-mediated or conjugative mechanisms has been extensively documented in the Lactobacillales and is a prominent process for niche-specific adaptation in the lactococci (27). Noteworthy is the potential for such transfer in Lc. lactis ssp. cremoris SK11 that harbors a conjugative plasmid (pLAC3) and several additional plasmids encoding genes related to growth in milk (28).
Reconstruction of Gene Gain and Loss in the Evolution of Lactobacillales.
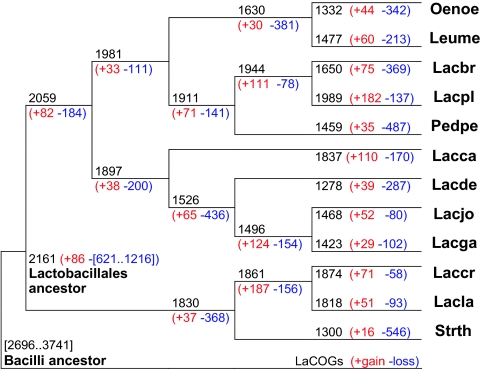
We used a version of the weighted parsimony algorithm (29) to reconstruct the events that occurred during the evolution of this group after its divergence from the common ancestor of all Bacilli (see Materials and Methods). This reconstruction suggests that the common ancestor of Lactobacillales had at least ≈2,100–2,200 genes, losing 600–1,200 genes (≈25–30%) and gaining <100 genes compared with the ancestor of all Bacilli, for which the genome size of ≈2,700–3,700 genes is predicted (Fig. 3). Thus, the origin of Lactobacillales involved extensive loss of ancestral genes (Table 5, which is published as supporting information on the PNAS web site). Many of the changes mapped to this stage of evolution seem to be related to the transition to life in a nutritionally rich medium. Thus, a number of genes for biosynthesis of cofactors were lost; conversely, a variety of peptidases were acquired, apparently via HGT. The Lactobacillales ancestor was likely a microaerophile or an anaerobe, which is reflected in the loss of heme/copper-type cytochrome/quinol oxidase-related genes and catalase, characteristic enzymes of aerobic bacteria. In addition, several probable nonorthologous gene displacements via HGT were identified (Table 5). Furthermore, Lactobacillales (or the common ancestor of Bacilli) might have acquired the complete mevalonate pathway via HGT, possibly from an archaeal source (directly or through a bacterial intermediate). This pathway displaced the ancestral bacterial deoxyxylulose pathway of isoprenoid biosynthesis. The acquisition of the mevalonate pathway tree with a subsequent duplication of the mevalonate kinase gene is supported by the specific organization of the genes for four enzymes of this pathway [mevalonate and phosphomevalonate kinases, mevalonate pyrophosphate decarboxylase, and isopentenyl-diphosphate δ-isomerase (LaCOGs 296, 298, 297, and 299, respectively)] in a single operon that is conserved in most Lactobacillales genomes.
Fig. 3.
Reconstruction of gene content evolution in Lactobacillales. The tree topology is as in Fig. 1, rooted by using Baccillus subtilis as the outgroup. For each species and each internal node of tree, the inferred number of LaCOGs present, and the numbers of LaCOGs lost (blue) and gained (red) along the branch leading to the given node (species) are indicated. Abbreviations are as in Fig. 2.
In addition to the metabolic reduction, a major part of the gene loss in the common ancestor of Lactobacillales were sporulation-related functions encoded by the common ancestor of Bacilli.Despite the absence of genes for sporulation, catalase, and other key enzymes of oxidative stress response (e.g., superoxide dismutase) in 8 of the 12 genomes analyzed here (possibly multiple losses), at least some lactobacilli show enhanced stress resistance. This resistance is demonstrated by the increased recovery of live lactobacilli from vacuum-dried and irradiated food (30) by comparison to staphylococcal and Salmonella species. This resistance may be mediated in part by the low content of iron, a potent oxidant, which is accompanied by accumulation of manganese, a powerful antioxidant (31–33). Additional protection is likely to be provided by other antioxidants, including glutathione and γ-glutamylcysteine. Several Lactobacillus species encode a bifunctional glutathione synthetase (GshAB), whereas others have only γ-glutamylcysteine synthetase (GshA) (LaCOG01892). However, even lactococci that cannot synthesize glutathione have been shown to accumulate it, apparently via transport from the environment (34).
