Abstract
Soybean has undergone several genetic bottlenecks. These include domestication in Asia to produce numerous Asian landraces, introduction of relatively few landraces to North America, and then selective breeding over the past 75 years. It is presumed that these three human-mediated events have reduced genetic diversity. We sequenced 111 fragments from 102 genes in four soybean populations representing the populations before and after genetic bottlenecks. We show that soybean has lost many rare sequence variants and has undergone numerous allele frequency changes throughout its history. Although soybean genetic diversity has been eroded by human selection after domestication, it is notable that modern cultivars have retained 72% of the sequence diversity present in the Asian landraces but lost 79% of rare alleles (frequency ≤0.10) found in the Asian landraces. Simulations indicated that the diversity lost through the genetic bottlenecks of introduction and plant breeding was mostly due to the small number of Asian introductions and not the artificial selection subsequently imposed by selective breeding. The bottleneck with the most impact was domestication; when the low sequence diversity present in the wild species was halved, 81% of the rare alleles were lost, and 60% of the genes exhibited evidence of significant allele frequency changes.
Keywords: artificial selection, crop domestication, genetic diversity, SNPs
The world's food supply depends on a small number of crop species. Because high-yielding cultivars dominate production but are relatively few in number and are genetically similar, genetic diversity in these crops is presumed to have declined to alarmingly low levels (1, 2). The reduction of genetic diversity does not bode well for future genetic gains in crop productivity and could result in broad susceptibility to newly emerging diseases or insect pests, thereby threatening long-term food and feed security (1, 3). The North American soybean crop accounts for 47% of world production (4) and may now be at a critically low level of diversity because of a series of genetic bottlenecks and intensive selection for enhanced agronomic performance. The perception that modern soybean cultivars are exceptionally uniform is supported by data based on coefficient of parentage analyses and surveys of germplasm for differences in genetic marker alleles (5).
Like many of the world's most important crops, soybean is an autogamous species. Inbreeding is predicted to decrease diversity, because purging of deleterious mutations also results in the loss of nondeleterious alleles at linked loci. In addition, evolutionary events such as domestication, founding events, and selection can affect the level of sequence variation within a crop. Domestication occurs when humans exert artificial selection on a wild species. Such selection, both positive and negative, over hundreds of generations results in the creation of a cultivated species. Founding events occur in crops when only a few individuals are used to introduce a crop into a new region or when breeders use only a few cultivars for all subsequent crop improvement. Domestication and founding events create genetic bottlenecks that can decrease genetic diversity, change allele frequencies, increase linkage disequilibrium (LD), and eliminate rare alleles in the resulting population (6). The magnitude of these effects will depend on the number of individuals involved, the selection intensity, and the duration of the genetic bottleneck.
Current evidence indicates that the cultivated soybean was domesticated from its annual wild relative [Glycine soja (Sieb. and Zucc.)] in China ≈5,000 years ago (5). The fraction of G. soja diversity retained through the domestication bottleneck is undefined. Domestication resulted in a multitude of localized Glycine max landraces. An estimated 45,000 of these unique Asian landraces have been collected and are maintained in G. max germplasm collections around the world. Despite this seemingly vast reservoir of genetic diversity, just 80 (<0.02%) of those landraces account for 99% of the collective parentage of North American soybean cultivars released between 1947 and 1988 (5). Even then, the contribution of each landrace is unequal, because just 17 of these 80 account for 86% of the collective parentage, with the remaining 63 landraces contributing <1% each (7). The process by which a few landraces, introduced from Asia to North America during the first half of the last century, became the genetic base of North American cultivars is often described as a diversity-reducing introduction bottleneck. As has occurred in other crop species, the intensive selection applied to this founding stock and to its descendants to create the elite soybean cultivars that growers use today is presumed to have led to additional losses in genetic diversity (1, 3).
Thus, the report of relatively low sequence diversity in cultivated soybean (8) relative to that in cultivated maize (an allogamous species) is not unexpected. A fundamental underlying question in the case of the autogamous soybean is the degree to which genetic diversity throughout the genome has been impacted by the domestication and introduction bottlenecks and by the subsequent intensive selection imposed by plant breeding. To answer this question, we evaluated DNA sequence variation within and among four populations of genotypes: elite North American soybean cultivars, Asian landrace founders of those elite cultivars, Asian landraces (with no known relationship to the founding stock), and accessions of the wild progenitor species G. soja. Our objective was to assess how genetic diversity in annual Glycine, as measured by DNA sequence variation, was altered by the human activities of domestication and subsequent founding and intensive breeding over the past 5,000 years. Through an understanding of DNA diversity in these four distinct populations, we were able to assess how soybean genome diversity was impacted by its transit through three genetic bottlenecks, domestication, introduction, and 75 years of intense breeding effort from a wild species to the elite cultivated crop species now grown widely in North America.
