Abstract
The Ll.LtrB intron, from the low G+C gram-positive bacterium Lactococcus lactis, was the first bacterial group II intron shown to splice and mobilize in vivo. The detailed retrohoming and retrotransposition pathways of Ll.LtrB were studied in both L. lactis and Escherichia coli. This bacterial retroelement has many features that would make it a good gene delivery vector. Here we report that the mobility efficiency of Ll.LtrB expressing LtrA in trans is only slightly affected by the insertion of fragments <100 nucleotides within the loop region of domain IV. In contrast, Ll.LtrB mobility efficiency is drastically decreased by the insertion of foreign sequences >1 kb. We demonstrate that the inhibitory effect caused by the addition of expression cassettes on Ll.LtrB mobility efficiency is not sequence specific, and not due to the expression, or the toxicity, of the cargo genes. Using genetic screens, we demonstrate that in order to maintain intron mobility, the loop region of domain IV, more specifically domain IVb, is by far the best region to insert foreign sequences within Ll.LtrB. Poisoned primer extension and Northern blot analyses reveal that Ll.LtrB constructs harboring cargo sequences splice less efficiently, and show a significant reduction in lariat accumulation in L. lactis. This suggests that cargo-containing Ll.LtrB variants are less stable. These results reveal the potential, yet limitations, of the Ll.LtrB group II intron to be used as a gene delivery vector, and validate the random insertion approach described in this study to create cargo-containing Ll.LtrB variants that are mobile.
Keywords: group II intron, Lactococcus lactis, Ll.LtrB, splicing, gene delivery, Tn5
INTRODUCTION
Group II introns are large ribozymes that self-splice from pre-mRNAs while simultaneously ligating their flanking exons (Lambowitz and Zimmerly 2004). They are found in eubacteria, archaea, and eukaryotic organelles derived from bacteria, such as fungal and plant mitochondria, and plant chloroplasts (Belfort et al. 2002; Lambowitz and Zimmerly 2004). These RNA enzymes are not conserved at the level of the primary sequence, but can be folded in a very characteristic secondary structure. This secondary structure consists of six domains (DI–DVI) radiating from a central wheel (Belfort et al. 2002; Lambowitz and Zimmerly 2004). Domains V and VI are the most conserved domains of group II introns and are part of the catalytic core (de Lencastre et al. 2005). In contrast, domain IV is dispensable for splicing and varies greatly in length since several group II introns harbor an open reading frame (ORF) in the loop region of this domain. When present within group II introns, ORFs are always found in the loop region of domain IV. Therefore, domain IV is believed to lie outside the catalytic core of these ribozymes (Belfort et al. 2002; Lambowitz and Zimmerly 2004). Some group II introns that harbor an intron-encoded protein are also mobile retroelements that invade new DNA sites in a duplicative process using an RNA intermediate (Belfort et al. 2002; Lambowitz and Zimmerly 2004). These retroelements can invade both cognate intronless alleles (homing sites; HSs) and non-homologous, or ectopic, sites by retrohoming and retrotransposition, respectively (Belfort et al. 2002; Lambowitz and Zimmerly 2004).
The Ll.LtrB group II intron from the low G+C gram-positive bacterium Lactococcus lactis interrupts a putative relaxase gene (ltrB), which is present on three conjugative elements: the pRS01 (48.4 kb) (Mills et al. 1996) and pAH90 (26.5 kb) (O'Sullivan et al. 2001) plasmids, and an integrative and conjugative element called the sex factor (50 kb) (Shearman et al. 1996). L. lactis is a mesophilic lactic acid bacterium (LAB) that is widely used in the dairy industry, for instance, in the manufacturing of cheese. Ll.LtrB was the first bacterial group II intron that was shown to splice (Mills et al. 1996; Shearman et al. 1996) and invade new sites in vivo (Mills et al. 1997). The detailed retrohoming and retrotransposition pathways of Ll.LtrB were studied in L. lactis and Escherichia coli (Cousineau et al. 1998, 2000; Ichiyanagi et al. 2002, 2003; Coros et al. 2005; Smith et al. 2005). Furthermore, conjugative transfer of Ll.LtrB-harboring elements was shown to support dissemination of this intron between L. lactis strains by both retrohoming and retrotransposition (Belhocine et al. 2004, 2005). Lateral or horizontal transfer of the Ll.LtrB intron from L. lactis to Enterococcus faecalis was also demonstrated by retrohoming following its transfer by conjugation (Belhocine et al. 2004).
Ll.LtrB (2492 nucleotides [nt]) harbors, in domain IV, a 599 amino acid ORF called LtrA. This multifunctional intron-encoded protein, with reverse transcriptase, maturase, and endonuclease activities, is essential for Ll.LtrB mobility by retrohoming (Cousineau et al. 1998). Following translation, LtrA binds specifically to the Ll.LtrB intron as a dimer within the pre-mRNA (Saldanha et al. 1999). Through its maturase function, LtrA promotes self-splicing of Ll.LtrB and concurrent ligation of the flanking exons. After Ll.LtrB splicing, the LtrA dimer remains associated with the excised intron lariat as a ribonucleoprotein particle (RNP; intron RNA lariat+two LtrA proteins). Upon RNP recognition of the homing site, mainly through base-pairing interactions, the intron RNA reverse splices into the sense strand of its double-stranded DNA target. Then, the antisense strand is nicked 9 nt downstream from the intron insertion site by the endonuclease domain of LtrA. Next, using the free 3′ OH generated by the endonuclease cleavage as a primer, LtrA makes a cDNA copy of the intron RNA by a process termed target-primed reverse transcription (TPRT). The final steps of the Ll.LtrB retrohoming pathway are independent of the RecA-dependent homologous recombination pathway (Cousineau et al. 1998) and were shown to involve RNase H from the host along with different DNA repair enzymes (Smith et al. 2005).
