Abstract
Clathrin adaptors are key factors in clathrin-coated vesicle formation, coupling clathrin to cargo and/or the lipid bilayer. A physically interacting network of three classes of adaptors participate in clathrin-mediated traffic between the trans-Golgi network (TGN) and endosomes: AP-1, Gga proteins, and epsin-like proteins. Here we investigate functional relationships within this network through transport assays and protein localization analysis in living yeast cells. We observed that epsin-like protein Ent3p preferentially localized with Gga2p, whereas Ent5p distributed equally between AP-1 and Gga2p. Ent3p was mislocalized in Gga-deficient but not in AP-1–deficient cells. In contrast, Ent5p retained localization in cells lacking either or both AP-1 and Gga proteins. The Ent proteins were dispensable for AP-1 or Gga localization. Synthetic genetic growth and α-factor maturation defects were observed when ent5Δ but not ent3Δ was introduced together with deletions of the GGA genes. In AP-1–deficient cells, ent3Δ and to a lesser extent ent5Δ caused minor α-factor maturation defects, but together resulted in a near-lethal phenotype. Deletions of ENT3 and ENT5 also displayed synthetic defects similar to, but less severe than, synthetic effects of AP-1 and Gga inactivation. These results differentiate Ent3p and Ent5p function in vivo, suggesting that Ent3p acts primarily with Gga proteins, whereas Ent5p acts with both AP-1 and Gga proteins but is more critical for AP-1–mediated transport. The data also support a model in which the Ent adaptors provide important accessory functions to AP-1 and Gga proteins in TGN/endosome traffic.
INTRODUCTION
Intracellular vesicle-mediated traffic is fundamentally important for the distribution of proteins and lipids to membrane organelles of eukaryotic cells. Clathrin-coated vesicles (CCVs) are a well-characterized class of transport carriers that ferry cargo from the plasma membrane to endosomes and between the trans-Golgi network (TGN) and endosomes. CCVs are distinguished by a polyhedral shell of clathrin that coats the vesicle membrane. This coat is assembled during vesicle formation from individual clathrin molecules containing three heavy- and three light-chain subunits (Brodsky et al., 2001). Because clathrin does not bind directly to lipids, coat assembly involves clathrin-binding adaptors that anchor the nascent coat to the membrane through association with lipids and/or the cytoplasmic domains of cargo proteins. By coupling cargo to coat assembly, such adaptors are central to the process of vesicle formation (Kirchhausen, 2000; Bonifacino and Traub, 2003; Owen et al., 2004). In addition to adaptors, a growing number of accessory factors have been identified that contribute to coated vesicle biogenesis (Traub, 2005).
Three classes of adaptors have been described that participate in CCV-mediated traffic between the TGN and endosomes in mammalian and yeast cells: AP-1, Gga proteins and epsin-like proteins. AP-1 is a heterotetrameric complex composed of two large subunits (β1 and γ), one medium subunit (μ1), and a small subunit (σ1). The two large subunits contain C-terminal regions that extend from the core of the complex as flexible linkers connected to globular domains known as “ears.” These hinge/ear regions in both yeast and mammalian AP-1 large subunits interact with clathrin, accessory factors, and other adaptors (Robinson and Bonifacino, 2001; Traub, 2005). Regions in the mammalian AP-1 core bind to phosphoinositides and an activated form of the small GTPase Arf, which promote AP-1 recruitment to membranes (Austin et al., 2002; Wang et al., 2003; Heldwein et al., 2004). The core also binds to sorting signals in cytoplasmic domains of transmembrane cargo proteins thereby collecting the cargo into nascent vesicles (Bonifacino and Traub, 2003; Traub, 2005). Yeast AP-1 is also dependent on activated Arf for localization but the lipid and cargo binding properties have not been characterized (Fernandez and Payne, 2006).
Gga proteins are monomeric adaptors first identified on the basis of sequence similarity to the C-terminal ear domain of the AP-1 γ subunit (Bonifacino, 2004). There are two Gga proteins in yeast and three in animal cells with similar architectures and probably analogous functions, although mammalian Gga proteins have been characterized in greater detail. Gga proteins are organized into distinct domains that carry out many of the same tasks as AP-1. The amino-terminal VHS domain mediates binding to sorting signals in protein cargo. A central GAT domain interacts with activated Arf, involved in membrane association, and ubiquitin, which functions as a posttranslationally added sorting signal (Puertollano and Bonifacino, 2004; Scott et al., 2004; Shiba et al., 2004). A hinge region following the GAT domain contains motifs for binding clathrin and AP-1. The hinge connects to the C-terminal γ-ear domain that associates with accessory proteins and epsin-like adaptors.
The functional relationships of AP-1 and Gga adaptors have not been well established. Both localize to the TGN and endosomes, but the extent of colocalization reported by different groups has varied (Dell’Angelica et al., 2000; Hirst et al., 2000, 2001; Puertollano et al., 2001; Doray et al., 2002; Fernandez and Payne, 2006). In mammalian and yeast cells, AP-1 is implicated in both anterograde and retrograde CCV-mediated traffic between the TGN and endosomes. In contrast, current evidence favors a primary role for Gga proteins in anterograde TGN-to-endosome traffic (Hinners and Tooze, 2003). Both yeast and mammalian Gga proteins associate with AP-1 (Costaguta et al., 2001; Doray et al., 2002; Bai et al., 2004), and this interaction has been proposed to mediate sequential roles for the two adaptors in mannose-6-phosphate receptor transport from the TGN to endosomes in mammalian fibroblasts (Doray et al., 2002). However, results from gene deletion studies in yeast are consistent with at least some separate function of AP-1 and Gga proteins in pathways between the TGN and endosomes (Black and Pelham, 2000; Costaguta et al., 2001; Hirst et al., 2001; Ha et al., 2003). Whether these pathways involve distinct AP-1 and Gga vesicle populations and target different endosomal compartments remains unresolved.
Recently, epsin-like proteins have been recognized as a new class of monomeric TGN/endosome adaptors (Duncan and Payne, 2003; Legendre-Guillemin et al., 2004). The founding member of the epsin family, epsin1, is an endocytic clathrin adaptor that contains a signature N-terminal ENTH (epsin N-terminal homology) domain that binds phosphoinositides. The epsin1 ENTH domain is necessary for membrane localization and also promotes membrane curvature (Ford et al., 2002; Stahelin et al., 2003). The C-terminal region of epsin contains multiple binding sites for clathrin and the endocytic AP-2 adaptor complex α subunit ear. Another monomeric endocytic adaptor, AP180, has a similar modular design with an N-terminal domain related to the ENTH domain and a C-terminal region with α-ear– and clathrin-binding sites. However, the AP180 N-terminal domain binds phosphoinositides in a different manner and does not promote membrane curvature (Ford et al., 2001; Stahelin et al., 2003). This has led to a division of the family into ENTH and ANTH (AP180 N-terminal homology) domain subclasses.
A single epsin-like protein that localizes to the TGN and endosomes has been identified in mammalian cells, EpsinR/Enthoprotin/Clint (hereafter referred to as EpsinR; Kalthoff et al., 2002b; Wasiak et al., 2002; Hirst et al., 2003; Mills et al., 2003). Like epsin1, epsinR contains an N-terminal ENTH domain and clathrin-binding sites in the C-terminal region. Unlike epsin1 binding to AP-2, epsinR preferentially binds to the γ-ears of AP-1 and, to a lesser extent Gga proteins, through specific motifs in the C-terminal region. Consistent with its interactions and localization to the TGN/endosome, epsinR depletion or overexpression alters clathrin-dependent traffic of certain cargo between these compartments (Mills et al., 2003; Hirst et al., 2004; Saint-Pol et al., 2004).
We previously identified two epsin-like TGN/endosome adaptors in yeast, Ent3p and Ent5p, as interaction partners of AP-1 and Gga γ-ear domains (Duncan et al., 2003). Ent3p is an ENTH domain protein with three γ-ear–binding motifs and a weak match to a consensus clathrin-binding sequence in the C-terminal region. Ent5p contains an N-terminal ANTH domain, followed by a C-terminal region with two γ-ear–binding motifs and two consensus clathrin-binding sites. The domain and motif differences between Ent3p and Ent5p suggest divergence in function. In support of this view, coimmunoprecipitations revealed differential clathrin and adaptor binding by Ent3p and Ent5p: Ent5p associated with clathrin, AP-1, and Gga2p, whereas Ent3p displayed robust interaction only with Gga2p (Duncan et al., 2003). Additionally, the SNARE Vti1p binds to the Ent3p ENTH domain but not the Ent5p ANTH domain (Chidambaram et al., 2004). This Ent3p property is shared by epsinR, which appears to serve as a cargo-selective adaptor for Vti1b transport in mammalian fibroblasts (Hirst et al., 2004).
Despite the differences between Ent3p and Ent5p, analysis of single and double mutant deletion strains have not revealed functional distinctions. Strains lacking both Ent3p and Ent5p, but not single mutant strains, exhibit defects in TGN/endosome traffic, clathrin localization, and protein sorting into the vacuole lumen (Duncan et al., 2003; Friant et al., 2003; Chidambaram et al., 2004; Eugster et al., 2004). Thus, the functional consequences of physical differences between Ent3p and Ent5p are unclear.
Taken together, studies of TGN/endosome clathrin-mediated traffic in both mammalian cells and yeast have uncovered a network of physically interacting adaptors, with as yet incompletely defined functional relationships. To further probe this network in vivo, we have carried out an extensive analysis of genetic interactions between adaptor mutations, assessing effects on adaptor localization and TGN-endosome protein traffic. Our results indicate that AP-1, Gga, and Ent5p adaptors are localized independently of each other and Ent3p. In contrast, Ent3p localization depends on Gga proteins. Analysis of TGN/endosome traffic in Gga- or AP-1–deficient strains also carrying deletions of ENT3 or ENT5 support a preferential function for Ent3p with Gga adaptors. These results are concordant with the specific physical interaction of Ent3p with Gga proteins and, considered with the wider spectrum of Ent5p interaction partners, support a model in which Ent3p primarily functions in Gga-dependent transport, whereas Ent5p plays a more ubiquitous role in TGN/endosome trafficking pathways.
