Abstract
The pregnancy-associated glycoproteins (PAG) are putative peptide-binding proteins and products of a large family of genes whose expression is localized to the placental surface epithelium of artiodactyl species. We have tested the hypothesis that natural selection has favored diversification of these genes by examining patterns of nucleotide substitution in a sample of 28 closely related bovine, caprine, and ovine family members that are expressed only in trophoblast binucleate cells. Three observations were made. First, in codons encoding highly variable domains of the proteins, there was a greater accumulation of both synonymous and nonsynonymous mutations than in the more conserved regions of the genes. Second, in the variable regions, the mean number of nonsynonymous nucleotide substitutions per site was significantly greater than the mean number of synonymous substitutions per site. Third, nonsynonymous changes affecting amino acid charge occurred more frequently than expected under random substitution. This unusual pattern of nucleotide substitution implies that natural selection has acted to diversify these PAG molecules at the amino acid level, which in turn suggests that these molecules have undergone functional diversification. We estimate that the binucleate cell-expressed PAG originated 52 ± 6 million years ago, soon after the divergence of the ruminant lineage. Thus, rapid functional diversification of PAG expressed in trophoblast binucleate cells seems to have been associated with the origin of this unique placental adaptation.
Keywords: aspartic proteinase, multigene families, positive selection, trophoblast, pregnancy-associated glycoprotein
The pregnancy-associated glycoproteins (PAGs) constitute an extensive family of recently duplicated genes within the aspartic proteinase superfamily. In cattle and sheep and probably also in other pecoran mammals, there may be as many as 100 or more genes (1) encoding these molecules. Thus far, 21 different cDNAs, differing by at least 5% in nucleotide sequence, have been cloned from the placenta of cattle (ref. 1 and J.A.G. and R.M.R., unpublished data), and another 11 have been cloned from sheep (1). PAGs have also been identified in pigs, a nonruminant species (2), and seem to occur throughout the Artiodactyla order (1). Only a single representative PAG-like gene has been found in the horse and zebra (order Perissodactyla; ref. 3), in the cat (Carnivora; ref. 4), and in the mouse (Rodentia; ref. 5), suggesting that the duplications that gave rise to the numerous ruminant PAG were initiated relatively recently and certainly since the separation of the Artiodactyla and Perissodactyla. In artiodactyls, the PAG are expressed in the outer epithelial cell layer of the placenta, which is in direct contact with the uterine endometrium. Their function at this interface between fetal and maternal tissue remains unknown; however, they are not believed to act primarily as proteinases, because many, possibly the majority, possess mutations that are likely to render them inactive as enzymes (6, 7). Structural modeling, however, suggests that they have retained the peptide-binding cleft of aspartic proteinases (7), and recent work has indicated that they are capable of binding short peptides (L. Landon, J.A.G., and R.M.R., unpublished data). The available PAG sequences from sheep and cow suggest that members of this gene family may have differentiated functionally (1).
The evolution of new protein function after gene duplication is believed to have played a major role in the evolution of life on earth (8, 9), but the mechanism by which this process occurs remains controversial. The most widely cited model for the evolution of new protein function assumes that new function arises as a result of selectively neutral mutations that are fixed by chance in a redundant gene copy and fortuitously provide it with a new function (10). However, molecular studies have provided several lines of evidence suggesting that new gene functions rarely, if ever, evolve in this way (9, 11). For example, several studies have shown that when genes differentiate functionally, gene duplication is often followed by positive Darwinian selection acting on functionally important regions of proteins. Positive selection is revealed by comparing the patterns of synonymous and nonsynonymous (amino acid-altering) nucleotide substitution. In most genes, the number of synonymous substitutions per synonymous site exceeds the number of nonsynonymous substitutions per nonsynonymous site (12). This pattern occurs, because most amino acid changes are selectively disadvantageous and thus are removed by natural selection (so-called “purifying” selection; ref. 13). The opposite pattern is evidence that natural selection has acted to favor amino acid changes (9). Such evidence has been obtained for comparisons between recently duplicated members of a number of multigene families (e.g., refs. 9 and 14–17). The present paper describes the use of this approach to test for evidence of adaptive diversification in the PAG gene family of Bovidae.
Methods
DNA Sequences Analyzed.