The Ll.LtrB group II intron, essentially a bacterial retrotransposon, has several interesting features that make it a potential gene delivery vector. First, this mobile element is highly specific, recognizing a 35-nt sequence (Guo et al. 2000; Mohr et al. 2000; Singh and Lambowitz 2001). Second, since the rules of Ll.LtrB target recognition are relatively well known, mostly involving a series of Watson–Crick base-pair interactions, Ll.LtrB can be retargeted to insert specifically into almost any sequence (Frazier et al. 2003; Zhong et al. 2003; Perutka et al. 2004, Jones et al. 2005; Yao et al. 2005, 2006). Third, the very high retrohoming efficiency of Ll.LtrB in L. lactis enables easy identification of chromosomal insertions without using selection markers. Fourth, the Ll.LtrB retrohoming pathway is RecA-independent (Cousineau et al. 1998); thus, it does not require a proficient homologous recombination system to support gene delivery. Fifth, Ll.LtrB inserts into DNA without exchanging flanking markers (RecA independent), therefore limiting the potential undesirable changes to both the target sequence and the neighboring genes (Cousineau et al. 1998). Moreover, in comparison to other retroviral vectors, Ll.LtrB does not harbor promoter sequences, again limiting its effect on the expression of genes flanking intron insertion. Sixth, Ll.LtrB was shown to be active in a wide range of organisms from bacteria (Gram-negative and Gram-positive) to human cells (Cousineau et al. 1998; Guo et al. 2000). Finally, Ll.LtrB was previously shown to tolerate the addition of foreign sequences within domain IV (Matsuura et al. 1997; Cousineau et al. 1998, 2000). Two markers, one for retromobility (td group I intron) and one for selection (kanamycin), were inserted within domain IV immediately downstream of the ltrA gene. This Ll.LtrB variant was shown to still be mobile, albeit at a reduced rate. Therefore, it was suggested that this bacterial retroelement might be exploited to deliver foreign sequences within genomes (Matsuura et al. 1997; Cousineau et al. 1998). We thus decided to investigate the potential of Ll.LtrB as a gene delivery vector by analyzing the ability of this intron to carry and deliver foreign sequences to its homing site.
Here we show, using an intron expressing LtrA in trans, that the mobility efficiency of Ll.LtrB is only slightly affected by the insertion of fragments <100 nt within the loop region of domain IV. In contrast, Ll.LtrB mobility efficiency is drastically decreased by the insertion of foreign sequences >1 kb. We demonstrate that the inhibitory effect caused by the addition of different expression cassettes on Ll.LtrB mobility efficiency is not sequence specific and is not due to the expression, or the toxicity, of the cargo genes. Using Tn5-based genetic screens, we demonstrate that in order to keep mobility, the loop region of domain IV, more specifically domain IVb, is by far the best region in which to insert foreign sequences within Ll.LtrB. Poisoned primer extension and Northern blot analyses reveal that Ll.LtrB constructs harboring cargo sequence splice less efficiently and show a significant reduction in lariat accumulation in L. lactis. This suggests that cargo-containing Ll.LtrB variants are less stable. These results reveal the potential, and limitations, of the Ll.LtrB group II intron to be used as a gene delivery vector, and validate the random insertion approach described in this study to create proficient cargo-containing Ll.LtrB variants.
RESULTS
Ll.LtrB expressing LtrA in trans has a significantly higher mobility efficiency than the wild-type intron in L. lactis
Since Ll.LtrB is a potential gene delivery vector, we first wanted to identify the most efficient intron variant for this purpose. Our goal was to improve its basal mobility efficiency in order to start with the most efficient intron possible. We almost completely removed the ORF, coding for the LtrA protein, by deleting 1596 nt from the loop region of domain IV (Guo et al. 2000). The resulting intron is 64% smaller but still retains 117 and 87 nt from the 5′ and 3′ end of ltrA, respectively (Figs. 1A, donor plasmid; 4). Since LtrA is essential for Ll.LtrB mobility (Cousineau et al. 1998), the entire ltrA gene was cloned upstream from the intron, between the nisin-inducible promoter and exon I (Fig. 1A, donor plasmid). We hypothesized that a significantly smaller intron would be able to accommodate larger expression cassettes while keeping its mobility proficiency, in comparison to the wild-type intron. Moreover, a similar Ll.LtrB variant was previously created in which ltrA was cloned downstream of the intron, after exon II rather than before exon I (Guo et al. 2000). This variant was shown to have a significant increase in mobility efficiency when compared with wild-type Ll.LtrB in E. coli (Guo et al. 2000). The enhanced mobility efficiency of this construct is thought to be due to a reduction in its susceptibility to host RNAses (Matsuura et al. 1997; Guo et al. 2000). However, the insertion of large expression cassettes within this intron variant would position ltrA quite far from the nisin-inducible promoter. This could potentially reduce levels of LtrA expression and affect Ll.LtrB splicing and mobility efficiency. Therefore, we decided to clone ltrA upstream rather than downstream from the intron to insure that LtrA expression is optimal in all Ll.LtrB variants.
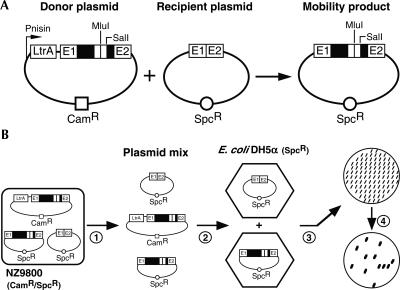
FIGURE 1.
Ll.LtrB mobility assay and mobility efficiency scoring. (A) Two-plasmid Ll.LtrB mobility assay. The assay consists of co-transforming both intron donor and recipient plasmids into NZ9800 L. lactis cells and monitoring the appearance of mobility products. The intron donor plasmid (pLE1-based, CamR) harbors various Ll.LtrB constructs, expressing LtrA in trans, under the control of the nisin-inducible promoter (Pnis). The recipient plasmid (pMNHS, SpcR) contains the intron homing site (E1/E2). Upon nisin induction, the Ll.LtrB intron can move from the donor to the recipient plasmid, generating mobility products. The positions of the engineered SalI and MluI restriction sites, present within the intron, are denoted. The remaining 5′ and 3′ ltrA sequences within Ll.LtrB are represented by empty boxes. (B) Scoring of intron mobility efficiency by colony patch hybridization. Plasmid mixes (donor plasmid, recipient plasmid, and mobility products) from independent mobility assays (panel A) are recovered (step 1) and transformed into E. coli DH5α cells (LB/Spc) (step 2). For each mobility assay, 100 colonies are patched on an LB/Spc plate (step 3), transferred onto a nylon membrane, and hybridized with an intron-specific 32P-labeled probe (step 4). Therefore, the ratio of recipient plasmids that received the intron is calculated.
Using a two-plasmid mobility assay (Fig. 1A), we assessed the mobility efficiency of the Ll.LtrB variant that expresses LtrA in trans in L. lactis, along with other variants as controls (Fig. 2, numbers 1–4). The assay consists of co-transforming the NZ9800 L. lactis strain with a donor plasmid (pLE-based), where LtrA and the intron are under the control of the nisin promoter (Kuipers et al. 1993), and a recipient plasmid (pMNHS) that harbors the intron recognition site (homing site, HS) (Mills et al. 1997; Cousineau et al. 1998). After Ll.LtrB expression, upon nisin induction, the plasmid mix (donor plasmid, recipient plasmid, mobility products) is recovered from L. lactis and transformed into E. coli DH5α cells, selecting for cells that received the recipient plasmid (LB/Spc). One hundred colonies are then patched onto an LB/Spc plate, transferred to a nylon membrane, and hybridized with an intron-specific probe. Therefore, this procedure reveals the percentage of recipient plasmids that were invaded by Ll.LtrB and the mobility efficiency of the intron (Fig. 1B) (Cousineau et al. 1998; Belhocine et al. 2004, 2005).