Results
Sequence Diversity in Wild and Cultivated Soybean.
We sequenced a total of 6.3 Mbp of DNA, which consisted of 111 fragments from 102 randomly selected genes (Table 3, which is published as supporting information on the PNAS web site) in 120 soybean genotypes. These genotypes were representative members of four distinct populations: (i) 25 diverse G. max cultivars developed in the 1980s, hereafter termed Elite Cultivars; (ii) 17 G. max Asian accessions that were the primary founders of the North American cultivars, hereafter termed North American Ancestors; (iii) 52 diverse G. max Asian accessions representing descendant products of domestication, hereafter termed Landraces; and (iv) 26 diverse accessions of G. soja (Table 4, which is published as supporting information on the PNAS web site). The sequence data set was mostly complete, with only 0.5% missing data (Data Set 1, which is published as supporting information on the PNAS web site). The amount of aligned sequence in these 120 soybean genotypes included 22 kb of coding sequence, 11 kb of 5′ and 3′ UTR sequence, 18 kb of intron sequence, and 2 kb of perigenic genomic sequence, totaling 53 kb (Data Set 2, which is published as supporting information on the PNAS web site). A total of 438 single base changes plus 58 single or multiple base insertion-deletions, all collectively referred to as SNPs, were identified. Of the 496 total SNPs, 84 were nonsynonymous (i.e., the encoded amino acid was altered by the SNP), whereas 59 were synonymous (i.e., the encoded amino acid was not altered); the other 353 SNPs occurred in noncoding DNA (Data Set 1).
Effect of Selection on Diversity.
Two common measures used to describe sequence variation or nucleotide diversity are π (pi), the expected heterozygosity per nucleotide site (9); and θ (theta), the number of polymorphic sites in a genotypic sample corrected for sample size (10). The Elite Cultivars had a π value of 0.00111 and a θ of 0.00083 (Table 1), which are very similar to earlier estimates of soybean diversity (8). When compared with other organisms, soybean nucleotide diversity is similar to the θ = 0.00053–0.00083 values reported for humans (11, 12) but lower than the θ = 0.0023 reported for Sorghum bicolor (13) and ≈1 order of magnitude lower than the θ = 0.00627 reported for modern maize inbred lines (14). It is generally presumed that intensive artificial selection imposed in modern plant-breeding programs over the last half century has reduced genetic diversity from that present in the founding stock. A comparison of the Elite Cultivars with the founding North American Ancestors indicated no significant reduction (P > 0.05) in nucleotide diversity. Indeed, the Elite Cultivars retained 97% (π) and 83% (θ) of the diversity present in the North American Ancestors. The Fst value between the North American Ancestors and the Elite Cultivars was 0.005, further supporting the lack of appreciable divergence between the two populations.
Table 1.
Nucleotide diversity per base pair ×103 in coding and noncoding regions within the four soybean populations
| Population | Coding sequence diversity |
Noncoding sequence diversity |
Total |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonymous |
Nonsynonymous |
Total coding |
UTR |
Intron |
Total noncoding |
|||||||||
| π | θ | π | θ | π | θ | π | θ | π | θ | π | θ | π | θ | |
| G. soja | 4.73a* | 3.15a | 0.96a | 1.20a | 1.05a | 1.63a | 3.18a | 3.24a | 2.34a | 2.65a | 2.76a | 3.06a | 2.17a | 2.35a |
| Landraces | 1.84b | 1.18b | 0.74ab | 0.72b | 0.70b | 0.81b | 2.02b | 1.43b | 1.55b | 1.35b | 1.77b | 1.36b | 1.43b | 1.15b |
| N. Am. Ancestors | 1.21b | 1.29b | 0.56b | 0.58b | 0.60b | 0.73b | 1.28b | 1.07b | 1.14c | 1.07bc | 1.36b | 1.16bc | 1.14c | 1.00bc |
| Elite Cultivars | 1.22b | 0.77b | 0.60b | 0.54b | 0.61b | 0.59b | 1.10b | 0.86b | 0.96c | 0.76c | 1.22c | 0.92c | 1.11c | 0.83c |
The UTR sequence includes 5′ and 3′ UTR. N. Am., North American.