Loss of ancestral genes seems to be the prevailing trend in the evolution of Lactobacillales, as in other bacteria (35). Like all other bacterial lineages (36), lactobacilli also have a significant number of expanded gene families that evolved either by lineage-specific gene duplication or by acquisition of paralogous genes via HGT (37). A closer examination of these families indicates that adaptation to growth in nutrient-rich environments was the major driving force behind the fixation of duplications during the evolution of the Lactobacillales (Table 6, which is published as supporting information on the PNAS web site). An interesting case of ancient gene acquisition is the second enolase, which is characteristic of the Lactobacillales. All other bacteria have a single copy of this nearly ubiquitous glycolytic enzyme, but most of the Lactobacillales have two (with some differential gene loss). Phylogenetic analysis shows that one of these copies is the ancestral version in Gram-positive bacteria, whereas the other copy had been acquired by the ancestor of the Lactobacillales from a different bacterial lineage, most likely, Actinobacteria. The evolution rate of both enolases seems to be increased in the Lactobacillales branches of the phylogenetic tree (Fig. 6, which is published as supporting information on the PNAS web site). It has been shown that both enolases of Lc. lactis ssp. lactis have enzymatic activity (38); however, their specific physiological functions remain unknown. Many other genes for proteins involved in sugar metabolism and transport were duplicated early in the evolution of the Lactobacillales, including phosphoenolpyruvate phosphotransferase systems, β-galactosidase, GpmB family sugar phosphatases, galactose mutarotase, and l-lactate dehydrogenases of two distinct classes. In addition to the apparent acquisition of new peptidases via HGT, duplications of several lineage-specific genes for these enzymes and for amino acid transporters were detected. Several paralogous expansions include genes for putative proteins related to those involved in antibiotic resistance in other bacteria, such as β-lactamases and penicillin V acylase. However, most LAB species analyzed here have been shown to be sensitive to common antibiotics and, after centuries of consumption by humans, have been, accordingly, “generally recognized as safe” (39, 40). Conceivably, the homologs of antibiotic-resistance genes are involved in normal cell-wall biosynthesis in the Lactobacillales. In the same context, expansion of a distinct family of tyrosine/serine phosphatases, which are often localized in the same operon with a serine/threonine protein kinase fused to several β-lactam-binding domains, is likely to be important for the regulation of cell-wall biosynthesis (41). Furthermore, Lactobacillales encode a paralog of class II lysyl-tRNA synthetase, which is fused to a membrane-associated domain (COG2898) implicated in oxacillin-like antibiotic resistance (42) and is probably involved in cell-wall biosynthesis.