Bovine PAG cDNAs were isolated from cDNA libraries prepared at day 19 (whole concepti), day 25 (dissected trophoblast; ref. 1), and day 260 of pregnancy (whole placental tissue; ref. 6) as described elsewhere (see ref. 1). Additional bovine transcripts were cloned by reverse transcription–PCR procedures from RNA isolated at day 19 (1), at day 25, and at term.
The primers used were a boPAG1 5′ oligonucleotide (5′-AGGAAAGAAGCATGAAGTGGCT3-′, with the start codon underlined; ref. 6), whose sequence is completely or almost completely conserved in all the PAGs cloned thus far, and one of the following 3′ oligonucleotides: 5′-GCGCTCGAGTTACACTGCCCGTGCCAGGC-3′, 5′-GTTCTCGAGCTTACACAGCAGGAGCCAAGCC-3′, 5′-GCGCTCGAGTTGAAGCAGCTCCAGCATTTA-3′, and 5′-GTTCTCGAGCTTATACCGCAGTAGCCAGTCC-3′. The latter four oligonucleotides represented sequences encompassing the end of the ORFs of known PAG genes (the stop codon is underlined). The reverse transcription–PCR products were cloned into the pGEM-T Easy vector (Promega). The positive clones were isolated, analyzed for size by restriction mapping, and partially sequenced. Those cDNAs sharing less than 95% identity with other bovine PAG were fully sequenced in both directions.
The cloning of the ovine PAG transcripts has been described in detail elsewhere (1).
To clone caprine PAG transcripts, RNA from day 45, 65, 87, and 115 placental tissue was reverse transcribed by using the same primers in a manner identical to that described above for boPAG transcripts.
Statistical Methods.
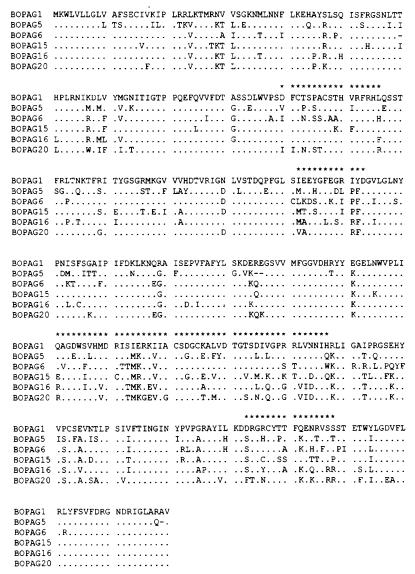
PAG sequences were aligned at the amino acid level by using the clustal w program (18). Fig. 1 shows a partial alignment; the complete alignment is available from the authors on request. In any set of pairwise comparisons among sequences, any amino acid position at which the alignment postulated a gap was excluded from all pairwise comparisons so that the same set of positions was compared in each case. By inspection of the aligned amino acid sequences, we identified four highly variable regions (Fig. 1). The third of these regions includes the B lobe catalytic domain of aspartic proteinases and its conserved signature motif (ref. 1; residues 268–273 in Fig. 1). The fourth region includes an invariant phenylalanine residue (position 341 in Fig. 1), which is involved in bridging between two loops (1).
Figure 1.
Alignment of selected PAG. Variable regions are marked with asterisks.
Phylogenetic trees were constructed by means of the maximum parsimony method (19) and the neighbor-joining method (20). Neighbor-joining trees were constructed on the basis of the following distances: the uncorrected proportion of amino acid difference (p), the γ-corrected estimate of the number of amino acid replacements per site (21), and the number of nonsynonymous nucleotide substitutions per site (dN; ref. 22). Synonymous sites were not used because synonymous sites were saturated with changes in certain comparisons among these sequences. The reliability of branching patterns in phylogenetic trees was tested by bootstrapping, which involved repeated pseudosampling from the data (with replacement; ref. 23); 1,000 bootstrap pseudosamples were used. All of these methods yielded essentially the same results; therefore, only the neighbor-joining tree based on p is presented here.
Numbers of synonymous (dS) and nonsynonymous (dN) nucleotide substitutions per site were estimated by Nei and Gojobori's method (22), modified as recommended by Zhang et al. (17) to take into account different frequencies of transitions and transversions. The original method (22) assumes equal usage of all four nucleotides in counting fractional synonymous and nonsynonymous sites. However, this method will lead to an underestimation of the number of synonymous sites, particularly in the case of 2-fold degenerate sites at which transitions are synonymous and transversions are nonsynonymous (23). The observed frequency of transitions and transversions at 2-fold degenerate sites is expected to reflect the effects both of mutational bias and of purifying selection, which is expected to eliminate many synonymous mutations. Therefore, it is inappropriate to estimate the transition:transversion ratio from such sites. Because all mutations at 4-fold degenerate sites are synonymous, these sites are most appropriate for estimating the transition:transversion ratio. Therefore, we estimated the transition:transversion ratio from 4-fold degenerate sites.