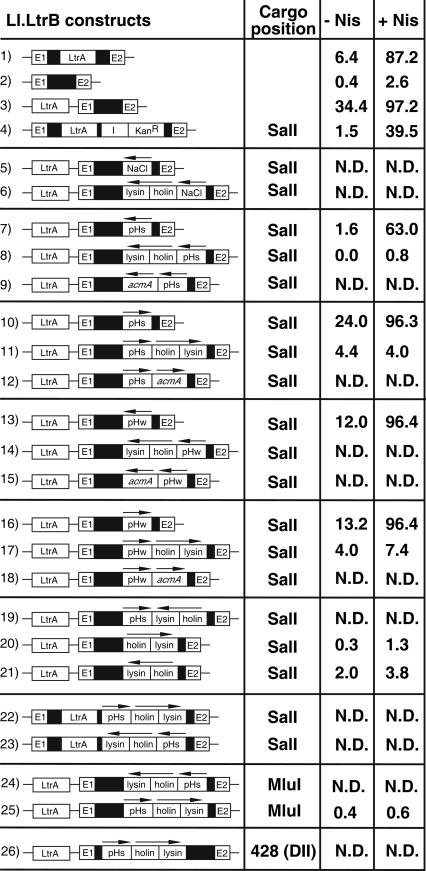
FIGURE 2.
Mobility efficiencies of different Ll.LtrB variants. Mobility efficiencies of different Ll.LtrB constructs were assessed either in the presence (+ Nis) or absence (− Nis) of nisin induction. Two-plasmid mobility assays (Fig. 1A) were performed for all Ll.LtrB constructs presented. However, in cases where the mobility products were not detectable on an agarose gel, the precise mobility efficiency was not scored (N.D. = not determined). Mobility efficiencies represent the average of four or five independent mobility assays. For clarity, the remaining 5′ and 3′ ltrA sequences are not denoted. (Arrows) Orientation of promoters and cargo genes. (I) td group I intron from bacteriophage T4 (Belfort et al. 1987; Cousineau et al. 1998), (DII) Ll.LtrB domain II.
By means of this mobility assay, we demonstrate that the Ll.LtrB construct, which expresses the LtrA protein in trans, is more proficient than the wild-type Ll.LtrB intron in L. lactis (97.2% versus 87.2%) (Fig. 2, cf. numbers 3 and 1). Also, we confirmed that LtrA is essential for Ll.LtrB mobility (87.2% versus 2.6%) (Fig. 2, cf. numbers 1 and 2). We believe that this 10% increase in mobility efficiency, between the construct expressing LtrA in trans and wild-type Ll.LtrB, is in reality more significant. For some highly proficient Ll.LtrB constructs, our two-plasmid mobility assay appears to be saturated in conditions of nisin induction (Fig. 2, numbers 3,10,13,16), therefore resulting in the underestimation of the actual increase in Ll.LtrB mobility. For example, the mobility efficiency of Ll.LtrB expressing LtrA in trans is 5.4-fold higher than wild-type Ll.LtrB in the absence of nisin (Fig. 2, 34.4% versus 6.4%). This indicates that the increase in mobility efficiency of the Ll.LtrB construct that expresses LtrA in trans in comparison to the wild-type intron is significant, and confirms that this new Ll.LtrB variant is a better vector to be used for gene delivery.
Ll.LtrB mobility is almost unaffected by small foreign sequences inserted in the loop region of domain IV
Two versions of a pH/growth-phase-inducible promoter (pHs, strong; pHw, weak) (Madsen et al. 1999) and a chloride (NaCl)-sensitive promoter (Sanders et al. 1997) were inserted into the loop region of domain IV of the Ll.LtrB construct expressing LtrA in trans (Fig. 2, numbers 5,7,10,13,16). These promoters were cloned at the engineered SalI site located immediately downstream from the LtrA stop codon (Figs. 1A, 4). The short fragments corresponding to the strong (79-nt) and weak (64-nt) pH promoters were introduced in both the sense and antisense orientations with reference to the intron orientation. On the other hand, the large chloride-inducible promoter (1251 nt) was cloned in the antisense orientation only. Three of the four pH promoters did not affect Ll.LtrB mobility upon nisin induction (Fig. 2, cf. numbers 3 and 10,13,16), while the pHs promoter in the antisense orientation reduced Ll.LtrB mobility by ∼34% (Fig. 2, cf. number 3 and 7). Furthermore, the three Ll.LtrB constructs unaffected following nisin induction were nevertheless somewhat affected by the presence of their cargos. Significant decreases of 10.4%, 22.4%, and 21.2% of their mobility efficiencies can be noticed in uninduced conditions (Fig. 2, cf. numbers 3 and 10,13,16). In contrast, the Ll.LtrB variant carrying the chloride-inducible promoter was found to be completely immobile in both induced and uninduced conditions (Fig. 2, number 5). These results suggest that the presence of large inserts within Ll.LtrB can significantly hinder Ll.LtrB mobility efficiency, while foreign sequences <100 nt have less important effects.
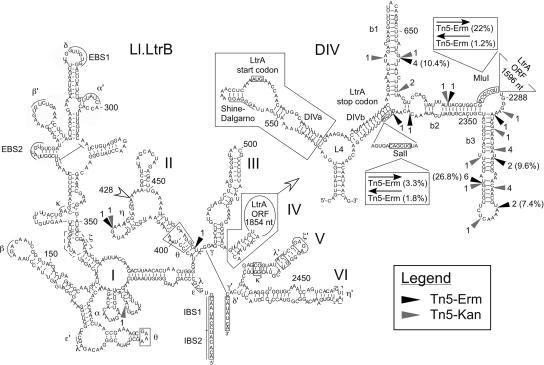
FIGURE 4.
Tn5 insertion sites within Ll.LtrB. The Tn5-Erm (black arrowhead) and Tn5-Kan (gray arrowhead) insertion sites and the frequency of occurrence are mapped on the Ll.LtrB secondary structure. The location of the pHs/holin/lysin expression cassette, within domain II at position 428, is also denoted (open arrowhead). The mobility efficiencies of some transposon-containing Ll.LtrB introns are shown in parentheses. The six domains of Ll.LtrB (I-VI) are indicated, and the detailed secondary structure of domain IV (DIV; DIVa and DIVb) is presented in the upper right corner. The engineered SalI and MluI restriction sites, the LtrA start and stop codons, and the exons are boxed. The LtrA Shine-Dalgarno sequence is underlined. The LtrA binding structure (DIVa) is also boxed. Greek letters correspond to previously identify long-range tertiary interactions between different regions of Ll.LtrB. Please note that during insertion, Tn5 creates a 9-nt direct repeat; the 9 nt located upstream from each insertion site are thus also present downstream form the inserted transposons (Goryshin and Reznikoff 1998). (EBS1 and EBS2) Exon binding sites 1 and 2, (IBS1 and IBS2) intron binding sites 1 and 2.