The subsequent evolution of the Lactobacillales reveals ancestral gene loss and metabolic simplification but also a considerable number of lineage-specific duplications and acquisitions of unique genes. Numerous parallel gene losses, especially of genes coding for biosynthetic enzymes, were detected in the major branches of Lactobacillales, which presumably reflects similar environmental pressures. For instance, genes for serine and glycine biosynthesis were lost in the common ancestor of Lactobacillaceae and Leuconostocaceae; genes for biosynthesis of arginine and aromatic amino acids were lost independently in Lb. brevis, P. pentosaceus, O. oeni, and the Lb. casei–Lb. delbrueckii group; and several fatty-acid-biosynthesis genes were lost in the Lb. gasseri and Lb. johnsonii branch (Table 2). Lineage-specific gene loss was extensive in the evolution of all lineages of Lactobacillales, but several species were especially notable “losers.” In particular, S. thermophilus not only lost numerous genes but also exhibited many fresh pseudogenes, suggesting an active and ongoing process of genome decay, similarly reported for two different strains of the same species (5). Moreover, substantial gene loss (368 genes according to the present reconstruction) also occurred at the base of the streptococci–lactococci branch (including several genes involved in cell division that are conserved in most bacteria, such as crcB, mreB, mreC, and MinD). Other lineages particularly prone to gene loss are P. pentosaceus (487 genes lost) and the Leuconostoc and Oenococcus branch, with 381 genes lost at the base of the branch and considerable additional loss in each species. Substantial gene loss also occurred during the evolution of the Lb. delbrueckii group (Lb. delbrueckii, Lb. gasseri, and Lb. johnsonii), leading to additional genome reduction in Lb. gasseri and Lb. johnsonii (Fig. 3). In the species with larger genomes, such as Lb. plantarum and Lb. casei, the loss of ancestral genes was counterbalanced by the emergence of many new genes via duplication and HGT (Fig. 3).
Comparison of the number of genes lost or gained on a particular tree branch and the length of the corresponding branch reveals a pattern similar to that described previously for Proteobacteria (43). The number of gene losses (even when normalized by the size of the ancestral genome) strongly and significantly correlates with the branch length determined from sequence divergence (R = 0.68; P < 5 × 10−4), whereas the number of gene gains (again, regardless of normalization) does not show such a correlation (R = 0.16; P > 0.1). The clock-like behavior of gene loss is consistent with a large number of small-scale events, which are randomly distributed along the evolutionary path. This pattern suggests evolution under purifying selection. In contrast, the lack of such correlation for gene gain appears to involve relatively large batches of genes acquired at a time, with longer intervals between the acquisition events, perhaps because of positive selection.
In addition to the reconstruction of the ancestral gene sets, we compared the genome organizations of all of the sequenced Lactobacillales genomes with previously developed computational methods (44). Only closely related species showed significant genome colinearity above the level of individual operons, and there was virtually no large-scale conservation of gene order between the four major groups of the Lactobacillales (data not shown). Thus, the processes of gene loss and acquisition during the evolution of these bacteria were accompanied by extensive genome rearrangements.
Phyletic Patterns and Central Metabolism Reconstruction.
Given the prominence of sugar metabolism and energy conversion systems in Lactobacillales, we examined the evolution of these systems through phyletic patterns, reflecting the presence or absence of genes in individual genomes in a manner similar to that described in ref. 45. Most of the genes involved in these functions are represented in all species (Fig. 7 and Table 7, which are published as supporting information on the PNAS web site). These genes include those coding for the downstream part of glycolysis, from glyceraldehyde-3P to pyruvate and pyruvate conversion to lactate and 2,3-butandiol; acetate formation from acetyl-CoA; several reactions of the pentose–phosphate pathway; and the mannose-specific phosphotransferase system. Clearly, these enzymes are insufficient to completely define the metabolism of any individual species, and several reactions are specific to individual lineages. The presence/absence patterns of key enzymes involved in lactate fermentation poorly correlate with the phenotypes of the Lactobacillales (Table 7). However, it has been shown that under certain conditions Lactobacillales can switch between sole production of lactic acid and the production of mixed end products, including acetic acid, lactic acid, ethanol, and CO2 (46, 47).
The metabolic potential of the Lactobacillales is complemented by its predicted transport capabilities. In particular, amino acid uptake systems dominate over sugar and peptide uptake systems. Among the detected sugar uptake systems, those specific for oligosaccharides and glycosides outnumber those for free sugars. In addition, Lactobacillales encode a variety of predicted drug, peptide, and macromolecular efflux pumps, some of which are likely to be involved in intercellular signaling.