Hughes et al. (24) developed a method of testing the hypothesis that nonsynonymous differences occur in such a way as to change some amino acid property of interest to a greater extent than is expected under random substitution. This method involves computing the proportion of conservative nonsynonymous difference per conservative nonsynonymous site (pNC) and the proportion of radical nonsynonymous difference per radical nonsynonymous site (pNR). When pNC > pNR, nonsynonymous differences occur in such a way as to conserve the property of interest. When pNC = pNR, nonsynonymous differences occur at random with respect to the property of interest. When pNR > pNC, nonsynonymous differences occur in such a way as to change the property of interest to a greater extent than expected at random. Because this last pattern indicates a directional change in amino acid sequence, it is suggestive of positive selection (24).
For use in the present analyses, we developed a modified version of the earlier method (24) that takes into account transition:transversion ratio. Taking transitional bias into account is important in such analyses, because transitional mutations in nonsynonymous sites are often conservative with respect to amino acid properties such as charge or polarity, whereas transversional mutations are frequently radical. (A computer program for this method is available from A.L.H. on request.) We applied this method with respect to two amino acid properties, charge and polarity. With respect to charge, amino acid residues were categorized as positive (H, K, and R), negative (D and E), or neutral (remainder). With respect to polarity, residues were categorized as polar (H, K, R, D, E, G, S, T, C, Y, N, and Q) or nonpolar (remainder). Any difference causing a change of category was counted as a radical difference.
For estimation of dS, dN, pNC, and pNR, we estimated the ratio of transitional to transversional mutations by comparing eight phylogenetically independent pairs of closely related sequences. These were pairs of sequences that clustered together in the phylogeny (Fig. 2): caPAG3 and ovPAG3, ovPAG4 and ovPAG7, boPAG20 and boPAG7, caPAG9 and ovPAG1, caPAG11 and ovPAG6, boPAG4 and boPAG6, caPAG1 and caPAG6, and boPAG1 and boPAG3. The estimated transition to transversion ratio at 4-fold degenerate sites was 2.4.
Figure 2.
Phylogenetic tree of PAG, constructed by the neighbor-joining method (20) based on the proportion of amino acid differences (p). Numbers on the branches are bootstrap percentages; only values ≥ 60% are shown. The bracket marks a group of 28 closely related PAGs expressed in trophoblast binucleate cells. Species abbreviations are as follows: bo-, bovine; ca-, goat; ov-, sheep.
SEMs for dS, dN, pNC, and pNR were estimated according to the method described by Nei and Jin (25).
Results
In the phylogenetic tree (Fig. 2), sequences from bovine, sheep, and goat PAGs are intermixed, indicating that many gene duplications in this family took place before these three species diverged. The phylogeny enabled us to identify a subgroup of closely related sequences from the three species (marked by bracket in Fig. 2) in comparisons among which synonymous sites were well below saturation. We analyzed this group of 28 sequences in testing for positive selection.
In pairwise comparisons among these 28 closely related sequences, mean dN, was significantly greater than mean dS in the variable regions (Table 1). In the remainder of the gene, mean dS was higher than mean dN, although the difference was not statistically significant (Table 1). Both mean dS and mean dN in the variable regions were significantly greater than the corresponding values for the remainder of the gene (Table 1).
Table 1.
Mean numbers of synonymous (dS) and nonsynonymous (dN) nucleotide substitutions per 100 sites in variable regions and remainders of PAG genes
| Domain | dS | dN |
|---|---|---|
| Variable | 19.5 ± 2.1 | 29.4 ± 1.7*** |
| Remainder | 12.8 ± 0.7 | 11.2 ± 0.5 |
Values are shown as means ± SEM. Tests of the hypothesis that dS = dN: ***, P < 0.001.