Insertion of long foreign sequences within Ll.LtrB significantly reduces mobility by retrohoming
The holin/lysin cassette (1180 nt) from the L. lactis bacteriophage r1t (van Sinderen et al. 1996), and the L. lactis autolysis gene acmA (1390 nt) (Buist et al. 1997), were both cloned downstream of the four versions of the pH promoter within Ll.LtrB. The importance of these genes in the induction and control of L. lactis cell lysis during cheese ripening was previously demonstrated and discussed (de Ruyter et al. 1997; Sanders et al. 1997). In all cases, the insertion of these genes within Ll.LtrB completely abolished mobility in both induced and uninduced conditions (Fig. 2, numbers 8,9,11,12,14,15,17,18). These results suggest that the hindrance of Ll.LtrB mobility caused by some inserts is not sequence specific, but rather is size related. Different long inserts (1.0–1.5 kb; chloride-inducible promoter, holin/lysin, acmA) present in both orientations, which lead to completely different RNA sequences following Ll.LtrB transcription, all have severe effects on intron mobility efficiency. Furthermore, this size limitation for fore-ign sequences inserted within Ll.LtrB is supported by the decrease in mobility efficiency observed with the wild-type intron harboring the td group I intron from bacteriophage T4, as well as a gene conferring kanamycin resistance (Fig. 2, cf. numbers 1 and 4). The insertion of these two fragments (1500 nt) at the engineered SalI site within domain IV of the intron caused a 48% decrease in its mobility efficiency, supporting the hypothesis that foreign sequences >1 kb can significantly affect Ll.LtrB mobility efficiency.
Reduction in Ll.LtrB mobility efficiency is not caused by the expression, or toxicity, of the cargo gene
The presence of different expression cassettes in the sense orientation within Ll.LtrB could possibly recruit the translation machinery to the unspliced transcripts and/or the spliced intron lariats. This can have drastic effects on splicing and/or mobility efficiency of these intron variants. If associated to unspliced transcripts, the translation machinery would likely prevent proper folding of the intron and interfere with splicing. Moreover, if the spliced intron is recognized by ribosomes, the translation machinery would sequester the intron lariat and greatly hinder reverse splicing of the intron into the homing site. Additionally, the promoter of the expression cassette may be recognized and initiate transcription from within the Ll.LtrB intron. This would decrease the overall transcription level of the intron. Therefore, these situations could ultimately have severe effects on the mobility efficiency of Ll.LtrB constructs that carry expression cassettes.
Some Ll.LtrB variants were constructed to address the effect of gene expression from within the intron on its mobility proficiency. The holin/lysin genes were cloned in the reverse orientation downstream from the sense pHs promoter within Ll.LtrB (Fig. 2, number 19). The mobility efficiency of this Ll.LtrB construct, which does not express the holin/lysin genes, is as severely affected as the other expression cassettes previously studied (Fig. 2). In addition, the holin/lysin genes were inserted at the same site (SalI) within Ll.LtrB in both orientations, but without any promoter. These Ll.LtrB constructs showed a similar reduction in their mobility efficiencies (Fig. 2, numbers 20,21). Therefore, this suggests that expression of the cargo gene from within the intron is unlikely to be the cause of the reduction in Ll.LtrB mobility efficiency. These results also rule out the possibility that the toxicity of the cargo genes may indirectly affect Ll.LtrB mobility efficiency, since the holin/lysin genes, which induce L. lactis lysis, are not expressed from the two Ll.LtrB variants that harbor these genes in the antisense orientation (Fig. 2, numbers 19,21).
Furthermore, a similar reduction in Ll.LtrB mobility efficiency was observed when the pHs/holin/lysin expression cassette was cloned, in both orientations, within the wild-type intron at the same position (SalI) (Fig. 2, numbers 22,23). This suggests that the effect of large inserts on Ll.LtrB mobility is not specific to the construct expressing LtrA in trans, since the same effect is seen with the wild-type Ll.LtrB intron. In addition, the same expression cassette (pHs/holin/lysin) was inserted within Ll.LtrB expressing LtrA in trans at a second restriction site (MluI), also engineered in the loop region of domain IV (Figs. 1A, 4). The mobility efficiencies of these Ll.LtrB constructs were also severely impaired, and essentially they were shown to be completely immobile (Fig. 2, numbers 24,25). These results show that the SalI and MluI unique restriction sites, both engineered in the loop region of domain IV, are not favorable positions in which to insert foreign sequences within Ll.LtrB.
Genetic screen to identify potential insertion sites within Ll.LtrB that do not hinder mobility
Because of the complex secondary structure of the Ll.LtrB group II intron (Fig. 4), it was difficult to predict what other sites within the intron could be used to insert additional DNA fragments without affecting mobility. In fact, we tried without success to insert the pHs/holin/lysin cassette in domain II at position 428 in the sense orientation (Figs. 2, number 26; 4). Looking at general group II intron secondary structures, this seemed to be a good position in which to insert foreign sequences within Ll.LtrB without affecting its mobility efficiency.
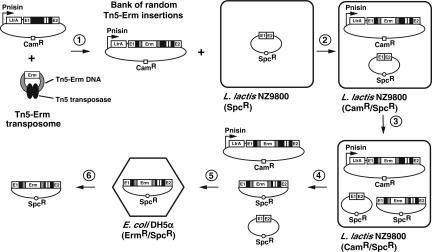
In order to find potential insertion sites within the Ll.LtrB construct expressing LtrA in trans that would still allow the intron to retrohome efficiently, we designed the following screen (Fig. 3). Using the transposome technology from Epicentre (Goryshin and Reznikoff 1998), we constructed a Tn5 transposon that carries an erythromycin resistance gene (Tn5-Erm). In the presence of recombinant Tn5 transposase, the Tn5-Erm double-stranded DNA was incubated with the donor plasmid carrying the Ll.LtrB construct expressing LtrA in trans (Fig. 3, step 1). This in vitro reaction created a saturated bank of random Tn5-Erm insertions. The bank was generated to contain single insertions of the Tn5-Erm transposon between every nucleotide of the intron. Next, to find which insertions within Ll.LtrB would still allow mobilization, we subjected the bank of Tn5-Erm insertions to a mobility assay in L. lactis (Fig. 3, steps 2–4). Then, the recipient plasmids (Spc) that received a marked copy of Ll.LtrB (Erm) were selected from the plasmid mix (Spc/Erm) (Fig. 3, step 5). Recipient plasmids conferring resistance to both spectinomycin and erythromycin are thus the ones that received a mobile copy of Ll.LtrB containing a Tn5-Erm transposon (Fig. 3, step 5).