Other metabolic capabilities of Lactobacillales are listed in Table 8, which is published as supporting information on the PNAS web site. Generally, Lb. brevis, Lb. johnsonii, Lb. gasseri, Lb. delbrueckii, and P. pentosaceus have extremely narrow repertoires of biosynthetic pathways, whereas Lc. lactis ssp. lactis, Lc. lactis ssp. cremoris, Lb. plantarum, and Le. mesenteroides retain a much broader biosynthetic repertoire.
The Bacteriocins.
Lactobacillales are known for producing specific antimicrobial peptides, the bacteriocins (48, 49). Several proteins are responsible for the modification, export, and regulation of bacteriocin production and are often encoded in the same operon with the bacteriocins (48, 49). Because bacteriocins are small proteins with highly diverged sequences, they are often hard to identify by amino acid conservation. Therefore, genome context analysis is required for a more complete characterization of the bacteriocin repertoire. Among the Lactobacillales genomes analyzed here, seven have clustered genes for (putative) bacteriocins and associated proteins. Within these regions, we identified two prebacteriocin families. One family consists of precursors of a known bacteriocin, pediocin from P. pentosaceus, homologs of which also are present in Le. mesenteroides and Lb. casei (LaCOG01709). The second family consists of previously unidentified putative bacteriocin precursors distantly related to Divercin V41 (50) and present in P. pentosaceus and Lb. johnsonii (LaCOG03352). In addition, numerous small ORFs located in the immediate vicinity of the putative genes for bacteriocins and associated proteins might encode novel peptides (Fig. 8, which is published as supporting information on the PNAS web site). Bacteriocin-production-related genes seem to be among those that are often transferred horizontally as indicated by the analysis of the respective phylogenetic trees and differences in the operon organization, even in closely related genomes.
Concluding Remarks.
This work is an extensive comparative analysis of a compact group of relatively closely related prokaryotic genomes that show a gradient of sequence conservation. Loss of ancestral genes and metabolic simplification are the central trends of LAB evolution. Major gene loss already occurred at the stage of the common ancestor of Lactobacillales, which indicates early adaptation to nutritionally rich environments. However, genome degradation appears to be an ongoing process given that all species of Lactobacillales show loss of specific genes, and many possess numerous pseudogenes. Beyond gene loss, Lactobacillales have clear ancestral adaptations for nutritionally rich, microaerophilic environments, which include acquisition via HGT and duplication of genes for various enzymes and transporters of sugar and amino acid metabolism. The molecular systems responsible for the production of specific antimicrobials, such as the bacteriocins, are among other adaptations that become apparent through comparative genomic analysis, probably reflecting the long-term existence of Lactobacillales in complex microbial communities. Comparison of the genomes of Lactobacillales suggests that the milk-digesting phenotype evolved independently in different bacterial lineages. This phenotype apparently does not require a unique set of genes but rather emerged through assortment and adaptation of enzymes shared with other bacteria.
The comparative genomic analysis described here also suggests a revision of the taxonomy of the Lactobacillales. Phylogenetic analysis of multiple protein sequences showed that the streptococci–lactococci branch is basal in the Lactobacillales tree and that the Pediococcus group is a sister to the Leuconostoc group, which supports the paraphyly of the Lactobacillus genus. Furthermore, Lb. casei is confidently placed at the base of the Lb. delbrueckii group, which contradicts the earlier classification.
Materials and Methods
Whole-genome shotgun sequencing was carried out at the U.S. Department of Energy Joint Genome Institute. Genomes were sequenced to ≈8× depth and assembled by using Jazz, the Joint Genome Institute assembler (51). Gap closure was carried out at Fidelity Systems, Inc., by using direct genomic sequencing (52).
ORFs were identified with the GeneMarkS program (53). Gene functions were predicted by assigning predicted genes to COGs (www.ncbi.nlm.nih.gov/COG) by using the COGNITOR method (24) and by database searches conducted with the PSI-BLAST program (54). Transfer RNAs were predicted with the tRNAscan-SE program (55). LaCOGs were constructed by using previously described procedures (23, 56). Phylogenetic analysis was performed by using the least-square or maximum-likelihood methods, and gene gain/loss scenarios were reconstructed with a version of the weighted parsimony algorithm (29).