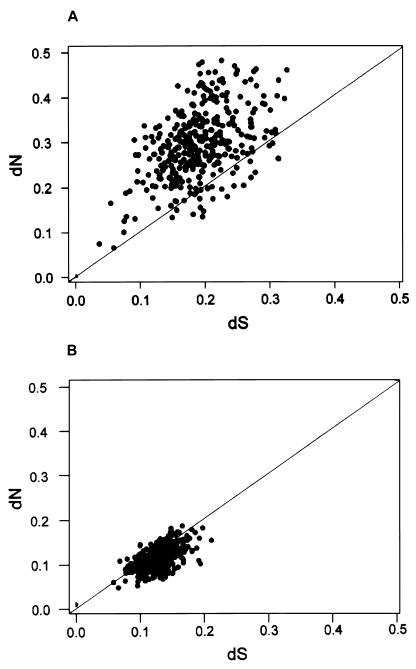
The pattern of synonymous and nonsynonymous nucleotide substitution is illustrated further in Fig. 3, where dN is plotted against dS for each pairwise comparison. In the variable regions, dN exceeded dS in 92.3% of pairwise comparisons (349 of 378), as indicated by points above the 45° line in Fig. 3A. The value dN was particularly likely to exceed dS, when dS was relatively low (Fig. 3A). By contrast, in the remainder of PAG genes, dS exceeded dN in 74.1% (280 of 378) of pairwise comparisons (Fig. 3B).
Figure 3.
Number of nonsynonymous substitutions per site (dN) plotted against the number of synonymous substitutions per site (dS) in all pairwise comparisons among variable regions (A) and the remainder of 28 closely related PAG genes (B; see Fig. 2). In each case, a 45° line is drawn.
When pNC and pNR were estimated with respect to amino acid residue charge, pNR exceeded pNC in the variable regions (Table 2). Thus, in the variable regions, amino acid changes occur disproportionately in such a way as to cause residue charge change. By contrast, in the remainder of PAG genes, pNC and pNR were not significantly different with respect to residue charge (Table 2). In neither the variable regions nor the remainder of PAG genes was there a significant difference between pNC and pNR with respect to residue polarity (Table 2).
Table 2.
Mean percentages of conservative (pNC) and radical (pNR) nonsynonymous differences per site, with respect to amino acid residue charge and polarity in variable regions and remainders of PAG genes
| Property | pNC, % | pNR, % |
|---|---|---|
| Charge | ||
| Variable | 20.6 ± 1.5 | 30.3 ± 2.6** |
| Remainder | 10.0 ± 0.6 | 11.2 ± 0.9 |
| Polarity | ||
| Variable | 24.5 ± 1.7 | 23.0 ± 2.3 |
| Remainder | 10.9 ± 0.6 | 9.2 ± 0.8 |
Values are shown as mean percentages ± SEM. Tests of the hypothesis that pNC = pNR: **, P < 0.01.
Mean dS values were used to estimate the times of origin of ruminant PAG genes. To obtain a calibration, apparently orthologous pairs of genes were compared between cattle (subfamily Bovine) and sheep and goats (subfamily Caprinae), as indicated in the phylogenetic tree in Fig. 2. Comparisons were made between boPAG8 and caPAG8 and between boPAG11, caPAG2, and ovPAG2. Mean dS for these comparisons was 0.082 ± 0.016. Assuming that the two lineages diverged 20 million years ago (26), the divergence time between the 28 trophoblast binucleate cell-expressed genes and the nearest out group (ovPAG5) was estimated at 52 ± 6 million years ago (mean dS = 0.214 ± 0.023). By using the same calibration, the deepest branch point in the phylogeny of Fig. 2 was estimated at 87 ± 6 million years ago (mean dS = 0.359 ± 0.023).
Discussion
These results indicate that positive Darwinian selection has acted to promote diversity at the amino acid level in the variable regions of duplicated PAG genes. Furthermore, this selection has acted to promote amino acid residue charge changes to a greater extent than expected under random substitution. The results thus suggest that the variable regions are important for functional specificity of PAG proteins and that the pattern of amino acid residue charges in these regions may play a particularly important role in functional differences among members of this family. These variable regions represent surface-exposed loops (1), and we have suggested earlier that small additive changes in the packing of these loop regions influence ligand binding within the substrate binding clefts, thereby providing PAGs with a considerable potential range of peptide-binding specificities (1).