FIGURE 3.
Genetic screen to identify potential insertion sites within Ll.LtrB. In the presence of the Tn5 transposase (black ovals), the Tn5-Erm transposon DNA (Tn5-Erm DNA) was mixed in a 1:1 molar ratio with a plasmid harboring the Ll.LtrB construct that expresses LtrA in trans (step 1). This in vitro reaction created a saturated bank of random Tn5 insertions around the donor plasmid, consisting of insertions between every nucleotide within the intron. This bank of random Tn5-Erm insertions was transformed in NZ9800 L. lactis cells, which contained the recipient plasmid pMNHS (step 2). Intron expression was induced with nisin (step 3), the plasmid mix (donor plasmid, recipient plasmid, and mobility products) was recovered (step 4), and then retransformed in E. coli DH5α cells (step 5). E. coli cells were plated on LB/Erm/Spc plates to select for Ll.LtrB mobility products that carried a copy of the Tn5-Erm transposon. Only introns that were still mobile following the insertion of a Tn5-Erm transposon could invade the recipient plasmid. Twenty independent Ll.LtrB mobility products were recuperated (step 6), and the locations of Tn5 insertions were analyzed by sequencing the intron–transposon junctions from both ends of the transposon. This screen was repeated with a Tn5-Kan transposon.
Twenty independent Tn5-Erm insertion sites were identified within Ll.LtrB mobility products, and are mapped in Figure 4 (black arrowhead). Surprisingly, 18 of 20 Tn5-Erm transposons were found located in a short region composed of the remaining ltrA sequence within domain IV of Ll.LtrB (Fig. 4). This region of 124 nt, containing 80% of the Tn5-Erm insertions, corresponds to only 13% of the ORF-less Ll.LtrB (900 nt). Moreover, these Tn5-Erm insertions are highly condensed within subregion b of domain IV (DIVb), outside the primary LtrA binding site, DIVa (Fig. 4, DIVa, boxed area) (Wank et al. 1999; Singh et al. 2002; Watanabe and Lambowitz 2004). The two Tn5-Erm insertions outside domain IVb were found in domain II at positions 418 and 466 (Fig. 4). Furthermore, four of the 18 Tn5-Erm insertions located within domain IVb were found more than once within the Ll.LtrB mobility products recovered. This suggests two possibilities: first, these sites are Tn5 insertion hot spots, or, second, these insertion sites were found repeatedly because more of these mobility products were produced during the screen. Mobility of the intron was thus less affected by the insertion of the Tn5-Erm transposon at these particular sites. In order to address this question, we cloned the Ll.LtrB introns interrupted by the Tn5-Erm transposon, which were found more than once, from mobility products back into our donor plasmid (Fig. 4, position 656 [four times], position 2303 [two times], position 2316 [two times], position 2329 [six times]) to perform mobility assays. Correspondingly, the mobility efficiency of these constructs was proportional to the number of times they were isolated from the mix of selected mobility products. These four Ll.LtrB constructs, which were represented two (twice), four, or six times in our pool of 20 independent mobility products, showed mobility efficiencies of 7.4%, 9.6%, 10.4%, and 26.8%, respectively (Fig. 4). This confirms the correlation observed between their relative representation in our pool of mobility products and their mobility efficiencies. Therefore, these Ll.LtrB introns are significantly more mobile than the constructs harboring foreign sequences of similar size at the SalI site (Fig. 2).
In order to challenge the results obtained with Tn5-Erm, we repeated the same screen with a Tn5-Kan transposon (Epicentre) and analyzed 20 independent Ll.LtrB mobility products to locate the Tn5-Kan insertion sites (Fig. 4, gray arrowhead). Remarkably, we found essentially the same distribution of Tn5-Kan insertion sites within the intron that we previously observed with the Tn5-Erm transposon. The majority of the insertions were found within domain IVb (18 of 20), while one was found in domain II, at exactly the same position as a Tn5-Erm insertion (position 418), and one was in domain I (position 39) (Fig. 4). This shows that the Tn5 insertion sites found from these two independent screens are not biased by the sequence of the antibiotic resistance gene present within the Tn5 transposon; we obtained essentially the same distribution of insertion sites with both screens. Moreover, we conclude that the engineered SalI site (Fig. 4), used previously to insert different expression cassettes, is a poor position in which to add foreign sequences within domain IVb since only two Tn5 transposons were found in its vicinity (Fig. 4). In comparison, the engineered MluI site appears to be a better site for the insertion of foreign sequences within Ll.LtrB since it lies in the middle of the Tn5 insertion area (Fig. 4). In order to test this hypothesis, we directly cloned the Tn5-Erm transposon, in both orientations, at the SalI and MluI sites within Ll.LtrB (Fig. 4). Only the sense Tn5-Erm transposon at the MluI site showed relatively good mobility efficiency at 22%; the other three Ll.LtrB constructs showed very little mobility (Fig. 4).
Taken together, these results demonstrate that domain IVb is by far the best region to insert foreign sequences within Ll.LtrB in order to keep its mobility properties, and suggest that the engineered MluI site is a better location to insert foreign sequences within Ll.LtrB than the SalI site.
Accumulation of the intron lariat is severely reduced in cells expressing Ll.LtrB introns that are less mobile
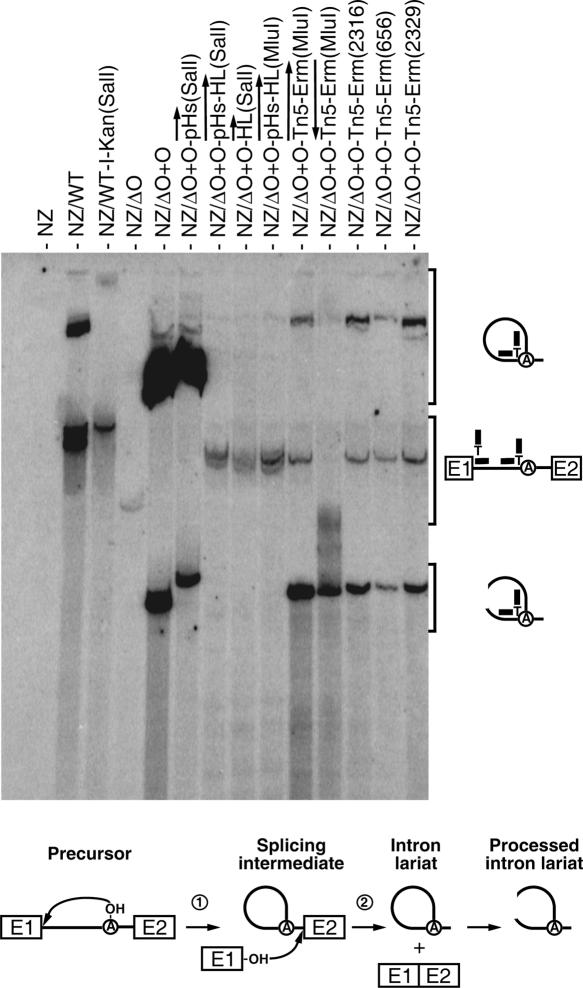
The presence of additional sequences within Ll.LtrB may affect splicing competence and/or lariat stability, and consequently affect mobility efficiency. Thus, we performed Northern blot analyses on total RNA from cells harboring some of our donor plasmids to assess the amount of lariat produced by these Ll.LtrB variants. We used a branch point probe that can specifically recognize excised intron lariats (Fig. 5). This 25-nt probe was designed to base pair with 12 nt on each side of the branch point on the intron lariat. Moreover, to allow for a continuous recognition of the branch site of spliced introns, a T was placed at the junction between the two 12-nt recognition sequences (Fig. 5). This additional nucleotide is thought to recognize the branch point nucleotide (A). The branch point probe was shown to be specific since signals from Northern hybridizations were detected only in L. lactis cells that express the intron (Fig. 5). Accordingly, no lariats were detected in cells that do not overexpress the intron (Fig. 5, NZ) or the nonsplicing ORF-less Ll.LtrB (Fig. 5, NZΔO). However, at the hybridization conditions used, a faint signal that corresponds to the precursor can be detected in cells expressing the nonsplicing ORF-less Ll.LtrB (Fig. 5, NZΔO). Indeed, both halves of this probe can partially hybridize with the pre-mRNA precursors at both the 5′ end of the intron and upstream from the branch point (Fig. 5).