Additional methodological details and a detailed list of data deposition numbers are provided in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We dedicate this work to the career and many contributions of Prof. Larry McKay, a pioneering researcher on the genetics of lactic acid bacteria. This work was supported by the Intramural Program of the National Library of Medicine; the U.S. Department of Energy Office of Science, Biological, and Environmental Research Program and the University of California, Lawrence Livermore National Laboratory under Contract W-7405-Eng-48; the Lawrence Berkeley National Laboratory under Contract DE-AC03-76SF00098l; and the Los Alamos National Laboratory under Contract W-7405-ENG-36. Genome closure was supported by Department of Energy Small Business Innovative Research Grants DE-FG02-98ER82577 and DE-FG02-00ER83009 (to A. Slesarev and S.K.). Additional funding came from the American Vineyard Foundation (to D.M.); the California Competitive Grants Program for Research in Viticulture and Enology (to D.M.); the Western Dairy Center (to B.W.); the Utah Agricultural Experiment Station (to B.W.); Dairy Management, Inc. (to B.W., T.K., and D.O.); Danisco, Inc. (to T.K.); the Southeast Dairy Foods Research Center (to T.K.); the North Carolina Dairy Foundation (to T.K.); the North Carolina Agricultural Research Service (to T.K.); National Institutes of Health Grant R01 GM55434 (to M.S.); the U.S. Department of Agriculture Agricultural Research Service Food Science Research Unit (to F.B.); Pickle Packers International, Inc. (to F.B.); the Utah State Community University Research Initiative (to J.B.); the Nebraska American Dairy Association (to R.H.); Minnesota Agriculture Experiment Station Project 18-062 (to K.B.); the Minnesota–South Dakota Dairy Foods Research Center (to D.O.); and the University of Wisconsin College of Agriculuture and Life Science (to J.S.).
Abbreviations
- LAB
lactic acid bacteria
- COG
cluster of orthologous genes
- LaCOG
Lactobacillales-specific COG
- HGT
horizontal gene transfer
Footnotes
References
- 1.Miller N, Wetterstrom W. In: The Cambridge World History of Food. Kiple K, Ornelas K, editors. Vol 2. Cambridge, UK: Cambridge Univ Press; 2000. pp. 1123–1139. [Google Scholar]
- 2.Wood B. Microbiology of Fermented Foods. London: Blackie; 1998. [Google Scholar]
- 3.Kandler O. Antonie van Leeuwenhoek. 1983;49:209–224. doi: 10.1007/BF00399499. [DOI] [PubMed] [Google Scholar]
- 4.Altermann E, Russell W, Azcarate-Peril M, Barrangou R, Buck B, McAuliffe O, Souther N, Dobson A, Duong T, Callanan M, et al. Proc Natl Acad Sci USA. 2005;102:3906–3912. doi: 10.1073/pnas.0409188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolotin A, Quinquis B, Renault P, Sorokin A, Ehrlich S, Kulakauskas S, Lapidus A, Goltsman E, Mazur M, Pusch G, et al. Nat Biotechnol. 2004;22:1554–1558. doi: 10.1038/nbt1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, Ehrlich S, Sorokin A. Genome Res. 2001;11:731–753. doi: 10.1101/gr.169701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaillou S, Champomier-Verges M, Cornet M, Crutz-Le Coq A, Dudez AM, Martin V, Beaufils S, Darbon-Rongere E, Bossy R, Loux V, Zagorec M. Nat Biotechnol. 2005;23:1527–1533. doi: 10.1038/nbt1160. [DOI] [PubMed] [Google Scholar]