MATERIALS AND METHODS
General Methods and Media
Restriction endonucleases were from New England Biolabs (Beverly, MA). Unless noted, all reagents were from Sigma (St. Louis, MO). Antibodies against α-factor and ALP have been described previously (Seeger and Payne, 1992); the antibody against carboxypeptidase S (CPS) was a gift from Steven Nothwehr (University of Missouri, MO). Sequences of oligonucleotides used in this study are available on request.
YP media is 1% Bacto-yeast extract (Difco, Detroit, MI) and 2% Bacto-peptone (Difco). YPD is YP with 2% dextrose. SD media is 0.67% of yeast nitrogen base without amino acids (Difco) and 2% dextrose. Supplemented SD media contains 20 μg/ml l-histidine, uracil, and l-tryptophan, and 30 μg/ml l-leucine, adenine, and l-lysine. SDYE is supplemented SD plus 0.2% yeast extract. YPG and SG media contain 2% galactose in place of dextrose. Solid media contains 2% agar. Cell densities were measured by spectrophotometry.
Plasmids and Strains
Plasmids.
pFA6a-mRFP-kanMX6 (Huh et al., 2003), which contains an integrative version of monomeric RFP (Campbell et al., 2002) was used to create pFA6a-mRFP-HIS3MX6 and pFA6a-mRFP-TRP1 through swapping BglII/PmeI fragments with pFA6a-3HA-HIS3MX6 and pFA6a-TRP1 (Longtine et al., 1998), respectively. To generate pRS315-GGA2 a DraI/HpaI fragment of genomic GGA2 was converted to blunt ends with T4 DNA Polymerase before ligation into the SmaI site of pRS315 (Sikorski and Hieter, 1989). For the GGA2 thermosensitive (ts) allele (gga2-ts) screen, pRS315-GGA2 was cleaved with AvrII/BsmI, releasing a fragment encompassing the VHS domain. The gapped plasmid was purified and cotransformed with overlapping VHS-encoding DNA fragments obtained by PCR mutagenesis (Muhlrad et al., 1992). Plasmids carrying gga2-33 and gga2-47 alleles were cleaved with AvrII/BsmI, and the fragment encoding mutagenized VHS domain was ligated into the corresponding sites in pRS315-GGA2, generating pRS315-gga2-33 and pRS315-gga2-47. To construct integrative pRS306-gga2-33 and pRS306-gga2-47, an HindIII/BbsI fragment from pRS315-gga2-33 or pRS315-gga2-47 was inserted into the HindIII/SmaI sites of pRS306 to generate pRS306-gga2-33* and pRS306-gga2-47*. Then, an XbaI/AseI fragment from pRS315-gga2-33 or pRS315-gga2-47 was ligated into pRS306-gga2-33* and pRS306-gga2-47* digested with XbaI/SacII to generate pRS306-gga2–33 and pRS306-gga2-47, respectively. In these constructions, BbsI, AseI, and SacII sites were converted to blunt ends with T4 DNA Polymerase. PCR was used to clone genomic copies of ENT3 and ENT5 genes into pBluescriptKS (Stratagene, La Jolla, CA) to generate pMD170 (ENT3 [−306 to +1665] introduced into BamHI/XbaI sites) and pMD171 (ENT5 [−466 to +1493] introduced into BamHI/HindIII sites). From pMD170, a BamHI/SacI fragment was subcloned into pRS425 to generate p425-ENT3. From pMD171, an XhoI/BamHI fragment was subcloned into pRS425 to generate p425-ENT5.
Strains.
Table 1lists the strains used in this study. In general, deletions, fluorescent tags, and the GAL1 promoter were introduced by standard PCR-based methods (Sikorski and Hieter, 1989; Longtine et al., 1998). GPY2507 and GPY2555 were generated by successive PCR-based gene replacement. GPY3900, GPY3907, GPY3912, GPY3913, GPY3890, and GPY4106 were obtained by transformation of haploid cells with PCR-generated fragments as described (Longtine et al., 1998). All other strains were generated from diploid cells obtained by mating, sporulation, and isolation of haploid spores. MunI-cleaved pRS306-gga2-33 and pRS306-gga2-47 were used to integrate gga2-33 and gga2-47, respectively. Deletion of APL2 was carried out with a SacI/BamHI fragment from papl2-Δ6 (Yeung et al., 1999).
Table 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| SEY6210 | MATα ura3–52 leu2-3112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | Robinson et al. (1988) |
| FY1679-18B | MATα ura3-52 trp1-Δ63 leu2Δ1 his3Δ200 GAL2+ | Thierry et al. (1990) |
| GPY2149 | SEY6210 gga2Δ::HIS3 | Costaguta et al. (2001) |
| GPY2151 | SEY6210 gga1Δ::HIS3 | Costaguta et al. (2001) |
| GPY2507 | FY1679-18B GAL1-APL2::TRP1 gga1Δ::HIS3 gga2Δ::URA3 | This study |
| GPY2555 | FY1679-18B GAL1-GGA2::HIS3MX6 apl2Δ::URA3 gga1Δ::TRP1 | This study |
| GPY2728 | SEY6210 ent3Δ::TRP1 | Duncan et al. (2003) |
| GPY2731 | SEY6210 ent5Δ::TRP1 | Duncan et al. (2003) |
| GPY2734 | SEY6210 ent3Δ::TRP1 ent5Δ::TRP1 | Duncan et al. (2003) |
| GPY3100-11A | SEY6210 apl2Δ-6::TRP1 gga1Δ::HIS3 gga2-47::pRS306 | This study |
| GPY3100-4B | SEY6210 apl2Δ-6::TRP1 | This study |
| GPY3101-5A | SEY6210 apl2Δ-6::TRP1 gga1Δ::HIS3 | This study |
| GPY3101-6D | SEY6210 apl2Δ-6::TRP1 gga1Δ::HIS3 gga2-33::pRS306 | This study |
| GPY3206 | SEY6210 ENT3-GFP(S65T)::kanMX6 | This study |
| GPY3213 | SEY6210 ENT5-GFP(S65T)::kanMX6 | This study |
| GPY3217-7A | SEY6210 apl2Δ-6::TRP1 gga1Δ::HIS3 gga2–33::pRS306 ENT5-GFP(S65T)::kanMX6 | This study |
| GPY3298-13A | SEY6210 ent3Δ::TRP1 ent5Δ::TRP1 APL2-GFP(S65T)::HIS3MX6 | This study |
| GPY3298-27C | SEY6210 APL2-GFP(S65T)::HIS3MX6 | This study |
| GPY3298-29C | SEY6210 APL2-GFP(S65T)::HIS3MX6 GGA2-mRFP::kanMX6 | This study |
| GPY3425 | SEY6210 apl2Δ-6::TRP1 ENT3-GFP(S65T)::kanMX6 | This study |
| GPY3429 | SEY6210 MATa gga1Δ::HIS3 gga2Δ::HIS3 ENT3-GFP(S65T)::kanMX6 | This study |
| GPY3431 | SEY6210 gga1Δ::HIS3 gga2Δ::HIS3 | This study |
| GPY3434 | SEY6210 apl2Δ-6::TRP1 ENT5-GFP(S65T)::kanMX6 | This study |
| GPY3440 | SEY6210 gga1Δ::HIS3 gga2Δ::HIS3 ENT5-GFP(S65T)::kanMX6 | This study |
| GPY3630 | SEY6210 gga1Δ::HIS3 gga2-33::pRS306 ENT3-GFP(S65T)::kanMX6 | This study |
| GPY3669 | SEY6210 gga1Δ::HIS3 gga2-33-GFP(S65T)::pRS306::kanMX6 | This study |
| GPY3683 | SEY6210 gga1Δ::HIS3 gga2Δ::HIS3 ent3Δ::TRP1 | This study |
| GPY3685 | SEY6210 gga1Δ::HIS3 gga2Δ::HIS3 ent5Δ::TRP1 | This study |
| GPY3688 | SEY6210 apl2Δ-6::TRP1 ent3Δ::TRP1 | This study |
| GPY3690 | SEY6210 apl2Δ-6::TRP1 ent5Δ::TRP1 | This study |
| GPY3786 | SEY6210 apl2Δ-6::TRP1 GGA2-GFP(S65T)::kanMX6 | This study |
| GPY3802 | SEY6210 ent3Δ::TRP1 ent5Δ::TRP1 GGA2-GFP(S65T)::kanMX6 | This study |
| GPY3874 | SEY6210 GGA2-GFP(S65T)::kanMX6 | This study |
| GPY3890 | SEY6210 ent5(1–197)-GFP(S65T)::TRP1 | This study |
| GPY3900 | GPY3213 APL2-mRFP::TRP1 | This study |
| GPY3907 | GPY3431 APL2-GFP(S65T)::TRP1 | This study |
| GPY3912 | GPY3206 ENT5-mRFP::HIS3MX6 | This study |
| GPY3913 | GPY3425 ENT5-mRFP::HIS3MX6 | This study |
| GPY3954 | SEY6210 ENT3-GFP(S65T)::kanMX6 GGA2-mRFP::kanMX6 | This study |
| GPY3957 | SEY6210 apl2Δ-6::TRP1 ENT3-GFP(S65T)::kanMX6 GGA2-mRFP::kanMX6 | This study |
| GPY3962 | SEY6210 ENT5-GFP(S65T)::kanMX6 GGA2-mRFP::kanMX6 | This study |
| GPY3964 | SEY6210 apl2Δ-6::TRP1 ENT5-GFP(S65T)::kanMX6 GGA2-mRFP::kanMX6 | This study |
| GPY3974 | SEY6210 ENT3-GFP(S65T)::kanMX6 APL2-mRFP::TRP1 | This study |
| GPY4106 | SEY6210 ent3(1-176)-GFP(S65T)::kanMX6 | This study |
Gene disruptions, tags, and promoter integrations were confirmed by PCR amplification of the corresponding genomic loci.
Generation of GGA2 Thermosensitive Alleles
An AvrII/BsmI-gapped pRS315-GGA2 plasmid was cotransformed with overlapping regions of DNA generated by PCR mutagenesis into GPY2507 cells (see Plasmids for details) and plated at low density on supplemented SG lacking leucine (SG-Leu). After 3 to 4 d at 25°C, colonies (500–1000/plate) were replicated onto either SD-Leu or SG-Leu and subjected to incubation at 25 or 38°C. Plasmids were recovered from ts strains as described in Bensen et al. (2000) and transformed into GPY2555 to confirm ts growth phenotypes. Of ∼30000 colonies, a total of 12 ts alleles were obtained.