The pattern of synonymous and nonsynonymous substitution seen in the variable regions (Fig. 3A), where dN was particularly likely to exceed dS when the latter was low, seems to be characteristic of positive selection leading to diversification of genes within multigene families (9, 15, 16). This pattern evidently occurs, because there is a burst of positively selected nonsynonymous substitutions immediately after gene duplication, as the duplicate genes adapt to distinct functions. Because dS is relatively low in comparisons between closely related sequences, this burst of positively selected nonsynonymous substitutions is likely to cause dN to exceed dS in such comparisons. However, once each duplicate gene has adapted to its specific function, purifying selection is expected to predominate, allowing the number of synonymous substitutions per site to catch up to and eventually exceed the number of nonsynonymous substitutions per site (9). As a result, it is generally possible to detect such selection only within a certain relatively limited time after gene duplication, probably 30–50 million years (at most) in the case of eukaryotes (9). Because genes of the PAG family have duplicated repeatedly within the order Artiodactyla, they provide a family of relatively recently duplicated and functionally differentiated genes ideal for studying the process of adaptive diversification of duplicate genes.
One surprising result was the finding that mean dS in the variable regions was significantly higher than the corresponding value for the remainder of the genes (Table 1). Because dS is expected to reflect the mutation rate, it is predicted that dS will be relatively constant across different regions of a protein-coding gene (27). In most cases, analyses have supported this prediction (9). Exceptions to the rule can occur in the following circumstances: when the mutation rate varies over the length of a gene, as has sometimes been observed, especially in genes with long coding regions such as the cystic fibrosis transmembrane transporter genes (28), or alternatively, when different portions of two related genes differ in divergence time as a result of interlocus recombination or “gene conversion” (29). Given that the variable regions analyzed herein consist of four short isolated gene regions, it seems unlikely that these regions have higher mutation rates than the rest of the gene. Thus, it seems most plausible that dS in the variable region genes exceeds that in the remainder of the genes, because interlocus gene conversion events have caused some degree of homogenization of loci outside the variable regions.
It is interesting that the 28 closely related PAG genes analyzed in Fig. 2 are expressed predominantly in trophoblast binucleate cells (ref. 1 and J.A.G., J.M.G., and R.M.R., unpublished data). These large cells, which comprise 15–20% of postattachment trophectoderm (30), can migrate from the trophoblast and fuse with uterine epithelial cells to form either a syncytium (as in the sheep, in the goat, and in the early stages of placentation of the cow) or short-lived trinucleated cells (in established cow placenta; refs. 30 and 31). These cells constitute a unique feature of the synepitheliochorial placentation of the ruminant artiodactyl species. This type of placenta, with very limited invasive potential, probably developed from the noninvasive epitheliochorial placenta observed in nonruminant artiodactyls, such as camels (32) and pigs (33). The calculation of 52 ± 6 million years for the origin of the binucleate cell-specific PAGs is only slightly lower than recent estimates for the divergence of the Ruminantia and the Suidae (26). It is tempting to speculate that the burst of duplications that gave rise to the binucleate cell PAG group is linked to the emergence of this placental type within Ruminantia. Similarly, the origin of the Artiodactyla has been estimated at about 83 million years ago (26), a value that is very close to our estimate of when the PAG genes as a whole first began to duplicate.
Acknowledgments
We thank Dr. Joseph M. Quattro for carefully reading the manuscript and for providing comments. This work was supported by National Institutes of Health Grants GM34940 and HD35898 to A.L.H. and R.M.R., respectively, and Grant 96-35205-3257 from the U.S. Department of Agriculture Competitive Grants Program to R.M.R.