- 8.Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters S, Sandbrink H, Fiers M, et al. Proc Natl Acad Sci USA. 2003;100:1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pridmore R, Berger B, Desiere F, Vilanova D, Barretto C, Pittet A, Zwahlen M, Rouvet M, Altermann E, Barrangou R, et al. Proc Natl Acad Sci USA. 2004;101:2512–2517. doi: 10.1073/pnas.0307327101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claesson M, Li Y, Leahy S, Canchaya C, van Pijkeren J, Cerdeno-Tarraga A, Parkhill J, Flynn S, O'Sullivan G, Collins J, et al. Proc Natl Acad Sci USA. 2006;103:6718–6723. doi: 10.1073/pnas.0511060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van de Guchte M, Penaud S, Grimaldi C, Barbe V, Bryson K, Nicolas P, Robert C, Oztas S, Mangenot S, Couloux A, et al. Proc Natl Acad Sci USA. 2006;103:9274–9279. doi: 10.1073/pnas.0603024103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klappenbach J, Dunbar J, Schmidt T. Appl Environ Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Mattia E, Grego S, Cacciari I. Microb Ecol. 2002;43:34–43. doi: 10.1007/s00248-001-1035-3. [DOI] [PubMed] [Google Scholar]
- 14.McKay L, Baldwin K. FEMS Microbiol Rev. 1990;7:3–14. doi: 10.1111/j.1574-6968.1990.tb04876.x. [DOI] [PubMed] [Google Scholar]
- 15.Vandamme P, Pot B, Gillis M, de Vos P, Kersters K, Swings J. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wood B, Holzapfel W. The Genera of Lactic Acid Bacteria. Glasgow, UK: Blackie; 1995. [Google Scholar]
- 17.Wolf Y, Rogozin I, Grishin N, Tatusov R, Koonin E. BMC Evol Biol. 2001;1:8. doi: 10.1186/1471-2148-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siezen R, van Enckevort F, Kleerebezem M, Teusink B. Curr Opin Biotechnol. 2004;15:105–115. doi: 10.1016/j.copbio.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Takezaki N, Rzhetsky A, Nei M. Mol Biol Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- 20.Yang D, Woese C. Syst Appl Microbiol. 1989;12:145–149. [Google Scholar]
- 21.Jordan I, Rogozin I, Wolf Y, Koonin E. Theor Popul Biol. 2002;61:435–447. doi: 10.1006/tpbi.2002.1588. [DOI] [PubMed] [Google Scholar]
- 22.Lynch M, Conery J. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 23.Tatusov R, Koonin E, Lipman D. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 24.Tatusov R, Fedorova N, Jackson J, Jacobs A, Kiryutin B, Koonin E, Krylov D, Mazumder R, Mekhedov S, Nikolskaya A, et al. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leipuviene R, Qian Q, Bjork G. J Bacteriol. 2004;186:758–766. doi: 10.1128/JB.186.3.758-766.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novichkov P, Omelchenko M, Gelfand M, Mironov A, Wolf Y, Koonin E. J Bacteriol. 2004;186:6575–6585. doi: 10.1128/JB.186.19.6575-6585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood B, Warner P. Genetics of Lactic Acid Bacteria. New York: Kluwer Academic/Plenum; 2003. [Google Scholar]
- 28.Siezen R, Renckens B, van Swam I, Peters S, van Kranenburg R, Kleerebezem M, de Vos W. Appl Environ Microbiol. 2005;71:8371–8382. doi: 10.1128/AEM.71.12.8371-8382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirkin B, Fenner T, Galperin M, Koonin E. BMC Evol Biol. 2003;3:2. doi: 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hastings J, Holzapfel W, Niemand J. Appl Environ Microbiol. 1986;52:898–901. doi: 10.1128/aem.52.4.898-901.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horsburgh M, Wharton S, Karavolos M, Foster S. Trends Microbiol. 2002;10:496–501. doi: 10.1016/s0966-842x(02)02462-9. [DOI] [PubMed] [Google Scholar]