Metabolic Labeling and Immunoprecipitations
Metabolic labeling and immunoprecipitation of α-factor were performed as described (Yeung et al., 1999), except that the labeling media also contained 10 μg/ml α2-macroglobulin. ALP was analyzed by pulse-chase immunoprecipitation as described (Seeger and Payne, 1992). For analysis of CPS cells were concentrated to 2 × 107 cells/ml in supplemented SD and metabolically labeled for 20 min (25°C) or 10 min (38°C) with 100 μCi/ml Express-Tag (Perkin Elmer-Cetus, Boston, MA). Labeling was terminated by addition of 0.2% yeast extract, 2 mM l-methionine and 1.7 mM l-cysteine, and cells were allowed to grow for various chase periods. Cell extract preparation and immunoprecipitation were performed as in Seeger and Payne (1992), except that samples were treated with endoglycosidase H (Odorizzi et al., 1998). Image processing and quantitations were performed using a Molecular Dynamics PhosphorImager (Sunnyvale, CA) and ImageQuant software (Amersham/GE Healthcare, Chalfont St. Giles, United Kingdom).
Microscopy
Cells were grown to midlogarithmic phase on supplemented SD media at 25°C, sedimented for 3 min at 750 × g and resuspended at 2.5 × 107 cells/ml in fresh supplemented SD media, incubated for 30 min at 25 or 38°C, and then placed on ice water for 10 min before observation in the microscope. No more than four strains were processed at any given time. For a 2-h incubation at 38°C, the midlogarithmic culture was set at 38°C for 80 min and then concentrated, incubated at 38°C, and processed as described above.
Images were captured using a 100× α-Plan Fluar objective on a Zeiss Axiovert 200M microscope (Zeiss, Hallbegmoos, Germany) with a Hamamatsu ORCA-ER camera (Hamamatsu, Hamamatsu City, Japan).
Quantitations and Statistical Methods
Images were converted to TIFF format using AxioVision LE software (Zeiss).
To quantify colocalization, each image was processed with the SpotEnhancingFilter2D plug-in (Sage et al., 2005) for ImageJ (Rasband, 1997–2006), and masks were generated by thresholding the spot-enhanced image at 5 units intervals between 50 and 150 intensity units. The mask that best fit the spots in the original image was determined by visual inspection. Colocalization for a protein is expressed as the percentage of total mask (spot) area. Cells were counted using the corresponding DIC image for each field. Hypothesis testing for population proportions was used to calculate the statistical significance reported in Figure 6 (Daniel, 2005).
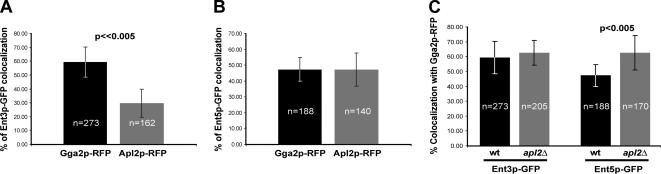
Figure 6.
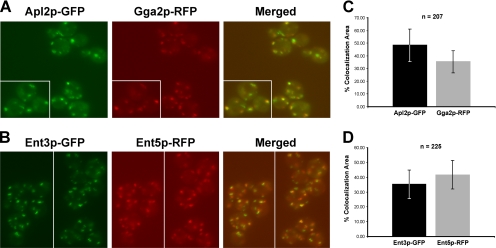
Quantitation of Ent3p preferential colocalization with Gga2p. (A) Percentage of Ent3p-GFP colocalized with Gga2p-RFP (■) and Ent3p-GFP colocalized with Apl2p-RFP (▩). (B) Percentage of Ent5p-GFP colocalized with Gga2p-RFP (■) and Ent5p-GFP colocalized with Apl2p-RFP (▩). (C) ENT3-GFP GGA2-RFP (■, wt, GPY3954), apl2Δ ENT3-GFP GGA2-RFP (▩, GPY3957), ENT5-GFP GGA2-RFP (■, wt, GPY3962) and apl2Δ ENT5-GFP GGA2-RFP (▩, GPY3964) were imaged as described for Figure 4 and colocalization of Ent3p and Ent5p with Gga2p was quantified. Colocalization was measured as described in Materials and Methods. n = the number of cells used for the quantitation. The level of significance (p value) was calculated as described in Material and Methods, and significant differences (p < 0.005) are indicated.
RESULTS
Differential Effects of ent3Δ or ent5Δ in GGA- and AP-1–deficient Strains
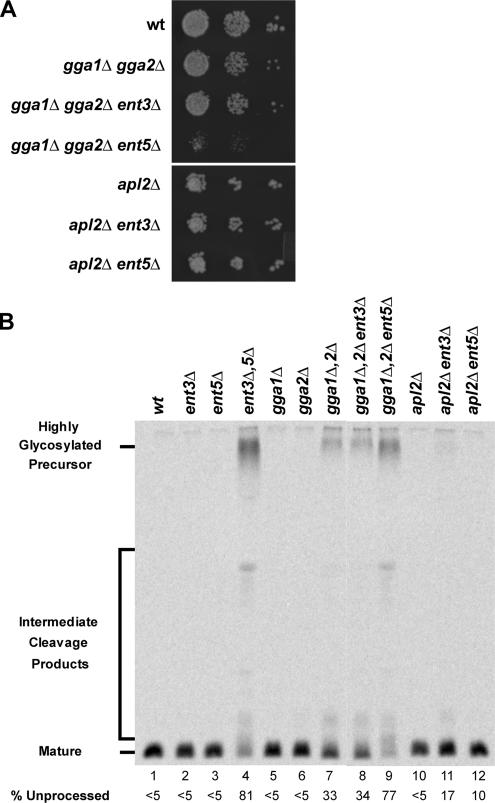
As an approach to assess preferential function of Ent3p or Ent5p with either AP-1 or Gga proteins, we carried out a synthetic genetic analysis. This strategy involves testing whether a combination of mutations in different genes is substantially more deleterious (“synthetic”) than individual mutations (Guarente, 1993). For example, mutational inactivation of AP-1 or both Gga proteins does not severely compromise cell growth, but inactivation of both adaptors has near-lethal effects (Costaguta et al., 2001). Thus, strains lacking one of the TGN/endosome adaptors are sensitized to perturbations in functions mediated by the other. To assess synthetic interactions, we carried out crosses of congenic strains harboring gene deletions in ENT3, ENT5, GGA1 and GGA2, and APL2. APL2 encodes the AP-1 β subunit and deletion of this gene inactivates AP-1 (Yeung et al., 1999). The resulting heterozygous diploid strains were induced to undergo meiosis and subjected to tetrad analysis. By growth assays, ent5Δ but not ent3Δ compromised the growth of gga1Δ gga2Δ cells (Figure 1A). Based on the synthetic genetic interactions of AP-1 and Gga mutations (Costaguta et al., 2001; Hirst et al., 2001), the synthetic growth defect in gga1Δ gga2Δ ent5Δ cells suggests a role for Ent5p in AP-1–mediated transport. Neither mutation alone caused growth defects in apl2Δ cells (Figure 1A). In haploids containing both ent3Δ and ent5Δ together with gga1Δ gga2Δ or apl2Δ, the cells formed only microcolonies. The growth effects observed in these strains indicate that neither Ent3p nor Ent5p is absolutely necessary for AP-1– or Gga-mediated transport. However, together they are required for AP-1–mediated traffic in the absence of Gga proteins and vice versa, suggesting that Ent3p and Ent5p can provide function in both AP-1– and Gga-dependent pathways.
Figure 1.
Synthetic genetic interactions between deletions of GGA, APL2, and ENT genes. (A) Synthetic growth defects. Wild-type (wt, SEY6210), gga1Δ gga2Δ (GPY3431), gga1Δ gga2Δ ent3Δ (GPY3683), gga1Δ gga2Δ ent5Δ (GPY3685), apl2Δ (GPY3100-4B), apl2Δ ent3Δ (GPY3688), and apl2Δ ent5Δ (GPY3690) cells were grown to midlogarithmic phase in YPD. Tenfold serial dilutions of cells were spotted onto solid YPD media and grown at 25°C for 4 d. (B) Synthetic α-factor maturation defects. Wild-type (wt, SEY6210; lane 1), ent3Δ (GPY2728; lane 2), ent5Δ (GPY2731; lane 3), ent3Δ ent5Δ (GPY2734; lane 4), gga1Δ (GPY2151; lane 5), gga2Δ (GPY2149; lane 6), gga1Δ gga2Δ (GPY3431; lane 7), gga1Δ gga2Δ ent3Δ (GPY3683; lane 8), gga1Δ gga2Δ ent5Δ (GPY3685; lane 9), apl2Δ (GPY3100-4B; lane 10), apl2Δ ent3Δ (GPY3688, lane 11), and apl2Δ ent5Δ (GPY3690; lane 12) cells were metabolically labeled for 45 min at 25°C and secreted α-factor was immunoprecipitated from the culture supernatants. Samples were analyzed by SDS-PAGE. Precursor levels of α-factor were quantified by phosphoimage analysis.
Another measure of synthetic genetic effects that more directly evaluates clathrin-mediated TGN/endosome traffic is the level of maturation of α-factor mating pheromone. This pheromone is synthesized as a precursor that is transported through the secretory pathway to the TGN, where it undergoes proteolytic maturation. The initiating protease in the maturation cascade is Kex2p, a furin-like endoprotease (Fuller et al., 1988). Kex2p localization in the TGN is maintained by cycling between the TGN and endosomes (Conibear and Stevens, 1998). In cells lacking clathrin, cycling is blocked, and Kex2p is mislocalized to the plasma membrane. The consequent depletion of Kex2p from the TGN reduces α-factor maturation leading to secretion of substantial levels of precursor forms that are easily distinguished from mature α-factor by SDS-PAGE (Payne and Schekman, 1989; Seeger and Payne, 1992). Previously we observed more severe defects in α-factor maturation in apl2Δ gga2Δ mutants than in single mutants or even gga1Δ gga2Δ mutants completely lacking Gga-mediated transport (Costaguta et al., 2001). Thus, in cells deficient for either AP-1 or GGA adaptors, synthetic effects on α-factor maturation represent a sensitive assay for inhibition of the extant adaptor-mediated pathway.