*Values within a column followed by the same letter are not significantly different based on Duncan's multiple range test (P > 0.05).
We also developed an alternative statistical means of evaluating this apparent lack of reduction in diversity and population divergence. Extensive pedigree information is available for nearly all North American soybean cultivar releases (15). It is thus possible to compute the mathematical contribution of each of the 17 North American Ancestors to the parentage of the 25 Elite Cultivars. Using the pedigree-based contribution data, plus the SNP allele data observed in the 17 North American Ancestors, a theoretical Elite Cultivar sample was created to simulate, with no selection after the founding event, the Elite Cultivar sample. The π and θ of the simulated Elite Cultivar sample were 0.00107 and 0.00091, respectively, which were very similar and not significantly different (P > 0.05) from the π and θ of the actual Elite Cultivar sample.
Tajima's D is often used to determine allele frequency changes by comparing two populations before and after a genetic bottleneck (14). However, the large number of monomorphic fragments for which Tajima's D cannot be calculated makes comparisons difficult and hard to interpret in our populations. However, we did develop a permutation test where alleles were randomly assigned to the two populations based on the combined population allele frequency to determine whether the allele frequency change between populations was significant. Only seven of the genes containing one or more SNPs exhibited a significant allele frequency change (P < 0.05) between the North American Ancestors and the Elite Cultivars (Data Set 3, which is published as supporting information on the PNAS web site). Although increased LD is another hallmark of a severe genetic bottleneck, the extensive LD reported previously over a 2- to 3-cM region in soybean (8) makes LD information on a per gene basis unlikely to be informative. In fact, we found complete LD, as indicated by D′ = 1, within all but one gene in the North American Ancestors and the Elite Cultivars, two genes in the Landraces, and three genes in the G. soja (data not shown).
Founding Effect of Soybean Introduction to North America.
Although we have shown that long-term selective breeding in soybean after the establishment of a founder population has slightly decreased sequence diversity in the Elite Cultivars and changed only a few allele frequencies, the relatively few North American Ancestors found in the pedigrees of the Elite Cultivars represent a very limited sampling of the Asian landraces from which they derive (1, 3). Using only a limited number of introductions from the center of origin would be expected to impose an introduction bottleneck, thereby restricting the genetic variation available for the subsequent creation of elite North American cultivars.
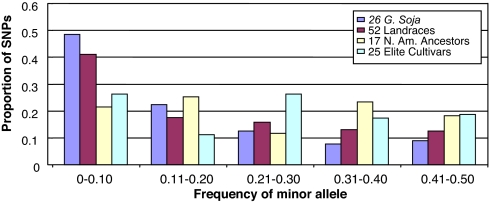
Overall, the founding stock of North American Ancestors retained 80% (π) and 87% (θ) of the nucleotide diversity of the Landraces (Table 1). These reductions in nucleotide diversity were not statistically significant (P > 0.05). Still, it is common for low-frequency alleles to be eliminated during a founding event. The proportion of SNPs with a minor allele frequency of ≤0.10 in the North American Ancestors was about half that of the Asian Landraces (Fig. 1). Of the 98 low-frequency SNP alleles in the Landrace population, 76 were not present in the North American Ancestors (Data Set 3). Thus, the impact of the limited number of founder genotypes was a 78% loss of the low-frequency alleles detected in the Landraces. Haplotype is a term used to designate a specific combination of linked alleles within a contiguous segment of DNA, and thus haplotype diversity (16) provides another measure of genetic diversity. Mean haplotype diversity in the 102 genes was 0.30 in the North American Ancestors, which was 94% of, and not significantly different (P > 0.05) from, the haplotype diversity in the Landraces. A total of 39 gene fragments were monomorphic in the North American Ancestors, whereas 14 of these 39 gene fragments were polymorphic in the Landraces (Table 2).
Fig. 1.
Distribution of the SNP minor allele frequencies for each of the four soybean populations.
Table 2.