- 32.Daly M, Gaidamakova E, Matrosova V, Vasilenko A, Zhai M, Venkateswaran A, Hess M, Omelchenko M, Kostandarithes H, Makarova KS, et al. Science. 2004;306:1025–1028. doi: 10.1126/science.1103185. [DOI] [PubMed] [Google Scholar]
- 33.Archibald F, Fridovich I. J Bacteriol. 1981;145:442–451. doi: 10.1128/jb.145.1.442-451.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Hugenholtz J, Abee T, Molenaar D. Appl Environ Microbiol. 2003;69:5739–5745. doi: 10.1128/AEM.69.10.5739-5745.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochman H. Proc Natl Acad Sci USA. 2005;102:11959–11960. doi: 10.1073/pnas.0505863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jordan I, Makarova K, Spouge J, Wolf Y, Koonin E. Genome Res. 2001;11:555–565. doi: 10.1101/gr.166001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koonin E, Makarova K, Aravind L. Annu Rev Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jamet E, Ehrlich S, Duperray F, Renault P. Lait. 2001;81:115–129. [Google Scholar]
- 39.Katla A, Kruse H, Johnsen G, Herikstad H. Int J Food Microbiol. 2001;67:147–152. doi: 10.1016/s0168-1605(00)00522-5. [DOI] [PubMed] [Google Scholar]
- 40.Teuber M, Meile L, Schwarz F. Antonie van Leeuwenhoek. 1999;76:115–137. [PubMed] [Google Scholar]
- 41.Yeats C, Finn RD, Bateman A. Trends Biochem Sci. 2002;27:438. doi: 10.1016/s0968-0004(02)02164-3. [DOI] [PubMed] [Google Scholar]
- 42.Nishi H, Komatsuzawa H, Fujiwara T, McCallum N, Sugai M. Antimicrob Agents Chemother. 2004;48:4800–4807. doi: 10.1128/AAC.48.12.4800-4807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snel B, Bork P, Huynen M. Genome Res. 2002;12:17–25. doi: 10.1101/gr.176501. [DOI] [PubMed] [Google Scholar]
- 44.Wolf Y, Rogozin I, Kondrashov A, Koonin E. Genome Res. 2001;11:356–372. doi: 10.1101/gr.gr-1619r. [DOI] [PubMed] [Google Scholar]
- 45.Koonin E, Galperin M. Sequence–Evolution–Function: Computational Approaches in Comparative Genomics. London: Kluwer Academic; 2003. [PubMed] [Google Scholar]
- 46.Hemme D, Foucaud-Scheunemann C. Int Dairy J. 2004;14:467–494. [Google Scholar]
- 47.Liu S-Q. Int J Food Microbiol. 2003;83:115–131. doi: 10.1016/s0168-1605(02)00366-5. [DOI] [PubMed] [Google Scholar]
- 48.Twomey D, Ross R, Ryan M, Meaney B, Hill C. Antonie van Leeuwenhoek. 2002;82:165–185. [PubMed] [Google Scholar]
- 49.Nes I, Johnsborg O. Curr Opin Biotechnol. 2004;15:100–104. doi: 10.1016/j.copbio.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 50.Metivier A, Pilet M, Dousset X, Sorokine O, Anglade P, Zagorec M, Piard JC, Marion D, Cenatiempo Y, Fremaux C. Microbiology. 1998;144:2837–2844. doi: 10.1099/00221287-144-10-2837. [DOI] [PubMed] [Google Scholar]
- 51.Aparicio S, Chapman J, Stupka E, Putnam N, Chia J, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, et al. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 52.Slesarev A, Mezhevaya K, Makarova K, Polushin N, Shcherbinina O, Shakhova V, Belova G, Aravind L, Natale D, Rogozin I, et al. Proc Natl Acad Sci USA. 2002;99:4644–4649. doi: 10.1073/pnas.032671499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Besemer J, Lomsadze A, Borodovsky M. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lowe T, Eddy S. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tatusov R, Natale D, Garkavtsev I, Tatusova T, Shankavaram U, Rao BS, Kiryutin B, Galperin M, Fedorova N, Koonin E. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.