To analyze Ent3p and Ent5p function in AP-1– and Gga-dependent transport, we monitored α-factor maturation in mutants carrying single, double, or triple combinations of ENT, GGA, and APL2 deletions. Cells were metabolically labeled and α-factor was immunoprecipitated from the culture supernatant. None of the single mutants displayed maturation defects (Figure 1B, lanes 1–3, 5–6, and 10). As reported previously (Duncan et al., 2003), ent3Δ ent5Δ cells exhibit a nearly complete inhibition of α-factor maturation, with 81% of the secreted pheromone in precursor form (Figure 1B, lane 4). This defect is more severe than that observed in gga1Δ gga2Δ cells (33%; Figure 1B, lane 7) or apl2Δ gga2Δ cells (61%; Costaguta et al., 2001), consistent with key roles of Ent3p and Ent5p in TGN/endosome traffic. Importantly, ent3Δ and ent5Δ had different effects on α-factor maturation when combined with gga1Δ gga2Δ or apl2Δ. Compared with the precursor levels secreted by gga1Δ gga2Δ cells (33%, Figure 1B, lane 7), gga1Δ gga2Δ ent5Δ cells exhibited a severe maturation defect (77%; Figure 1B, lane 9) equivalent to that of ent3Δ ent5Δ cells. In contrast, ent3Δ did not enhance the maturation defect of gga1Δ gga2Δ cells (34%; Figure 1B, lane 8). Conversely, ent3Δ elevated precursor levels in combination with apl2Δ, whereas ent5Δ combined with apl2Δ had a marginal but reproducible effect (Figure 1B, lanes 10–12). These differential genetic interactions detected by a functional assay for clathrin-dependent transport provide evidence that Ent3p functions primarily with Gga proteins, whereas Ent5p acts with both AP-1 and Gga adaptors, though it is more strictly required with AP-1.
Localization of AP-1 and Gga2p Is Not Dependent on Ent Proteins
It has been proposed that monomeric endocytic adaptors epsin1 and AP180 might collaborate to recruit AP-2 to incipient sites of endocytosis by interacting with the plasma membrane through their respective ENTH and ANTH domains and binding AP-2 through motifs located in the disordered and flexible C-termini (Kalthoff et al., 2002a). Ent3p and Ent5p represent a similar pair of ENTH/ANTH domain adaptors at the TGN/endosome with binding motifs for AP-1 and Gga rather than AP-2 (Duncan and Payne, 2003). We therefore considered the possibility that Ent3p and Ent5p are important for AP-1 and/or Gga protein localization.
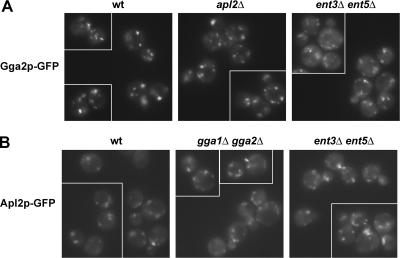
To address the role of different adaptors in AP-1 and Gga protein localization, we visualized adaptor distribution in living cells expressing functional forms of C-terminally GFP-tagged Apl2p and Gga2p from the normal chromosomal loci. Gga2p-GFP localizes as several discrete puncta in wild-type cells, likely representing the TGN and possibly endosomes (Hirst et al., 2001; Boman et al., 2002; Figure 2A, WT). This pattern was unaffected in AP-1–deficient cells (Figure 2A, apl2Δ) and in cells lacking Ent3p and Ent5p (Figure 2A, ent3Δ ent5Δ). Similarly, distribution of Apl2p-GFP was not altered significantly by deletion of the GGA genes or the ENT genes (Figure 2B). Thus, localization of AP-1 and Gga2p are independent of each other and the Ent adaptors.
Figure 2.
AP-1 and Gga2p localization is independent of other clathrin Golgi adaptors. (A) GGA2-GFP (wt, GPY3874), apl2Δ GGA2-GFP (GPY3786), ent3Δ ent5Δ GGA2-GFP (GPY3802), and (B) APL2-GFP (wt, 3298-27C), gga1Δ gga2Δ APL2-GFP (GPY3907), and ent3Δ ent5Δ APL2-GFP (3298-13A) cells were grown in supplemented SD media to midlogarithmic phase at 25°C and imaged by epifluorescence microscopy. Each panel contains a gallery of cells imaged from the same field.
Localization of Ent3p Requires Gga Proteins
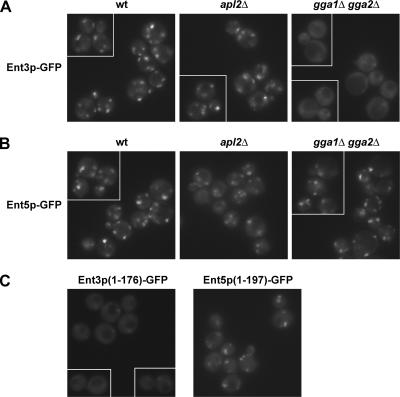
In a reciprocal set of experiments, C-terminally tagged forms of Ent3p and Ent5p were monitored in wild-type, apl2Δ, and gga1Δ gga2Δ cells. Like Gga proteins and AP-1, Ent3p-GFP, and Ent5p-GFP concentrate at cytoplasmic foci (Figure 3, A and B, WT). This pattern was largely unaltered in apl2Δ cells (Figure 3, A and B), although the overall intensity of Ent5p-GFP puncta was somewhat reduced (for example, the 1% brightest pixels decreased in intensity by 10–20%). Ent5p-GFP displayed a normal distribution in gga1Δ gga2Δ mutants (Figure 3B). In striking contrast, Ent3p-GFP was largely diffuse in the absence of Gga proteins, with only faint and infrequent puncta (Figure 3A). This change was not due to a decline in Ent3p-GFP levels as determined by immunoblotting (unpublished results). These results suggest that Ent3p localization requires interaction with Gga proteins, whereas Ent5p localization is largely independent of interaction with other adaptors.
Figure 3.
Ent3p localization requires Gga adaptors. (A) ENT3-GFP (wt, GPY3206), apl2Δ ENT3-GFP (GPY3425), and gga1Δ gga2Δ ENT3-GFP (GPY3429); (B) ENT5-GFP (wt, GPY3213), apl2Δ ENT5-GFP (GPY3434), gga1Δ gga2Δ ENT5-GFP (GPY3440), and (C) ent3(1-176)-GFP (GPY4106) and ent5(1-197)-GFP (GPY3890) cells were grown as described for Figure 2, and GFP was imaged by epifluorescence microscopy. Each panel contains a gallery of cells imaged from the same field.
Our interpretations based on localization in AP-1– or Gga-deficient cells were supported by analysis of wild-type cells expressing the Ent3p ENTH domain [Ent3p(1-176)] or the Ent5p ANTH domain [Ent5p(1-197)] tagged with GFP. These fusion proteins lack the C-terminal regions of Ent3p and Ent5p that encompass the γ-ear– and clathrin-binding motifs. Ent5p(1-197)-GFP distributed in a punctate pattern (Figure 3C), most similar to full-length Ent5p-GFP in apl2Δ cells (with substantial colocalization with clathrin; M. C. Duncan, unpublished results). Ent3p(1-176)-GFP was diffusely distributed (Figure 3C), resembling Ent3p-GFP localization in gga1Δ gga2Δ cells. Taken together our results indicate that the ANTH domain is sufficient for localization of Ent5p, whereas Ent3p is localized primarily by interaction with Gga proteins. These distinct localization features of Ent3p and Ent5p correlate well with the functional relationships defined by synthetic genetic analysis.
Relative Localization of TGN/Endosome Adaptors
We further explored the relationship between Ent, Gga, and AP-1 adaptors by examining colocalization of adaptor pairs in wild-type cells. Initially we analyzed the relative distributions of Apl2p compared with Gga2p and Ent5p compared with Ent3p. The overlap between Gga2p with Apl2p was extensive (Figure 4, A and C): 48 ± 13% of Apl2p-GFP puncta were coincident with Gga2p-RFP puncta and 35 ± 9% of the Gga2p-RFP puncta colocalized with Apl2p-GFP puncta. Similar levels of overlap were observed with the two Ent adaptors (Figure 4, B and D): 35 ± 10% of Ent3p-GFP colocalized with Ent5p-RFP, whereas 42 ± 9% of Ent5p-RFP coincided with Ent3p-GFP. Thus, these pairs of adaptors exhibit substantial overlap, consistent with their common participation in clathrin-mediated TGN/endosome transport. Nevertheless, individual adaptors are present at a substantial number of sites or at times when the other adaptor in the pair is absent.
Figure 4.
Partial colocalization of AP-1 with Gga2p and Ent3p with Ent5p. (A) APL2-GFP GGA2-RFP (GPY3298–29C) and (B) ENT3-GFP ENT5-RFP (GPY3912) cells were grown to midlogarithmic phase, and GFP and RFP were imaged by epifluorescence microscopy. Merged images are shown on the right panel. Insets represent cells from the same field. (C) Percentage of Apl2p-GFP that colocalizes with Gga2p-RFP (■) and Gga2p-RFP colocalized with Apl2p-GFP (▩). (D) Percentage of Ent3p-GFP colocalized with Ent5p-RFP (■) and Ent5p-RFP colocalized with Ent3p-GFP (▩). Colocalization was measured as described in Materials and Methods. n = the number of cells used for the quantitation.
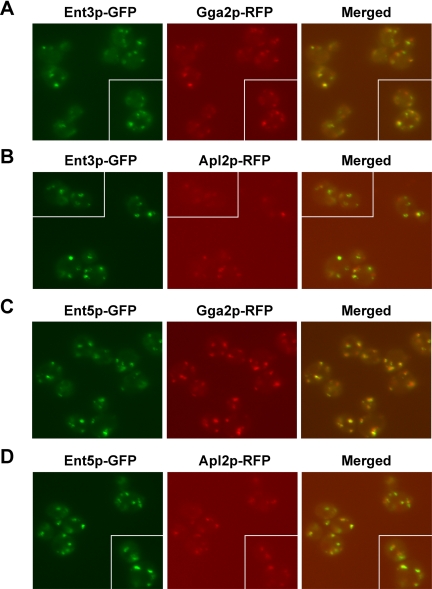
The limited overlap of AP-1 with Gga2p and Ent5p with Ent3p allowed us to determine whether Ent5p and Ent3p preferentially localized with AP-1 or Gga2p. In agreement with the functional and localization relationships between Ent3p and Gga2p, we observed a clear difference in the degree of Ent3p-GFP localization with Gga2p-RFP (59 ± 11%) compared with Apl2p-RFP (30% ± 10; Figures 5, A and B, and 6A). Ent5p-GFP displayed equivalent colocalization with Apl2p-RFP (47 ± 11%) and Gga2p-RFP (47 ± 7%; Figures 5, C and D, and 6B).