Number of loci fixed within the four soybean populations
| G. soja | Landraces | N. Am. Ancestors | Elite Cultivars | |
|---|---|---|---|---|
| No. loci fixed | 7 | 25 | 39 | 40 |
| Percent loci fixed | 6.8 | 24.5 | 38.2 | 39.2 |
N. Am., North American.
The Landraces had nucleotide diversity values of π = 0.00143 and θ = 0.00115 (Table 1). The Landraces were somewhat, but not significantly, more diverse than the North American Ancestors, and the latter were slightly, but not significantly, more diverse than the Elite Cultivars. However, the cumulative effect of both bottlenecks was consequential, in that the genetic diversity of the Landraces was significantly greater (P < 0.05) than that of the Elite Cultivars. The Elite Cultivars retained 78% (π) and 72% (θ) of the diversity present in the Landraces. This is surprisingly close to the 77% diversity that maize elite inbred lines retained across 21 loci relative to the diversity found in maize Landraces (17). Low-frequency variants were only minimally impacted by the improvement bottleneck. Although only 22 of the 98 low-frequency SNPs present in the Landraces were present in the North American Ancestors, 21 remained in the Elite Cultivar population.
The cumulative effect of the genetic bottleneck of introduction to North America and subsequent selective breeding also had a significant effect on allele frequency changes. In 28 genes containing at least one SNP, a significant (P < 0.05) allele frequency change was observed (Data Set 1). A total of 15 gene fragments, variable in the Landraces, were fixed in the Elite Cultivars, because of selection or genetic drift (Table 2). However, the haplotype diversity for the Elite Cultivars (0.28), the North American Ancestors (0.30), and the Landraces (0.32) were not significantly different (P > 0.05).
Domestication Bottleneck.
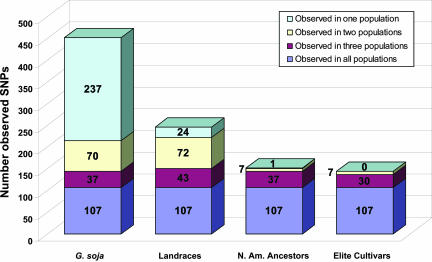
The domestication bottleneck is the most time-distant genetic constraint in the history of a crop and represents the first occurrence of human selection. We found that the Landraces retained 66% (π) and 49% (θ) of the nucleotide diversity found in G. soja (Table 1). The smallest reduction occurred in nonsynonymous sites, with the Landraces retaining 77% (π) of the diversity present in G. soja. The greatest number of allele frequency changes also occurred as a result of domestication with 61 genes having one or more SNPs with a significant (P < 0.05) allele frequency change (Data Set 3). Haplotype diversity was significantly lower in the Landraces (0.32) than in the wild soybean progenitor (0.51; P < 0.0001), which was consistent with the reduction in nucleotide diversity. The Landraces retained ≈63% of the haplotype diversity of G. soja. A total of 18 gene fragments that were variable in G. soja were fixed in the three G. max soybean populations (Table 5, which is published as supporting information on the PNAS web site). It is worth noting that in the comparison of unique SNPs among the four soybean populations, G. soja had the largest reservoir of unique sequence variants, with a total of 237 SNPs (Fig. 2). G. soja contained a total of 215 SNPs with a minor allele frequency ≤0.10, of which 175 were unique to G. soja and not found in any of the G. max populations (Data Set 3). The elimination of 81% of the low-frequency sequence variants in G. soja is consistent with the anticipated effects of a genetic bottleneck such as domestication.
Fig. 2.
Comparison of the number of unique and shared SNPs among four soybean populations.
Discussion
It has been well documented that selection targeted at individual loci will reduce genetic diversity within and around the selected loci (6). Conversely, it is assumed that selection in modern breeding programs acts simultaneously upon many loci controlling a variety of traits under selection. A logical conclusion would be that such selection would greatly reduce diversity throughout the genome, as has been predicted (3). This would be true for beneficial alleles present before the imposition of selection or de novo variation created during domestication and modern plant breeding. However, the lack of a statistically significant reduction in DNA sequence diversity and the lack of allele frequency changes between the Elite Cultivars vs. their North American Ancestors do not support this conclusion. Our data indicate that modern soybean breeding has minimally affected allelic structure of the genome compared with the other historical genetic bottlenecks. The only other major effect could be a significant increase of LD, which would be difficult to assess in the North American Ancestors, due to the small number of individuals in this group, which would inflate estimates of D′.
There are several factors that could be responsible for what seems to be a minimal amount of “genetic erosion” after the North American founding event. One explanation is that selection in modern soybean-breeding programs acts on only a small proportion of the genome. This selection would likely reduce the diversity and change allele frequencies in the sequence of DNA surrounding the loci that are targets of selection. Depending on how much LD is increased surrounding these loci, the effects of such selection might not extend far enough to affect overall genome diversity. Diversity loss also could have been mitigated by balancing selection and epistasis, given that the North American cultivated germplasm is comprised of 12 subpopulations (i.e., Maturity Groups 000–IX) adapted to a latitudinal gradient in photoperiod. Soybean breeders have had to develop cultivars for the specific photoperiod and production conditions encountered from Canada to Florida (5), and this would have maintained diversity for photoperiod response as well as combinations of numerous other region-specific biotic and abiotic stress resistance factors.