FIGURE 5.
Ll.LtrB lariat accumulation. Northern blot hybridizations were performed using total RNA extracted from NZ9800 L. lactis cells, which were induced with nisin for 3 h to express different variants of Ll.LtrB. The branch point probe used recognizes spliced Ll.LtrB lariats by base pairing with the 5′ extremity of the intron (12 nt) and the sequence upstream from the branch site (12 nt). The branch point probe (thick line), the intron (thin line), and exons (E1, E2) are depicted. The position of intron lariats, processed intron lariats, and precursor mRNA are denoted. A schematic of the Ll.LtrB splicing pathway is presented (bottom). The initial nucleophilic attack is made by the 2′ OH group of an internal adenosine residue (circled A, branch point) (step 1). The liberated 3′ OH group of exon I (E1) then attacks the phosphodiester bond located at the intron–exon II junction. This leads to the ligation of the two exons and liberation of the intron lariat (step 2). After splicing, the intron lariat accumulates and may be processed to create opened circular structures. (NZ) NZ9800, (WT) wild-type Ll.LtrB, (WT-I-Kan) wild-type Ll.LtrB harboring the td group I intron and a kanamycin resistance gene, (ΔO+O) Ll.LtrBΔORF+ORF construct. The position of the cargo within Ll.LtrB is denoted between parentheses.
Surprisingly, for mobile Ll.LtrB variants, the amount of excised intron lariat detected was found to be highly variable. The strongest signals were detected from the Ll.LtrB construct expressing LtrA in trans without any cargo gene, and from the construct carrying the small pH promoter (Fig. 5; NZΔO+O, NZΔO+O-pHs). These Ll.LtrB variants were shown to have mobility efficiencies of 97.2% and 96.3%, respectively (Fig. 2, numbers 3,10). Much less intron lariat was detected in cells where the wild-type intron was expressed, even though its mobility efficiency was shown to be quite high at 87.2% (Fig. 2, number 1). Therefore, a significant difference in accumulated intron lariat was observed between the Ll.LtrB construct expressing LtrA in trans without a cargo gene, harboring the small pH promoter, and the wild-type intron. This supports our hypothesis that upon nisin induction, our mobility assay is saturated, illustrated here by a significant difference in the accumulation of intron lariats (about sevenfold; Fig. 5), although the measured mobility efficiencies were similar (Fig. 2, 97.2% and 96.3% versus 87.2%). In contrast, no lariats were detected for the four constructs that were shown to be completely immobile (Figs. 5, NZΔO+O-pHs-HL, NZΔO+O-HL, NZΔO+O-pHs-HL[MluI], NZΔO+O-Tn5-Erm [antisense, MluI]; 2, numbers 11,20,25; 4). However, some lariats could be detected from all the constructs that were somehow affected by their cargo but still showed a reasonable level of mobility (Figs. 5, NZWT-I-Kan, NZΔO+O-Tn5-Erm [sense, MluI], NZΔO+O-Tn5-Erm [2316], NZΔO+O-Tn5-Erm [656], NZΔO+O-Tn5-Erm [2329]; 2, number 4; 4). As a direct comparison, the presence of lariat was detected only for Ll.LtrB harboring the Tn5-Erm transposon in the sense orientation at the MluI site, while no production of lariat was seen for the nonmobile Ll.LtrB carrying the Tn5-Erm in the reverse orientation [Fig. 5, cf. NZ/ΔO+O-Tn5-Erm(MluI) and Fig. 4].
The reduction of intron lariat observed alternatively may be caused by a shift in splicing to a hydrolytic pathway, which releases linear introns instead of lariats. However, using intron-specific probes that recognize both linear and lariat introns, no bands corresponding to excised linear introns were detected on Northern blots of the above-described Ll.LtrB constructs (Fig. 5; data not shown). We could therefore rule out the possibility that cargo genes shift Ll.LtrB splicing to a hydrolytic pathway.
These results show that the presence of cargo genes within domain IV of Ll.LtrB has a dramatic effect on the accumulation of intron lariats in L. lactis. This correlates with the significant effect of these cargo genes on Ll.LtrB mobility efficiency.
Foreign sequences within Ll.LtrB slightly affect intron splicing
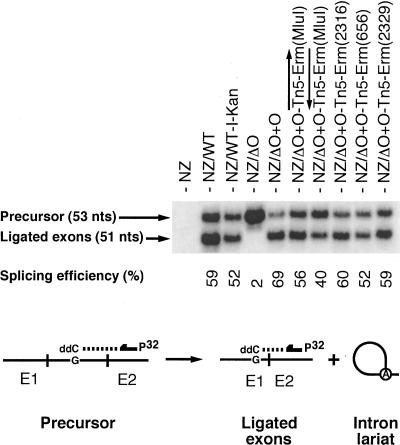
The reduction in lariat accumulation observed for some Ll.LtrB variants (Fig. 5) could be caused by a decrease in splicing efficiency and/or induction of lariat degradation. To determine if some of our cargo-containing Ll.LtrB variants are splicing less efficiently, we performed poisoned primer extension assays (Fig. 6). This assay monitors splicing efficiency by comparing the relative amounts of precursor and ligated exons from total RNA extracts, independently from the amount of accumulated intron lariat (Belfort et al. 1990; Watanabe and Lambowitz 2004). A labeled primer, complementary to exon II, is extended from both the precursor and ligated exons in the presence of a high concentration of ddCTP (Fig. 6). Since the sequences of these two RNAs are different following the exon II junction, the first encountered G residue is at a different distance from the primer. This generates differently sized bands for the precursor (53 nt) and the ligated exons (51 nt) (Fig. 6).