Figure 5.
Ent3p colocalizes preferentially with Gga2p. (A) ENT3-GFP GGA2-RFP (GPY3954), (B) ENT3-GFP APL2-RFP (GPY3974), (C) ENT5-GFP GGA2-RFP (GPY3962) and (D) ENT5-GFP APL2-RFP (GPY3900) cells were grown and imaged as described for Figure 4. Merged images are shown on the right panel. Each panel contains a gallery of cells imaged from the same field.
In Figure 3B we observed a subtle defect in Ent5p localization in apl2Δ cells, suggesting a role for AP-1 in Ent5p localization. As another means to evaluate the effects of apl2Δ on Ent5p distribution, colocalization of Ent5p and Ent3p with Gga2p was monitored in apl2Δ versus wild-type cells. In comparison to wild-type cells where 47 ± 7% Ent5p-GFP colocalized with Gga2p-RFP, in apl2Δ cells 63% ± 12 Ent5p-GFP localized together with Gga2p-RFP, a statistically significant increase in the extent of colocalization (Figure 6C). This was a specific effect on Ent5p because the level of Ent3p-GFP colocalization with Gga2p-RFP did not change upon deletion of APL2 (Figure 6C). Thus, although AP-1 is not a major determinant of Ent5p localization, our evidence indicates that the adaptor complex does play a role in establishing the normal distribution of Ent5p. In contrast, Ent3p localization appears to be independent of AP-1.
Thermosensitive Alleles of Gga2p with Mutations in the VHS Domain
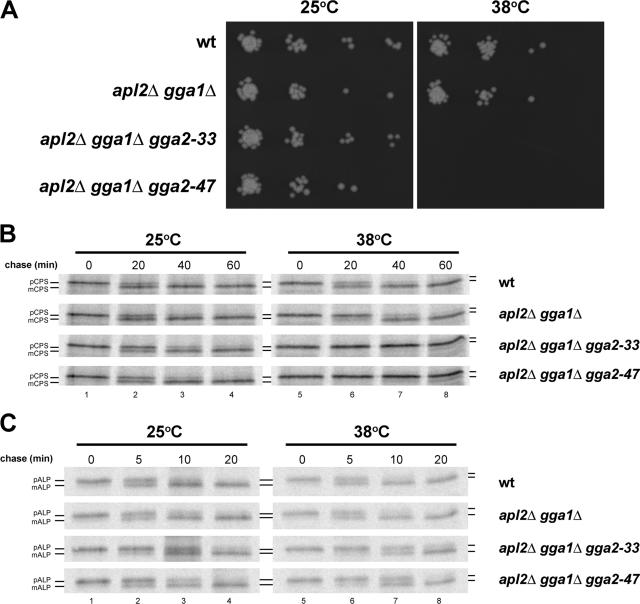
The severe growth defect of cells lacking Gga proteins and AP-1 (Costaguta et al., 2001) provided an opportunity to generate temperature-sensitive alleles of the Gga adaptors that would allow rapid inactivation of Gga function. Because the VHS domain of the yeast Gga proteins is critical for function (Boman et al., 2002), we used error-prone PCR to mutagenize the VHS domain of Gga2p and isolated a number of alleles that allowed growth of Gga- and AP-1–deficient cells at 25°C but not 38°C (see Materials and Methods for details). We analyzed two alleles in more detail, gga2-33 and gga2-47, which were introduced into the chromosomal GGA2 locus in apl2Δ gga1Δ cells. Figure 7A shows the temperature-sensitive growth defect of cells harboring these alleles.
Figure 7.
GGA2 thermosensitive alleles cause temperature-sensitive defects in growth and CPS maturation. Wild-type (wt, SEY6210), apl2Δ gga1Δ (GPY3101-5A), apl2Δ gga1Δ gga2-33 (GPY3101-6D), and apl2Δ gga1Δ gga2-47 (GPY3100-11A) cells were (A) grown to midlogarithmic phase on YPD media, and 10-fold serial dilutions were spotted onto solid YPD media and allowed to grow at 25 or 38°C for 3–4 d, (B) metabolically labeled for 20 min at 25°C or 10 min at 38°C and subjected to chase for the indicated time points. CPS was immunoprecipitated from cell lysates, processed with endoglycosidase H, and analyzed by SDS-PAGE, or (C) metabolically labeled for 10 min at 25 or 38°C and subjected to chase for the indicated time points. ALP was immunoprecipitated from cell lysates and analyzed by SDS-PAGE. Precursor (p) and mature (m) forms of CPS and ALP are indicated.
To examine effects of the two ts alleles on Gga-dependent trafficking, we monitored proteolytic maturation of the vacuolar hydrolase CPS in the apl2Δ gga1Δ gga2-ts strains. CPS is synthesized as an integral membrane precursor (pCPS) that is transported through the secretory pathway to the TGN. At the TGN, pCPS is directed by a Gga-dependent pathway to endosomes and then delivered to the vacuole where it undergoes proteolytic activation (Bowers and Stevens, 2005). Proteolytic maturation of pCPS is almost completely blocked by deletion of GGA1 and GGA2, making this a sensitive reporter for defects in Gga activity (Cowles et al., 1997b; Costaguta et al., 2001). CPS maturation was assayed by pulse-chase immunoprecipitation in wild-type cells and apl2Δ gga1Δ cells harboring either wild-type GGA2, gga2-33, or gga2-47. Cells were incubated at the permissive temperature (25°C) or shifted to the nonpermissive temperature (38°C) for 5 min before labeling. In wild-type cells CPS maturation is essentially complete after 40 min at both temperatures (Figure 7B, WT row). The same is true of the other three strains at 25°C, although there is a minor delay in the gga2-33 strain (Figure 7B, lanes 1–4). After shift to 38°C, the apl2Δ gga1Δ cells also exhibited a slight delay in CPS maturation, most evident as a shift in the ratio of pCPS to mCPS at the 20-min chase point (Figure 7B, row 2, lane 6). In contrast, CPS maturation was almost completely abolished in the gga2-ts strains (Figure 7B, rows 3–4, lanes 5–8), indicating a rapid inactivation of Gga2-dependent traffic. Similar results were obtained in gga1Δ gga2-ts strains expressing Apl2p (unpublished results). The effects of the ts alleles were relatively specific; shift to the nonpermissive temperature caused only minor delays (≈5 min) in transport and maturation of vacuolar ALP, which is delivered to the vacuole from the TGN through a clathrin- and Gga-independent pathway (Figure 7C; Cowles et al., 1997a; Panek et al., 1997; Piper et al., 1997; Vowels and Payne, 1998).
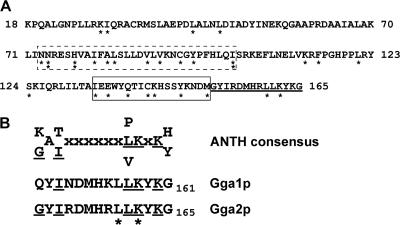
Sequencing of the gga2-33 and gga2-47 alleles revealed multiple mutations in each: H78R, F115Y, and S124F in gga2-33 and V91D and M151V in gga2-47. Sequence analysis of nine additional ts alleles revealed that multiple mutations were a common feature of the different alleles (Supplementary Table 1). Interestingly, mutations in the 11 alleles clustered in two regions in the VHS domain, aa 73–102 (Figure 8A, dashed box) and aa 134–151 (Figure 8A, solid box). The first of these regions is nearly identical to a segment (aa 70 to aa 97) that was previously shown by N-terminal deletion analysis to be important for Gga2p function in α-factor maturation and CPY transport (Mullins and Bonifacino, 2001). The second region has not been previously characterized but is adjacent to a sequence between aa 152 and aa 165 that bears strong resemblance to the consensus sequence for lipid binding in ANTH domains (Figure 8A, underlined, and 8B).
Figure 8.
GGA2 ts mutations in the VHS domain cluster in two regions. (A) Sequence of the VHS domain of Gga2p (amino acids 18–165). Positions of the mutations that confer temperature sensitivity are indicated by an asterisk (*) under the appropriate amino acid. The dashed and closed boxes delineate regions with clusters of mutations. The region similar to the ANTH consensus is underlined. (B) Alignment of the ANTH consensus with conserved regions in Gga1p and Gga2p. Identical amino acids are underlined. Temperature-sensitive mutations are indicated by an asterisk (*).
Ent3p Localization Defects in gga2-ts Cells
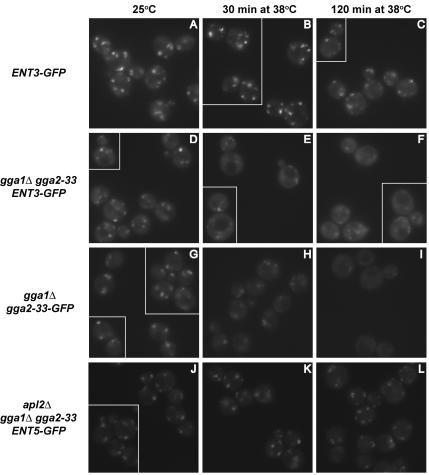
We used gga1Δ gga2-33 strains to analyze the immediate effects of Gga inactivation on Ent3p localization. Even at 25°C, gga1Δ gga2-33 cells exhibited fewer and less intense Ent3p-GFP foci than wild-type cells (Figure 9, A and D), indicating that the Gga2-33 protein is not completely functional at this temperature even though it supports relatively normal growth and CPS transport. After shifting gga1Δ gga2-33 cells to 38°C for 30 min most of the Ent3p was diffusely distributed (Figure 9, B and E), and by 120 min only residual localization was evident (Figure 9, C and F). The decreased levels of Ent3p localization in the mutant cells at the nonpermissive temperature corresponded closely to a decline in levels of Gga2-33p as measured by fluorescence microscopy of Gga2-33p-GFP (Figure 9, G–I) and immunoblotting (unpublished results). In contrast, levels of Ent3p-GFP did not decrease upon temperature shift as determined by immunoblotting (unpublished results). The rapid effect of Gga protein inactivation on Ent3p localization and the strong correlation between levels of Gga2 protein and the degree of punctate Ent3p provide further evidence that Gga proteins play a primary and direct role in Ent3p localization.