Besides intensive selection by modern plant breeding, the narrow genetic base is often cited as a contributing factor to low soybean diversity (1, 3). With only 17 North American Ancestors, of the many thousands that were available, contributing 86% of the parentage of modern cultivars, one would presume that only a small amount of diversity could have passed through the introduction bottleneck. Although the conventional population genetic measures of diversity (π and θ) suggested that the 17 North American Ancestors have almost as much diversity as the Asian Landraces, the 78% loss of rare alleles as a result of the introduction bottleneck agrees with theory that genetic bottlenecks can have little effect on diversity but still result in the loss of many rare alleles.
We found that 79% of the low-frequency sequence variants in the Landraces were not present in the Elite Cultivars. Thus, although there was a significant but relatively modest loss of diversity, as measured by θ, there was an extensive loss of rare sequence variants seen in the introduction bottleneck to North America. This suggests that the Elite Cultivars contain most of the common variation of the Asian Landrace collection and that variation useful for genetic improvement not present in the Elite Cultivars will be found at low frequency and require careful screening of the Asian Landrace collection. This conclusion is supported by the results of numerous attempts to discover traits of interest in the exotic soybean germplasm collection. For example, only 45 of the 9,153 genotypes screened for resistance to soybean cyst nematode (Heterodera glycine Ichinohe) race 3 possessed even moderate resistance (18). Van Duyn et al. (19) screened a large number of accessions in the U.S. Department of Agriculture, Agricultural Research Station National Soybean Germplasm Collection for resistance to foliar feeding insects, and found only three resistant genotypes. Chamberlain and Bernard (20) screened 2,060 Landrace accessions for brown stem rot (Phialophora gregata) resistance and found only one with resistance. Given the low frequency of useful variants for a given trait, it is unlikely that randomly adding even 100 new Asian Landraces (by recurrent introgressive matings) to the Elite germplasm pool would increase useful diversity beyond what is already present in the North American Elite Cultivars. Indeed, it is simply more effective to operate on a per need basis by screening the Asian Landrace germplasm to identify the few accessions that posses the desired variants, followed by the introgression of those variants into the Elite germplasm pool.
Overall, the effects of the domestication and introduction bottlenecks, combined with subsequent intensive selection in soybean, have resulted in sequence diversity losses in the Elite Cultivars vs. G. soja of 65, 49, and by 44%, as measured by θ, π, and haplotype diversity, respectively. Indeed, no allelic diversity was detected among the Elite Cultivars for ≈40% of the genes analyzed (Table 2). These bottlenecks have also significantly altered the allele frequencies of the genes we sampled. Wright et al. (14) sequenced 774 genes in a sampling of teosinte and modern maize inbred lines to determine the effects of domestication and modern breeding on diversity. They found that modern inbred maize lines have retained 57% of the diversity in teosinte. This reduction of 43% was due to the reduction in population size and selection during domestication and modern breeding, although the germplasm studied did not allow the authors to separate the effects of these two bottlenecks. Multiple studies have shown that >60% of the diversity is maintained after domestication of a number of grass species including: Zea mays, Sorghum bicolor, Orzya sativa, etc. (21). Buckler et al. (21) suggested that the large proportion of diversity maintained through the domestication bottleneck was due to the use of these crops as a basis for subsistence. This led to large quantities of these grass grains being grown during early cultivation, thereby maintaining large amounts of diversity. In soybean, the domestication bottleneck appears to have been somewhat more severe than the domestication bottlenecks of grasses. It is not known how many domestication events occurred in soybean (5). Our data do not reveal whether there was one or multiple domestications, but overall, the domestication bottleneck was responsible for a 50% reduction in diversity, the elimination of 81% of rare alleles present in G. soja, and a significant change in allele frequency in 60% of the genes analyzed.
Our data also indicate that G. soja, from which soybean was domesticated, has unusually low levels of sequence diversity for a wild crop species (π = 0.00217, θ = 0.00235). Loblolly pine (θ = 0.0041; ref. 22), Arabidopsis (θ = 0.0071; ref. 23), wild barley (θ = 0.0081; ref. 24), and teosinte (θ = 0.0109; ref. 14) have 2- to 5-fold greater nucleotide diversity than G. soja. Several factors may contribute to the lack of genetic diversity in G. soja, including effective population size, demography and autogamy (25).