FIGURE 6.
Ll.LtrB splicing efficiency. Poisoned primer extensions (bottom) were performed using different Ll.LtrB variants. This assay monitors splicing efficiency by comparing the relative amounts of precursor and ligated exons, from total RNA extracts, independently from the amount of accumulated intron lariat. A labeled primer, complementary to exon II (arrow), is extended from both the precursor and ligated exons in the presence of a high concentration of ddCTP. Since the sequences of these two RNAs are different past the exon II junction, the first encountered G residue is at a different distance from the primer. This generates differently sized bands for the precursor (53 nt) and the ligated exons (51 nt). The Ll.LtrB splicing efficiencies were calculated as the relative intensities of the precursor and ligated exons bands (ligated exons/precursor+ligated exons). (NZ) NZ9800, (WT) wild-type Ll.LtrB, (WT-I-Kan) wild-type Ll.LtrB harboring the td group I intron and a kanamycin resistance gene, (ΔO+O) Ll.LtrBΔORF+ORF construct. The position of the cargo within Ll.LtrB is denoted between parentheses.
We noticed that splicing efficiency varies from 40% to 69%, and, as a general trend, the most competent Ll.LtrB variants show the highest splicing efficiency. As expected, no ligated exons could be detected from cells overexpressing the nonsplicing Ll.LtrB variant (Fig. 6, NZΔO). The Ll.LtrB variant expressing LtrA in trans without cargo splices slightly more efficiently than the wild-type intron (69% versus 59%). This 10% increase in splicing efficiency does not correlate with the considerable difference observed in lariat accumulation between these two introns (about sevenfold; Fig. 5). Since splicing levels are similar, the level of lariat production and accumulation should also be similar. Therefore, this suggests that the wild-type intron lariat is less stable than its smaller counterpart in L. lactis. Similarly, the splicing efficiency of the Ll.LtrB variant harboring the retromobility and KanR markers is only 7% lower than the wild-type construct, while, again, a much fainter lariat signal is detected compared with the wild type (∼3.5-fold; Fig. 5). Correspondingly, the presence of different transposons within Ll.LtrB slightly reduces its splicing efficiency when compared with the variant expressing LtrA in trans. These results show that the presence of cargo genes within domain IV of Ll.LtrB hinder splicing.
Ll.LtrB splicing could be affected by cargo genes in two possible mechanisms. First, the cargo gene may interfere by allowing the formation of alternative structures that prevent splicing. Or, second, the cargo gene may also disrupt or prevent the formation of the LtrA binding structure within domain IV (Fig. 4, DIVa, boxed area). The latter possibility would prevent association of LtrA to the intron, thus affecting intron splicing and mobility.
However, in general, the reductions in splicing appear minor compared with the reduction of accumulated intron lariats observed (Fig. 5), which suggests again that the reduction in lariat accumulation is primarily due to intron lariat degradation. Moreover, a signal corresponding to processed intron lariat, which is in some cases more intense than the intact lariat, can also be observed from the Ll.LtrB variants harboring Tn5-Erm (Fig. 5). This supports the general idea that lariats of the Ll.LtrB variants harboring foreign sequences within domain IV are less stable.
Taken together, these results show that the minor reduction in splicing of cargo-containing Ll.LtrB introns is indeed contributing to the reduction of intron lariat accumulation, but also suggest that cargo genes are inducing lariat degradation.
DISCUSSION
In this study we examined the potential to use the Ll.LtrB group II intron from L. lactis as a gene delivery vector. Different expression cassettes were inserted at specific sites within the intron expressing LtrA in trans (DIV: SalI, MluI; DII: position 428), and the mobility efficiency of these cargo-containing introns was assessed. Then, genetic screens based on the Tn5 transposon were used to select introns that harbor a transposon at positions that do not drastically hinder mobility. These screens revealed that there is a strict limitation as to where foreign sequences can be inserted in Ll.LtrB without abolishing mobility. Using some Ll.LtrB variants, harboring either different expression cassettes or Tn5 transposon insertions, we studied intron splicing efficiency, stability of the intron lariat, and mobility efficiency.
We initially demonstrated that a variant of Ll.LtrB expressing the LtrA protein in trans is 5.4 times more efficient at invading its recognition site than the wild-type intron in L. lactis. This significant increase in mobility efficiency is probably the consequence of two cumulative factors. First, a significantly smaller intron could likely fold faster and more easily adopt its proficient splicing conformation leading to a higher splicing efficiency. Accordingly, we showed by poisoned primer extension that the Ll.LtrB intron expressing LtrA in trans, which is 64% smaller than wild-type Ll.LtrB, is splicing more efficiently (69% versus 59%). However, the position of the ltrA gene, directly downstream from the nisin promoter, could lead to higher expression levels of LtrA and also enhance Ll.LtrB splicing efficiency. Second, Northern blot analyses showed an appreciable increase of intron lariat accumulation of Ll.LtrB expressing LtrA in trans when compared with the wild-type intron. This suggests that, as previously suggested for E. coli (Matsuura et al. 1997; Guo et al. 2000), the LtrA coding region is a substrate for RNase degradation in L. lactis. Removal of the ltrA gene from the intron may therefore stabilize the excised intron lariat. Taken together, these results suggest that the mobility efficiency of Ll.LtrB expressing LtrA in trans is significantly higher than wild-type Ll.LtrB because this intron splices more efficiently, and its intron RNA lariat is more stable.
We found that mobility of the Ll.LtrB intron expressing LtrA in trans is almost unaffected by small foreign sequences (<100 nt) inserted in the loop region of domain IV (SalI), while large inserts (>1 kb) considerably hinder the invasion of its recognition site. Taken together, our mobility data suggest that reduction in Ll.LtrB mobility efficiency is not caused by the expression, or toxicity, of the cargo gene. Genetic screens using the Tn5 transposon successfully identified regions within Ll.LtrB where foreign sequences can be inserted while retaining some of the mobility proficiency of the intron. Using two independent screens (Tn5-Erm, Tn5-Kan), we demonstrated that domain IVb is by far the best region in which to insert additional sequences within Ll.LtrB, more particularly, stem b3. More than half (24 of 40) of the selected mobility events that harbor a transposon were found in this stem, while stems b1 and b2 contained a total of 12 Tn5 insertions. Some transposon insertion sites were identified more than once, and in both screens (Tn5-Erm, Tn5-Kan). We found that the most represented Tn5 insertion sites represent, as expected, the position that least affects the mobility of the intron. Our data also clearly show that the LtrA binding region within domain IV (DIVa) should be avoided to ensure that the intron retains LtrA-binding properties. Maintaining the ability to bind LtrA is an absolute prerequisite for Ll.LtrB mobility since LtrA is essential for numerous steps during retrohoming. For instance, it is required for splicing and reverse splicing of the intron, endonuclease cleavage of the bottom strand at the insertion site, and reverse transcription of the intron RNA (Cousineau et al. 1998). Since the majority of the Tn5 insertions were in stem b3, the furthest away from the LtrA binding region, this also suggests that insertions that are too close to DIVa could interfere with the proper folding of this domain and prevent LtrA binding.