Figure 9.
Gga2p inactivation causes a defect in Ent3p localization. ENT3-GFP (GPY3206, A–C), gga1Δ gga2-33 ENT3-GFP (GPY3630, D–F), gga1Δ gga2-33-GFP (GPY3669, G–I), and apl2Δ gga1Δ gga2-33 ENT5-GFP (GPY3217-7A, J–L) cells grown at 25°C were continued at 25°C or shifted to 38°C for the indicated times before epifluorescence microscopy. Each panel contains a gallery of cells imaged from the same field.
We also analyzed Ent5p localization in gga1Δ gga2-33 and apl2Δ gga1Δ gga2–33 cells shifted to the nonpermissive temperature. Ent5p-GFP retained a significant degree of punctate distribution in these cells supporting our conclusion that Ent5p can localize independently of AP-1 and Gga adaptors (Figure 9, J–L, and unpublished results).
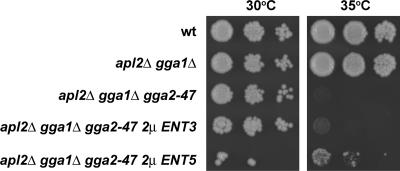
Ent5p Overexpression Partially Suppresses the Growth Defect of AP-1– and Gga-deficient Cells
Our results indicate that Ent3p depends on Gga proteins for localization and functions primarily in Gga-mediated traffic, whereas Ent5p localizes at least in part independently of Gga and AP-1 adaptors and appears to function with both adaptors. These findings raise the possibility that Ent5p might play a more independent role in TGN/endosome transport than Ent3p. As a test of this interpretation, we determined whether overexpression of Ent3p or Ent5p could suppress the growth defect of apl2Δ gga1Δ gga2-47 cells at elevated temperatures. At 38°C, neither Ent3p nor Ent5p overexpressed from a multicopy plasmid allowed growth of apl2Δ gga1Δ gga2-47 cells (unpublished results). However, at a slightly lower temperature, 35°C, where apl2Δ gga1Δ gga2-47 cells are still unable to grow, overexpression of Ent5p caused a clear, yet partial, suppression of the growth defect (Figure 10, rows 1–3 and 5). By comparison, overexpression of Ent3p was essentially ineffective (Figure 10, row 4). This difference was not due to differences in levels of Ent3p and Ent5p overexpression from the multicopy plasmids; both were expressed at roughly 30 times the endogenous levels (unpublished results). The ability of Ent5p but not Ent3p to partially compensate for loss of AP-1 and Gga function supports the proposal that Ent5p plays a more independent role than Ent3p in TGN/endosome traffic.
DISCUSSION
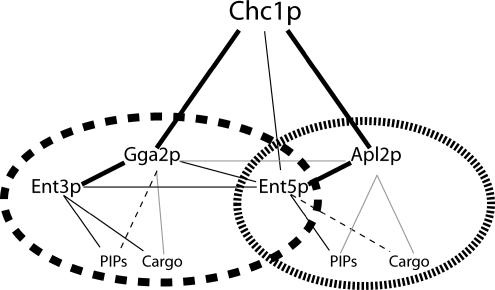
A physically interacting network of evolutionarily conserved clathrin adaptors are involved in traffic between TGN and endosomes. Here, as an approach to define functional relationships between these adaptors in vivo, we analyzed genetic interactions between mutations affecting different adaptors and determined whether particular adaptor localization is dependent on other members of the network. Our results support a model in which Ent3p function is primarily dedicated to Gga-mediated traffic, whereas Ent5p acts with both AP-1 and Gga proteins (Figure 11). Localization mechanisms contribute to this functional distinction; Ent3p requires Gga proteins for localization, but Ent5p can localize to a significant extent without either Gga proteins or AP-1.
Our previous coimmunoprecipitation analysis of Ent3p and Ent5p interaction partners revealed Ent5p association with Gga proteins and AP-1, but Ent3p association only with Gga proteins (Duncan et al., 2003). However, in vitro assays revealed similar levels of Ent3p and Ent5p binding by γ-ear domains of AP-1 and Gga proteins (Duncan et al., 2003). The synthetic genetic interactions reported here are entirely consistent with the coimmunoprecipitations, providing evidence for the functional significance of the difference in binding partners. Genetic interaction analysis of ent3Δ indicated primary function in Gga-mediated transport; ent3Δ with gga deletions did not exacerbate defects in α-factor maturation, but ent3Δ combined together with apl2Δ caused α-factor maturation defects not apparent in either single mutant. In contrast, ent5Δ interactions revealed that Ent5p acted to different extents in both AP-1– and Gga-mediated traffic. Introduction of ent5Δ into gga1Δ gga2Δ cells dramatically decreased α-factor maturation, consistent with strong inhibition of AP-1–mediated transport. A subtle but reproducible effect on α-factor maturation was also observed in apl2Δ ent5Δ cells, suggesting that Ent5p also provides some function in Gga-mediated transport. Overlapping function of Ent3p and Ent5p with Gga proteins could explain the mild synthetic phenotypes of individual ent mutations combined with apl2Δ. Supporting this possibility, the triple apl2Δ ent3Δ ent5Δ mutant was severely compromised for growth. The suggestion that both Ent3p and Ent5p act in vivo with Gga2p is also consistent with physical association of Ent3p and Ent5p with Gga2p detected by coimmunoprecipitations (Duncan et al., 2003).
It is somewhat less clear why introducing ent3Δ into gga1Δ gga2Δ ent5Δ further debilitated growth if Ent3p acts solely with Gga proteins. One possibility is that Ent3p does not normally function with AP-1 but can substitute to some extent when Ent5p is absent. Consistent with this notion, Ent3p has the ability to bind to the γ-ear domain of Apl4p in vitro (Duncan et al., 2003). Alternatively, Ent3p could function with AP-1 in a way that might not be revealed by assaying α-factor maturation, for example, as a cargo-specific adaptor.
The synthetic phenotypes of ent3Δ combined with ent5Δ also fit predictions from a model in which Ent3p function is primarily dedicated to Gga-mediated transport, whereas Ent5p acts with both Gga proteins and AP-1 (Black and Pelham, 2000; Costaguta et al., 2001). Neither deletion alone significantly affects growth or α-factor maturation, presumably because impact of individual deletions on the Gga pathway is compensated by functional redundancy between Ent3p and Ent5p, and inhibition of AP-1–mediated transport is innocuous. However, the double deletion would impede both Gga- and AP-1–mediated transport, thereby accounting for the growth inhibition and maturation defects that were stronger than those caused by complete inactivation of Gga function in gga1Δ gga2Δ cells. It is worth noting that the growth phenotype of ent3Δ ent5Δ cells is not as severe as full inactivation of both Gga proteins and AP-1 in the apl2Δ gga1Δ gga2Δ cells (Costaguta et al., 2001; Duncan et al., 2003), suggesting that some level of traffic can occur through Gga- and/or AP-1 in the absence of the Ent adaptors. This finding offers support for the view that AP-1 and Gga adaptors serve a more central role than Ent proteins in TGN/endosome traffic. The inability of overxexpressed Ent3p or Ent5p to suppress the growth phenotype of apl2Δ gga1Δ gga2-ts cells at the fully restrictive temperature of 38°C is also consistent with the primacy of AP-1 and Gga adaptors.
The different degrees of adaptor colocalization are concordant with the synthetic genetic interactions and the physical interactions. Ent3p preferentially colocalized with Gga proteins compared with AP-1. Furthermore, Ent3p was mislocalized in ggaΔ cells and dispersed rapidly after inactivation of a gga2-ts allele, indicating dependence on Gga proteins for localization. Consistent with these results, the Ent3p ENTH domain lacking the region with γ-ear–binding motifs was not localized. From these findings we propose that Ent3p binding to Gga γ-ear domains is necessary for proper recruitment of Ent3p to sites of CCV formation in vivo. Ent5p, on the other hand, colocalized equally with both adaptors and localized to a substantial extent in cells lacking both AP-1 and Gga proteins, or as a truncated protein lacking γ-ear– and clathrin-binding motifs. Thus, interactions mediated by the Ent5p ANTH domain appear particularly important for Ent5p localization.
We favor the idea that Gga-mediated localization of Ent3p involves recruitment of cytoplasmic Ent3p to membrane-associated Gga proteins. However, we have not reproducibly detected changes in membrane association of Ent3p in mutant cells by differential centrifugation, which we attribute to differences between conditions before and after cell lysis. Thus, it is possible that the changes in localization observed by microscopy reflect alterations in the distribution of Ent3p between different membrane structures rather than between membranes and the cytoplasm. Regardless, the data indicate that Gga proteins are key determinants in Ent3p but not Ent5p localization.
The similar overall architectures of Ent3p and Ent5p do not provide obvious explanations for the differences in localization mechanisms; both proteins contain N-terminal E/ANTH domains capable of lipid binding followed by a region with multiple γ-ear–binding sites (Duncan and Payne, 2003). In fact, Ent3p and Ent5p bind comparably to Gga and AP-1 γ-ears in vitro, suggesting that preferential association of Ent3p with Gga proteins is influenced by contributions from other factors in vivo (Duncan and Payne, 2003). One factor could be cargo binding. The SNARE protein Vti1p binds to the Ent3p ENTH domain but not to Ent5p (Chidambaram et al., 2004). Perhaps cooperative binding to Vti1p or other cargo and Gga γ-ears contributes to selective localization of Ent3p to Gga-containing structures.
Another factor contributing to differences in Ent3p and Ent5p localization could be phosphoinositide binding. Ent3p and Ent5p have been shown to bind phosphoinositides with some preference for PtdIns[3]P and PtdIns[3,5]P2 (Friant et al., 2003; Chidambaram et al., 2004; Eugster et al., 2004). The inability of the Ent3p ENTH domain alone to localize indicates that phosphoinositide binding by this domain is not sufficient for localization. However, if a particular phosphoinositide with affinity for the Ent3p ENTH domain is present at higher concentrations at membrane sites with Gga proteins than sites with AP-1, then Ent3p association with Gga-containing structures could be favored. Unlike the Ent3p ENTH domain, the Ent5p ANTH domain alone retains at least partial localization activity. Given the different modes of phosphoinositide binding by ENTH and ANTH domains, the differences between Ent3p ENTH domain and Ent5p ANTH domain could be attributable to either differences in affinity for a particular phosphoinositide or for different phosphoinositides. Another possibility is that protein binding by the Ent5p ANTH domain contributes to localization. Further analysis will be required to distinguish between these possibilities.