The widely held assumption that intensive modern crop breeding, when applied to the descendants of a small number of founder introductions collected from the center of crop origin, has resulted in a drastic reduction of genome diversity (1, 3) does not appear to be valid in soybean. Instead, it appears that the low nucleotide diversity in modern elite soybean cultivars is mainly due to an unusually low level of genetic variability in the wild progenitor, G. soja, followed by a 50% loss of diversity during the domestication bottleneck. The most significant loss of diversity occurred during domestication and the introduction bottleneck where there was a large loss of rare alleles present in G. soja and the Asian Landraces. These rare alleles are likely to benefit future soybean improvement. Expansion of the currently low number of G. soja accessions available to North American soybean geneticists and breeders should be considered a high priority, given the great amount of diversity in terms of the presence of rare and unique alleles not found in the available G. max germplasm collections and the Elite cultivars.
Materials and Methods
Plant Materials.
The plant material included genotypes listed in Table 4. The first population consisted of 26 G. soja plant introductions from China, Korea, Taiwan, Russia, and Japan collected from 23–50°N and 106–140°E. This population of accessions was selected to sample all of the geographical areas within the range of G. soja. Origin and maturity group of accessions were the primary selection criteria. The population of Landraces consisted of 52 Asian plant introductions from China, Korea, and Japan collected from 22–50°N and 104–140°E. More accessions were included from China, where domesticated soybean originated. Cluster analysis has previously determined that Landraces from Japan and Korea are similar but less diverse than and distinct from those originating in China (26). In addition, it has been shown that there was more diversity between Landraces from different Chinese provinces than among Landraces from the same province. Similar diversity differences were not apparent among Landraces from different Korean or Japanese provinces (26). To adequately represent this diversity, at least two Landraces were selected from each Chinese province in which soybean was grown before scientific plant breeding. Landraces within provinces were selected for extremes in maturity groups available to include Landraces that represent diverse geographical regions and/or cropping systems within provinces and for phenotypic differences. Landraces from Korea and Japan were selected to represent the range of diversity in maturity groups and phenotypic descriptors. The 17 North American Ancestors are G. max accessions from Asia that are estimated to contribute at least 86% of the genes present in the gene pool of North American soybean cultivars (7). The Elite Cultivars consisted of 25 North American cultivars publicly released between 1977 and 1990, selected to maximize diversity based upon an analysis of coefficient of parentage by Gizlice et al. (27). Pure line seeds of all accessions were obtained from the U.S. Department of Agriculture Soybean Germplasm Collection (U.S. Department of Agriculture, Agriculture Research Station, University of Illinois, Urbana, IL). DNA was extracted from bulked leaf tissue of 8–10 G. soja plants or 30–50 G. max plants, as described by Keim et al. (28).
PCR and Sequencing.
PCR primers were originally designed by Zhu et al. (8) to 178 randomly selected genes and cDNAs for which there was no prior information on sequence diversity. Zhu et al. (8) successfully obtained sequence data from 116 of the 178 genes and cDNAs. We screened all 116 genes in the four populations and obtained sequence data for all or most of the 120 G. soja and G. max genotypes for 102 genes from 111 PCR fragments with sequence lengths from 400 to 600 bp listed in Table 3. Subsequently, 37 of the 102 genes and cDNAs have been genetically mapped with the populations described by Song et al. (29) and are distributed throughout 15 of the 20 linkage groups in soybean (Table 5). PCR primers and amplification conditions were described by Zhu et al. (8). Forward and reverse sequencing reactions were performed on an ABI 3700 or ABI 3730 using ABI Prism BigDye Terminator (Version 3.1) cycle sequencing (Applied Biosystems, Foster City, CA). Sequence data from each amplicon were aligned and analyzed with the standard DNA analysis software Phred/Phrap, and SNP detection was carried out with a machine learning algorithm based on PolyBayes SNP discovery software (30, 31). The resulting alignments and SNP predictions were visually verified by using the Consed viewer (32). Fragments were resequenced if there was any ambiguity as to which allele was present.
Sequence Analysis.
Small insertions and deletions were recorded as a single SNP and included in all SNP sequence analysis. Nucleotide diversity estimates for π (9) and θ (10) were calculated for each of the 102 genes within each population. Each π and θ matrix consists of nG × nP observations, where nG is the number of genes, and nP is the number of populations. The total variation in the matrix was partitioned by PROC ANOVA (SAS Institute, Cary, NC) into population, gene, and population × gene sources of variation. The population × gene mean square was used to test differences among populations.