Abbreviation
- PAG
pregnancy-associated glycoprotein
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. M73961, L06153, AF020506–AF020514, AF192330–AF192338, AF191326–AF191336, M73962, U30251, and U94789–U94793).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050002797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050002797
References
- 1.Xie S, Green J, Bixby J B, Szafranska B, DeMartini J C, Hecht S, Roberts R M. Proc Natl Acad Sci USA. 1997;94:12809–12816. doi: 10.1073/pnas.94.24.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szafranska B, Xie S, Green J, Roberts R M. Biol Reprod. 1995;53:21–28. doi: 10.1095/biolreprod53.1.21. [DOI] [PubMed] [Google Scholar]
- 3.Green J A, Xie S, Szafranska B, Gan X, Newman A G, McDowell K, Roberts R M. Biol Reprod. 1999;60:1069–1077. doi: 10.1095/biolreprod60.5.1069. [DOI] [PubMed] [Google Scholar]
- 4.Gan X, Xie S, Green J, Roberts R M. Biol Reprod. 1997;56,Suppl. 1:191. (abstr.). [Google Scholar]
- 5.Chen X, Roberts R M, Green J A. Biol Reprod. 1999;60,Suppl. 1:216. (abstr.). [Google Scholar]
- 6.Xie S C, Low B G, Nagel R J, Kramer K K, Anthony R V, Zoli A P, Beckers J F, Roberts R M. Proc Natl Acad Sci USA. 1991;88:10247–10251. doi: 10.1073/pnas.88.22.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guruprasad K, Blundell T L, Xie S, Green J, Szafranska B, Nagel R J, McDowell K, Baker C B, Roberts R M. Protein Eng. 1996;9:849–856. doi: 10.1093/protein/9.10.849. [DOI] [PubMed] [Google Scholar]
- 8.Li W-H. In: Evolution of Genes and Proteins. Nei M, Koehn R K, editors. Sunderland, MA: Sinauer; 1983. pp. 14–37. [Google Scholar]
- 9.Hughes A L. Adaptive Evolution of Genes and Genomes. New York: Oxford Univ. Press; 1999. [Google Scholar]
- 10.Ohno S. Nature (London) 1973;244:259–262. doi: 10.1038/244259a0. [DOI] [PubMed] [Google Scholar]
- 11.Hughes A L. Proc R Soc London Ser B. 1994;23:119–124. [Google Scholar]
- 12.Li W-H. Molecular Evolution. Sunderland MA: Sinauer; 1997. [Google Scholar]
- 13.Kimura M. Nature (London) 1977;267:275–276. doi: 10.1038/267275a0. [DOI] [PubMed] [Google Scholar]
- 14.Hill R E, Hastie W D. Nature (London) 1987;326:96–99. doi: 10.1038/326096a0. [DOI] [PubMed] [Google Scholar]
- 15.Tanuka T, Nei M. Mol Biol Evol. 1989;6:447–459. doi: 10.1093/oxfordjournals.molbev.a040569. [DOI] [PubMed] [Google Scholar]
- 16.Hughes A L, Yeager M. J Mol Evol. 1997;44:675–682. doi: 10.1007/pl00006191. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Rosenberg H F, Nei M. Proc Natl Acad Sci USA. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson J D, Higgins D G, Gibson J J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swofford D L. paup, Phylogenetic Analysis Using Parsimony. Champaign, IL: Illinois Natural History Survey; 1990. , Version 3.0. [Google Scholar]
- 20.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 21.Ota T, Nei M. J Mol Evol. 1994;38:642–643. [Google Scholar]
- 22.Nei M, Gojobori T. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 23.Li W-H. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- 24.Hughes A L, Ota T, Nei M. Mol Biol Evol. 1990;7:515–524. doi: 10.1093/oxfordjournals.molbev.a040626. [DOI] [PubMed] [Google Scholar]
- 25.Nei M, Jin L. Mol Biol Evol. 1989;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Kumar S, Hedges S B. Nature (London) 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 27.Hughes A L, Nei M. Nature (London) 1988;355:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 28.Hughes A L. Mol Biol Evol. 1994;11:899–910. doi: 10.1093/oxfordjournals.molbev.a040163. [DOI] [PubMed] [Google Scholar]
- 29.Ohta T. Mol Phyl Evol. 1992;1:305–311. doi: 10.1016/1055-7903(92)90006-3. [DOI] [PubMed] [Google Scholar]
- 30.Wooding F B. Placenta. 1992;13:101–113. doi: 10.1016/0143-4004(92)90025-o. [DOI] [PubMed] [Google Scholar]
- 31.Hoffman L H, Wooding F B P. J Exp Zool. 1993;266:559–577. doi: 10.1002/jez.1402660607. [DOI] [PubMed] [Google Scholar]
- 32.Skidmore J A, Wooding F B P, Allen W R. Placenta. 1996;17:253–262. doi: 10.1016/s0143-4004(96)90046-6. [DOI] [PubMed] [Google Scholar]
- 33.Friess A E, Sinowatz F, Skolek-Winnisch R, Trautner W. Anat Embryol. 1980;158:179–191. doi: 10.1007/BF00315905. [DOI] [PubMed] [Google Scholar]