The different genetic and physical interactions we have described for the two TGN/endosome Ent adaptors imply functional differences. One likely distinction is cargo binding. Ent3p can provide a cargo-selective role for Vti1p (Chidambaram et al., 2004; Hirst et al., 2004) and perhaps other cargo in Gga-mediated transport. Another difference may lie in the ability of the ENTH domains to deform lipid bilayers. The epsin1 ENTH domain has a capacity to promote membrane curvature that is not observed with the AP180 ANTH domain (Ford et al., 2002; Stahelin et al., 2003). If the ENTH and ANTH domains of Ent3p and Ent5p mirror this difference then Ent3p may provide membrane deformation activity that is particularly important in Gga-mediated transport. In this scenario, such activity would not be as critical for AP-1–mediated traffic. Considering this view, it is intriguing that combinations of AP-2 and AP180 lead to curvature of lipid monolayers characteristic of invaginated clathrin-coated pits (Ford et al., 2001) that are not observed with either adaptor alone. Similarly, AP-1 and Ent5p may foster membrane curvature without substantial input from Ent3p. Ent3p and Ent5p also differ in clathrin-binding properties. Although Ent3p has been reported to bind clathrin (Friant et al., 2003), direct comparison of clathrin interaction by coimmunoprecipitation of Ent3p and Ent5p revealed substantially more robust binding by Ent5p (Duncan et al., 2003). We suggest that clathrin binding represents an important Ent5p activity contributed to both AP-1– and Gga-mediated transport processes that is not shared with Ent3p. The ability to bind clathrin and localize through the ANTH domain endows Ent5p with AP-1– and Gga-independent adaptor activity that is likely to account for the ability of overexpressed Ent5p to suppress the growth defect of apl2Δ gga1Δ gga2-47 cells.
Our characterization of gga2-ts alleles revealed clusters of mutations in two regions within the VHS domain. The first, aa 73–102, corresponds to a region defined by nested N-terminal deletions as particularly important for Gga2p function in CPY transport (Mullins and Bonifacino, 2001). Using SWISS-MODEL (Peitsch, 1996; Guex and Peitsch, 1997) and the crystal structure of the human GGA3 VHS domain (Misra et al., 2002) to model Gga2p, aa 73–102 of Gga2p are predicted to span helices 4 and 5 in the right-handed 8-helix superhelix (unpublished results). Given this positioning in the center of the superhelix, mutations in this region could affect the overall fold of the VHS domain and have a general effect on the domain structure. The second region, aa 134–151, mostly encompasses helix 7, which lays between helices 6 and 8 that directly contact cargo in GGA3. Thus, mutations in this region have the potential to disrupt cargo binding by Gga2p, although the identity of such cargo remains to be established. Another possible consequence of mutations in this region is suggested by adjacent sequences from 152 to 165 predicted to lie in helix 8. This stretch of amino acids contains 5/7 appropriately spaced residues matching the ANTH domain phosphoinositide-binding consensus sequence described by Ford et al. (2002). Four of the five conserved residues, including the two lysines, are predicted to lie in a surface-exposed patch based on the GGA3 structure. The importance of the helix 8 sequences is emphasized by the strong conservation of Gga1p and Gga2p in this region (11/14 identical residues; Figure 8B). Together these observations raise the possibility that Gga2p aa152–165 may interact with phosphoinositides and mutations in helix 8 or the adjacent helix 7 could disrupt this activity. Alternatively, as helix 8 in GGA3, the Gga2p helix 8 could be involved in cargo recognition. Future identification of Gga2p VHS-binding cargo and assays for lipid binding will address these possibilities and determine whether activities involving this region are shared with the less well-conserved sequences in human GGAs.
In summary, we have built a map of the physical and genetic interactions and characterized the localization relationships between members of the network of TGN/endosome clathrin adaptors. Our results support a model in which AP-1 and Gga proteins provide central adaptor function that is elaborated by Ent adaptors (Figure 11). Ent3p is recruited by, and interacts preferentially with, Gga proteins, whereas Ent5p functions with both AP-1 and Gga adaptors. This model provides a foundation to understand the contributions of specific adaptor functions to the process of CCV formation at the TGN and endosomes.
Supplementary Material
Figure 10.
Ent5p overexpression suppresses lethality of apl2Δ gga1Δ gga2-47 cells at 35°C. Wild-type pRS314 pRS425 (wt, SEY6210), apl2Δ gga1Δ pRS425 (GPY3101-5A), apl2Δ gga1Δ gga2-47 pRS425 (GPY3100-11A), apl2Δ gga1Δ gga2-47 p425-ENT3 (GPY3100-11A), and apl2Δ gga1Δ gga2-47 p425-ENT5 (GPY3100-11A) cells were grown on supplemented SD media lacking leucine to midlogarithmic phase and resuspended to 1 × 106 cells/ml. Tenfold serial dilutions of cells were spotted onto YPD solid media and incubated at 30 or 35°C for 3 d.
Figure 11.
The TGN/endosome clathrin adaptor network. Solid black lines indicate experimentally supported physical interactions with yeast adaptors. Thick black lines are physical associations supported by synthetic genetic data. Solid gray lines indicate interactions of mammalian AP-1 and Gga proteins that may apply to the yeast adaptors. Dashed lines indicate hypothetical interactions. Our data suggest that the network is functionally organized into two groups: one centered on Gga2p (long dash ellipse) and one centered on Apl2p (short dash ellipse).
ACKNOWLEDGMENTS
We thank Alex van der Bliek for use of the microscope and Steven Nothwehr for the gift of CPS antibody. We also thank members of the Payne laboratory for comments on the manuscript. This work was supported by National Institutes of Health (NIH) Grants GM39040 and GM67911 (G.S.P.), and partially by a UCLA Dissertation Year Fellowship (G.C.), and by NIH NRSA fellowships DK062608 (M.C.D.) and GM072119 (G.E.F.).
Abbreviations used:
- CCV
clathrin-coated vesicle
- TGN
trans-Golgi network
- CPS
carboxypeptidase S
- ALP
alkaline phosphatase
- ts
thermosensitive
- ENTH
epsin N-terminal homology
- ANTH
AP180 N-terminal homology
- mRFP
monomeric red fluorescent protein
- GFP
green fluorescent protein.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-05-0410) on June 21, 2006.
REFERENCES
- Austin C., Boehm M., Tooze S. A. Site-specific cross-linking reveals a differential direct interaction of Class 1, 2, and 3 ADP-ribosylation factors with adaptor protein complexes 1 and 3. Biochemistry. 2002;41:4669–4677. doi: 10.1021/bi016064j. [DOI] [PubMed] [Google Scholar]
- Bai H., Doray B., Kornfeld S. GGA1 interacts with the adaptor protein AP-1 through a WNSF sequence in its hinge region. J. Biol. Chem. 2004;279:17411–17417. doi: 10.1074/jbc.M401158200. [DOI] [PubMed] [Google Scholar]
- Bensen E. S., Costaguta G., Payne G. S. Synthetic genetic interactions with temperature-sensitive clathrin in Saccharomyces cerevisiae. Roles for synaptojanin-like Inp53p and dynamin-related Vps1p in clathrin-dependent protein sorting at the trans-Golgi network. Genetics. 2000;154:83–97. doi: 10.1093/genetics/154.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black M. W., Pelham H.R.B. A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J. Cell Biol. 2000;151:587–600. doi: 10.1083/jcb.151.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman A. L., Salo P. D., Hauglund M. J., Strand N. L., Rensink S. J., Zhdankina O. ADP-ribosylation factor (ARF) interaction is not sufficient for yeast GGA protein function or localization. Mol. Biol. Cell. 2002;13:3078–3095. doi: 10.1091/mbc.E02-02-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J. S. The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- Bonifacino J. S., Traub L. M. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- Bowers K., Stevens T. H. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta Mol. Cell Res. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Brodsky F. M., Chen C.-Y., Knuehl C., Towler M. C., Wakeham D. E. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu. Rev. Cell Dev. Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidambaram S., Mullers N., Wiederhold K., Haucke V., von Mollard G. F. Specific interaction between SNAREs and epsin N-terminal homology (ENTH) domains of epsin-related proteins in trans-Golgi network to endosome transport. J. Biol. Chem. 2004;279:4175–4179. doi: 10.1074/jbc.M308667200. [DOI] [PubMed] [Google Scholar]
- Conibear E., Stevens T. H. Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim. Biophys. Acta Mol. Cell Res. 1998;1404:211–230. doi: 10.1016/s0167-4889(98)00058-5. [DOI] [PubMed] [Google Scholar]
- Costaguta G., Stefan C. J., Bensen E. S., Emr S. D., Payne G. S. Yeast Gga coat proteins function with clathrin in Golgi to endosome transport. Mol. Biol. Cell. 2001;12:1885–1896. doi: 10.1091/mbc.12.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles C. R., Odorizzi G., Payne G. S., Emr S. D. The AP-3 Adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997a;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- Cowles C. R., Snyder W. B., Burd C. G., Emr S. D. Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J. 1997b;16:2769–2782. doi: 10.1093/emboj/16.10.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel W. W. Biostatistics: A Foundation for Analysis in the Health Sciences. Hoboken, NJ: Wiley; 2005. [Google Scholar]
- Dell’Angelica E. C., Puertollano R., Mullins C., Aguilar R. C., Vargas J. D., Hartnell L. M., Bonifacino J. S. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 2000;149:81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doray B., Ghosh P., Griffith J., Geuze H. J., Kornfeld S. Cooperation of GGAs and AP-1 in packaging MPRs at the trans-Golgi network. Science. 2002;297:1700–1703. doi: 10.1126/science.1075327. [DOI] [PubMed] [Google Scholar]