The number of synonymous and nonsynonymous sites was measured by using DnaSP sequence polymorphism software (Version 3.5) (33). Fst was calculated as described by Hudson et al. (34). Tajima's D was calculated without an outgroup as described by Tajima (35). Haplotype diversity was calculated as described by Weir (16) as 1−Σ P2ij, where ΣPij is the frequency of the jth haplotype for ith locus summed across all haplotypes in the locus.
Simulation Procedures.
The percentage of unique contribution of each North American Ancestor to the Elite Cultivars was obtained from Carter et al. (15). The percentage of unique contribution was converted to the number of contributed loci (NCL; or fragments) to each Elite Cultivar based on a total of 102 loci. For example, if ancestor A has a percentage of unique contribution of 50% to Elite Cultivar 1, then ancestor A's NCL would be equal to 51 loci. In some instances, the total contribution from the 17 North American Ancestors to each Elite Cultivar was <102, because other North American Ancestors aside from the 17 included in this study were present in the pedigree. In these cases, one or more of the 52 Landraces was randomly chosen to represent the ancestral contribution not accounted for by the 17 North American Ancestors (i.e., the number of Landraces randomly selected for any given Elite Cultivar was equal to 102 loci minus the sum of loci contributed to that cultivar by one or more of the 17 North American Ancestors).
The genotype of each Elite Cultivar was simulated based upon the calculated number of contributed loci from each North American Ancestor (or Landrace). The SNP genotypes of Elite Cultivars were then extracted randomly from the corresponding contributors where all 102 alleles were represented only once within each simulated Elite Cultivar. For each permutation, the program generated a SNP genotype matrix with 102 unique loci × 25 Elite Cultivars. π and θ were calculated for each locus at each permutation. A total of 5,000 permutations were performed; the means for each locus, together with the observed π and θ, were used for ANOVA and to test whether there was a significant difference between the observed and simulated values of π and θ of the Elite Cultivars.
Allele frequency differences of each SNP were calculated between G. soja and Landraces, Landraces and North American Ancestors, North American Ancestors and Elite Cultivars, and Landraces and Elite Cultivar populations. The frequency difference was defined as p12i = x1i/n1i−x2i/n2i, where x1i and x2i are the number of accessions with a given allele at a SNP locus i in populations 1 and 2, respectively; and n1i and n2i are the number of accessions in populations 1 and 2, respectively. The significance of the observed frequency differences was tested by permutation. First, the total number of accessions with the given SNP allele in the two populations being compared was counted (n12 = x1i + x2i); a total of n12 accessions in the two populations were randomly assigned the first allele, and the remaining accessions were assigned the second allele; and the permutated frequency difference under the assumption of no frequency difference between populations was calculated and compared with the observed frequency difference. The process was repeated 10,000 times for each locus. The measure of significance (p) is given by the ratio (N/10,000), where N is the number of times the expected absolute frequency difference between the populations was exceeded by the observed absolute frequency difference.
Supplementary Material
Acknowledgments
We thank Tina Sphon, Tad Sonstegard, and the Bovine Functional Genomics Laboratory Animal and Natural Resources Institute (Beltsville Agricultural Research Center East DNA Sequencing Facility) for assistance with the acquisition of sequence data. We thank Charles Fenster, William Kenworthy, Marla McIntosh, and two anonymous reviewers for helpful comments on this study. This work was supported in part by United Soybean Board Grants 4212 and 5212.
Abbreviation
- LD
linkage disequilibrium.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.National Research Council; Committee on Genetic Vulnerability of Major Crops. Genetic Vulnerability of Major Crops. Washington, DC: Natl Acad Sci; 1972. [Google Scholar]
- 2.Esquinas-Alcazar J. Nat Rev Genet. 2005;6:946. doi: 10.1038/nrg1729. [DOI] [PubMed] [Google Scholar]
- 3.Tanksley SD, McCouch SR. Science. 1997;277:1063–1066. doi: 10.1126/science.277.5329.1063. [DOI] [PubMed] [Google Scholar]
- 4.Wilcox JR. In: Soybeans: Improvement, Production, and Uses. Boerma HR, Specht JE, editors. Madison, WI: Am Soc of Agronomy, Crop Sci Soc of Am, Soil Sci Soc of Am; 2004. pp. 1–14. Vol Agronomy, no 16. [Google Scholar]
- 5.Carter TE, Nelson R, Sneller CH, Cui Z. In: Soybeans: Improvement, Production, and Uses. Boerma HR, Specht JE, editors. Madison, WI: Am Soc of Agronomy, Crop Sci Soc of Am, Soil Sci Soc of Am; 2004. pp. 303–416. Vol Agronomy, no 16. [Google Scholar]
- 6.Halliburton R. Upper Saddle River, NJ: Pearson/Prentice– Hall; 2004. Introduction to Population Genetics. [Google Scholar]
- 7.Gizlice Z, Carter TE, Jr, Burton JW. Crop Sci. 1994;34:1143–1151. [Google Scholar]
- 8.Zhu YL, Song QJ, Hyten DL, Van Tassell CP, Matukumalli LK, Grimm DR, Hyatt SM, Fickus EW, Young ND, Cregan PB. Genetics. 2003;163:1123–1134. doi: 10.1093/genetics/163.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tajima F. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watterson GA. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 11.Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A. Nat Genet. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- 12.Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, et al. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 13.Hamblin MT, Mitchell SE, White GM, Gallego J, Kukatla R, Wing RA, Paterson AH, Kresovich S. Genetics. 2004;167:471–483. doi: 10.1534/genetics.167.1.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD, Gaut BS. Science. 2005;308:1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- 15.Carter TE, Jr, Gizlice Z, Burton JW. Coefficient of Parentage and Genetic Similarity Estimates for 258 North American Soybean Cultivars Released byPublic Agencies During 1954–88. Washington, DC: US Government Printing Office; 1993. USDA Tech Bull 1814. [Google Scholar]
- 16.Weir BS. Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sunderland, MA: Sinauer; 1996. [Google Scholar]
- 17.Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS. Proc Natl Acad Sci USA. 2001;98:9161–9166. doi: 10.1073/pnas.151244298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anand SC, Gallo KM. Plant Dis. 1984;68:593–595. [Google Scholar]
- 19.Van Duyn JW, Turnipseed SG, Maxwell JD. Crop Sci. 1971;11:572–573. [Google Scholar]
- 20.Chamberlain DW, Bernard RL. Crop Sci. 1968;8:728–729. [Google Scholar]
- 21.Buckler ES IV, Thornsberry JM, Kresovich S. Genet Res. 2001;77:213–218. doi: 10.1017/s0016672301005158. [DOI] [PubMed] [Google Scholar]
- 22.Brown GR, Gill GP, Kuntz RJ, Langley CH, Neale DB. Proc Natl Acad Sci USA. 2004;101:15255–15260. doi: 10.1073/pnas.0404231101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmid KJ, Ramos-Onsins S, Ringys-Beckstein H, Weisshaar B, Mitchell-Olds T. Genetics. 2005;169:1601–1615. doi: 10.1534/genetics.104.033795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrell PL, Toleno DM, Lundy KE, Clegg MT. Proc Natl Acad Sci USA. 2005;102:2442–2447. doi: 10.1073/pnas.0409804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright SI, Gaut BS. Mol Biol Evol. 2005;22:506–519. doi: 10.1093/molbev/msi035. [DOI] [PubMed] [Google Scholar]
- 26.Li Z, Nelson RL. Crop Sci. 2001;41:1337–1347. [Google Scholar]
- 27.Gizlice Z, Carter TE, Jr, Gerig TM, Burton JW. Crop Sci. 1996;36:753–765. [Google Scholar]
- 28.Keim P, Olson TC, Shoemaker RC. Soybean Genet Newsl. 1988;15:150–152. [Google Scholar]
- 29.Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB. Theor Appl Genet. 2004;109:122–128. doi: 10.1007/s00122-004-1602-3. [DOI] [PubMed] [Google Scholar]
- 30.Marth GT, Korf I, Yandell MD, Yeh RT, Gu Z, Zakeri H, Stitziel NO, Hillier L, Kwok PY, Gish WR. Nat Genet. 1999;23:452–456. doi: 10.1038/70570. [DOI] [PubMed] [Google Scholar]
- 31.Matukumalli L, Grefenstette J, Hyten D, Choi I-Y, Cregan P, Van Tassell C. BMC Bioinformatics. 2006;7:4. doi: 10.1186/1471-2105-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordon D, Abajian C, Green P. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 33.Rozas J, Rozas R. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- 34.Hudson RR, Slatkin M, Maddison WP. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.