- Duncan M. C., Costaguta G., Payne G. S. Yeast epsin-related proteins required for Golgi-endosome traffic define a gamma-adaptin ear-binding motif. Nat. Cell Biol. 2003;5:77–81. doi: 10.1038/ncb901. [DOI] [PubMed] [Google Scholar]
- Duncan M. C., Payne G. S. ENTH/ANTH domains expand to the Golgi. Trends Cell Biol. 2003;13:211–215. doi: 10.1016/s0962-8924(03)00076-x. [DOI] [PubMed] [Google Scholar]
- Eugster A., Pecheur E. I., Michel F., Winsor B., Letourneur F., Friant S. Ent5p is required with Ent3p and Vps27p for ubiquitin-dependent protein sorting into the multivesicular body. Mol. Biol. Cell. 2004;15:3031–3041. doi: 10.1091/mbc.E03-11-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez G. E., Payne G. S. Laa1p, a conserved AP-1 accessory protein important for AP-1 localization in yeast. Mol. Biol. Cell. 2006;17:3304–3317. doi: 10.1091/mbc.E06-02-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford M.G.J., Mills I. G., Peter B. J., Vallis Y., Praefcke G.J.K., Evans P. R., McMahon H. T. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- Ford M.G.J., Pearse B.M.F., Higgins M. K., Vallis Y., Owen D. J., Gibson A., Hopkins C. R., Evans P. R., McMahon H. T. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- Friant S., Pecheur E. I., Eugster A., Michel F., Lefkir Y., Nourrisson D., Letourneur F. Ent3p is a PtdIns(3,5)P2 effector required for protein sorting to the multivesicular body. Dev. Cell. 2003;5:499–511. doi: 10.1016/s1534-5807(03)00238-7. [DOI] [PubMed] [Google Scholar]
- Fuller R. S., Sterne R. E., Thorner J. Enzymes required for yeast prohormone processing. Annu. Rev. Physiol. 1988;50:345–362. doi: 10.1146/annurev.ph.50.030188.002021. [DOI] [PubMed] [Google Scholar]
- Guarente L. Synthetic enhancement in gene interaction: a genetic tool come of age. Trends Genet. 1993;9:362–366. doi: 10.1016/0168-9525(93)90042-g. [DOI] [PubMed] [Google Scholar]
- Guex N., Peitsch M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Ha S.-A., Torabinejad J., DeWald D. B., Wenk M. R., Lucast L., De Camilli P., Newitt R. A., Aebersold R., Nothwehr S. F. The synaptojanin-like protein Inp53/Sjl3 functions with clathrin in a yeast TGN-to-endosome pathway distinct from the GGA protein-dependent pathway. Mol. Biol. Cell. 2003;14:1319–1333. doi: 10.1091/mbc.E02-10-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldwein E. E., Macia E., Wang J., Yin H. L., Kirchhausen T., Harrison S. C. Crystal structure of the clathrin adaptor protein 1 core. Proc. Natl. Acad. Sci. USA. 2004;101:14108–14113. doi: 10.1073/pnas.0406102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinners I., Tooze S. A. Changing directions: clathrin-mediated transport between the Golgi and endosomes. J. Cell Sci. 2003;116:763–771. doi: 10.1242/jcs.00270. [DOI] [PubMed] [Google Scholar]
- Hirst J., Lindsay M. R., Robinson M. S. Golgi-localized, gamma-ear-containing, ADP-ribosylation factor-binding proteins: roles of the different domains and comparison with AP-1 and clathrin. Mol. Biol. Cell. 2001;12:3573–3588. doi: 10.1091/mbc.12.11.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Lui W. W., Bright N. A., Totty N., Seaman M. N., Robinson M. S. A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J. Cell Biol. 2000;149:67–80. doi: 10.1083/jcb.149.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Miller S. E., Taylor M. J., von Mollard G. F., Robinson M. S. EpsinR is an adaptor for the SNARE protein Vti1b. Mol. Biol. Cell. 2004;15:5593–5602. doi: 10.1091/mbc.E04-06-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Motley A., Harasaki K., Peak Chew S. Y., Robinson M. S. EpsinR: an ENTH domain-containing protein that interacts with AP-1. Mol. Biol. Cell. 2003;14:625–641. doi: 10.1091/mbc.E02-09-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W.-K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O’Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Kalthoff C., Alves J., Urbanke C., Knorr R., Ungewickell E. J. Unusual structural organization of the endocytic proteins AP180 and epsin 1. J. Biol. Chem. 2002a;277:8209–8216. doi: 10.1074/jbc.M111587200. [DOI] [PubMed] [Google Scholar]
- Kalthoff C., Groos S., Kohl R., Mahrhold S., Ungewickell E. J. Clint: a novel clathrin-binding ENTH-domain protein at the Golgi. Mol. Biol. Cell. 2002b;13:4060–4073. doi: 10.1091/mbc.E02-03-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T. Three ways to make a vesicle. Nat. Rev. Mol. Cell Biol. 2000;1:187–198. doi: 10.1038/35043117. [DOI] [PubMed] [Google Scholar]
- Legendre-Guillemin V., Wasiak S., Hussain N. K., Angers A., McPherson P. S. ENTH/ANTH proteins and clathrin-mediated membrane budding. J. Cell Sci. 2004;117:9–18. doi: 10.1242/jcs.00928. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mills I. G., Praefcke G. J., Vallis Y., Peter B. J., Olesen L. E., Gallop J. L., Butler P. J., Evans P. R., McMahon H. T. EpsinR: an AP1/clathrin interacting protein involved in vesicle trafficking. J. Cell Biol. 2003;160:213–222. doi: 10.1083/jcb.200208023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S., Puertollano R., Kato Y., Bonifacino J. S., Hurley J. H. Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature. 2002;415:933–937. doi: 10.1038/415933a. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Hunter R., Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Mullins C., Bonifacino J. S. Structural requirements for function of yeast GGAs in vacuolar protein sorting, alpha-factor maturation, and interactions with clathrin. Mol. Cell. Biol. 2001;21:7981–7994. doi: 10.1128/MCB.21.23.7981-7994.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G., Babst M., Emr S. D. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Owen D. J., Collins B. M., Evans P. R. Adaptors for clathrin coats: structure and function. Annu. Rev. Cell Dev. Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- Panek H. R., Stepp J. D., Engle H. M., Marks K. M., Tan P. K., Lemmon S. K., Robinson L. C. Suppressors of YCK-encoded yeast casein kinase 1 deficiency define the four subunits of a novel clathrin AP-like complex. EMBO J. 1997;16:4194–4204. doi: 10.1093/emboj/16.14.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne G. S., Schekman R. Clathrin: a role in the intracellular retention of a Golgi membrane protein. Science. 1989;245:1358–1365. doi: 10.1126/science.2675311. [DOI] [PubMed] [Google Scholar]
- Peitsch M. C. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- Piper R. C., Bryant N. J., Stevens T. H. The membrane protein alkaline phosphatase is delivered to the vacuole by a route that is distinct from the VPS-dependent pathway. J. Cell Biol. 1997;138:531–545. doi: 10.1083/jcb.138.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R., Aguilar R. C., Gorshkova I., Crouch R. J., Bonifacino J. S. Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science. 2001;292:1712–1716. doi: 10.1126/science.1060750. [DOI] [PubMed] [Google Scholar]
- Puertollano R., Bonifacino J. S. Interactions of GGA3 with the ubiquitin sorting machinery. Nat. Cell Biol. 2004;6:244–251. doi: 10.1038/ncb1106. [DOI] [PubMed] [Google Scholar]
- Rasband W. S. Image J: U.S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/ 1997–2006 [Google Scholar]
- Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. S., Bonifacino J. S. Adaptor-related proteins. Curr. Opin. Cell Biol. 2001;13:444–453. doi: 10.1016/s0955-0674(00)00235-0. [DOI] [PubMed] [Google Scholar]
- Sage D., Neumann F. R., Hediger F., Gasser S. M., Unser M. Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics. IEEE Trans. Image Process. 2005;14:1372–1383. doi: 10.1109/tip.2005.852787. [DOI] [PubMed] [Google Scholar]
- Saint-Pol A. Clathrin adaptor epsinR is required for retrograde sorting on early endosomal membranes. Dev. Cell. 2004;6:525–538. doi: 10.1016/s1534-5807(04)00100-5. [DOI] [PubMed] [Google Scholar]
- Scott P. M. GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat. Cell Biol. 2004;6:252–259. doi: 10.1038/ncb1107. [DOI] [PubMed] [Google Scholar]
- Seeger M., Payne G. S. Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in Saccharomyces cerevisiae. J. Cell Biol. 1992;118:531–540. doi: 10.1083/jcb.118.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba Y., Katoh Y., Shiba T., Yoshino K., Takatsu H., Kobayashi H., Shin H.-W., Wakatsuki S., Nakayama K. GAT (GGA and Tom1) domain responsible for ubiquitin binding and ubiquitination. J. Biol. Chem. 2004;279:7105–7111. doi: 10.1074/jbc.M311702200. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahelin R. V., Long F., Peter B. J., Murray D., De Camilli P., McMahon H. T., Cho W. Contrasting membrane interaction mechanisms of AP180 N-terminal homology (ANTH) and epsin N-terminal homology (ENTH) domains. J. Biol. Chem. 2003;278:28993–28999. doi: 10.1074/jbc.M302865200. [DOI] [PubMed] [Google Scholar]
- Thierry A., Fairhead C., Dujon B. The complete sequence of the 8.2 kb segment left of MAT on chromosome III reveals five ORFs, including a gene for a yeast ribokinase. Yeast. 1990;6:521–534. doi: 10.1002/yea.320060609. [DOI] [PubMed] [Google Scholar]
- Traub L. M. Common principles in clathrin-mediated sorting at the Golgi and the plasma membrane. Biochim. Biophys. Acta Mol. Cell Res. 2005;1744:415–437. doi: 10.1016/j.bbamcr.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Vowels J. J., Payne G. S. A dileucine-like sorting signal directs transport into an AP-3-dependent, clathrin-independent pathway to the yeast vacuole. EMBO J. 1998;17:2482–2493. doi: 10.1093/emboj/17.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. J., Wang J., Sun H. Q., Martinez M., Sun Y. X., Macia E., Kirchhausen T., Albanesi J. P., Roth M. G., Yin H. L. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]
- Wasiak S., Legendre-Guillemin V., Puertollano R., Blondeau F., Girard M., de Heuvel E., Boismenu D., Bell A. W., Bonifacino J. S., McPherson P. S. Enthoprotin: a novel clathrin-associated protein identified through subcellular proteomics. J. Cell Biol. 2002;158:855–862. doi: 10.1083/jcb.200205078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung B. G., Phan H. L., Payne G. S. Adaptor complex-independent clathrin function in yeast. Mol. Biol. Cell. 1999;10:3643–3659. doi: 10.1091/mbc.10.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.