We demonstrated by poisoned primer extension assays that the presence of expression cassettes within Ll.LtrB expressing LtrA in trans only slightly affects the intron splicing efficiency, which was between 9% and 29%. On the other hand, Northern blot experiments revealed that the presence of expression cassettes within Ll.LtrB expressing LtrA in trans reduce considerably the amount of accumulated intron lariats. However, some lariat could be detected for all the Ll.LtrB variants that show reasonable mobility efficiencies. Taken together, these results suggest that the significant reduction in mobility efficiency of cargo-containing Ll.LtrB is mainly due to the instability of the lariat, ultimately leading to its degradation. When lying outside the compact fold of the intron catalytic core, the cargo sequences could be directly recognized and cleaved by host RNases targeting the intron lariat for degradation. An alternative scenario could be that cargo sequences interfere with the proper folding of the intron, which may alter the LtrA recognition structure and LtrA binding. This situation would also make this intron more susceptible to host RNases.
The strong bias for the insertion of the Tn5-Erm and Tn5-Kan transposons within DIVb of Ll.LtrB also has evolutionary implications. Two different theories were put forward to explain how mobile group II introns originated. First, ORF-less group II introns could have become mobile following the acquisition of an ORF, with reverse transcriptase and endonuclease activities, from another retroelement (Kennel et al. 1993; Wank et al. 1999). Co-evolution of the invaded group II intron and its newly acquired ORF would have eventually allowed the intron to take advantage of the reverse transcriptase and endonuclease activities of its encoded protein to become mobile and invade new sites. On the other hand, mobile group II introns could have been derived from a retroelement that acquired self-splicing capabilities, which minimized their impact following insertion (Curcio and Belfort 1996). Removal of the newly inserted mobile element, at the RNA level, would be less deleterious to the host than other elements that cannot be removed following their insertion. This scenario is supported by the retroelement ancestor hypothesis recently proposed for the evolution of group II introns (Toor et al. 2001). This study unveiled a high degree of co-evolution between the different group II intron RNA structures and the ORFs they encode. However, it did not rule out the possibility that the ancestral mobile group II introns that gave rise to the different classes of group II intron observed today were generated by the invasion of an ORF in their respective domain IV. The former hypothesis implies that the invasion of an ORF-less intron by an ORF occurred at a position that would not abolish splicing, since group II introns need to splice to keep their interrupted genes active. Moreover, in order to create a mobile intron, splicing proficiency should be maintained since intron splicing and reverse splicing are essential steps in the mobility pathway. Taken together, our Tn5 transposon results do not rule out the possibility that mobile group II introns were derived from retroelements that would have acquired self-splicing capabilities, but support the hypothesis that mobile group II introns gained the ability to move to new sites by acquiring an ORF with reverse transcriptase and endonuclease activities from other retromobile elements. The tight restriction for transposon insertion in Ll.LtrB predicts the loop region of domain IV as the primary insertion site for ORFs in group II introns. These results correlate with the observation that all group II intron-encoded proteins are found within the loop region of domain IV.
Finally, this study indicates that it is difficult to find a single position to insert different expression cassettes within DIVb of the Ll.LtrB intron. Therefore, the random insertion strategy used here is the best approach to select for the most appropriate insertion site that would interfere the least with the mobility efficiency of the intron.
MATERIALS AND METHODS
Strains and plasmids
The L. lactis subsp. cremoris R1, MG1363, IL1403, and NZ9800 L. lactis strains were grown without shaking in M17 medium supplemented with 0.5% glucose (GM17) at 30°C. E. coli strains (DH5α, DH10β), used for cloning and mobility scoring purposes, were grown with shaking at 37°C in LB broth.
Two 64-mer oligodeoxyribonucleotides were annealed to create the weak pH promoter (pHw): 5′-ATTTTTGGTTGCCATTTGTTAACGCTGCCTCCTCTCCCTAGTGCTATAATAATGCGGATCCGAG-3′ and 5′-CTCGGATCCGCATTATTATAGCACTAGGGAGAGGAGGCAGCGTTAACAAATGGCAACCAAAAAT-3′. The strong pH (pHs) and chloride (NaCl) promoters were both amplified from MG1363 genomic DNA using the following pairs of primers, respectively: 5′-ATTTTTGGTTGCCATTTGTTAACGC-3′ and 5′-CTCGGATCCGTTTTTTTTTGGCCATTTTTATTATAG-3′; 5′-AGCTGATCATGTAGTCGACGAGCGTTGTATAAGCTTTTATG-3′ and 5′-GCTAGTACGCACGGTCGACTTGGATCCCCTCCTTATATTTATGATTGAATT-3′. The acmA (Buist et al. 1997) and holin/lysin (van Sinderen et al. 1996) genes were amplified by PCR from IL1403 and L. lactis subsp. cremoris R1 genomic DNA, respectively, using the following primer pairs: 5′-CGCGGATCCTATGCCAGTTTCTCGTATTAAAGT-3′ and 5′-CGCGGATCCTTTATTTAATACGAAGATATTGACC-3′; 5′-CGCGGATCCATGAAAACATTTTTTAAAGATATGGC-3′ and 5′-CGCGGATCCTTAATGGATACGAGGATGTTGTC-3′. The Ll.LtrB construct missing a large portion of the ltrA gene (1596 nt) (Guo et al. 2000), and the construct containing the td group I intron from bacteriophage T4 and a kanamycin resistance marker (Cousineau et al. 1998), were previously described. The Ll.LtrB construct expressing LtrA in trans was generated by cloning the ltrA gene (SacI/SalI) between the nisin promoter and exon I (PstI) in the pLE-based plasmid harboring the ORF-less intron. The different inducible promoters (pHs, pHw, NaCl) were cloned in both orientations at the engineered SalI site of the Ll.LtrB intron expressing LtrA in trans. The acmA and holin/lysin genes were cloned at the BamHI sites engineered at the 3′ end of each inducible promoters. The insertion of the pHs/holin/lysin expression cassette in domain II was made by replacement cloning (BsrGI/BsiWI) of a PCR fragment obtained by mega primer amplification (three fragments—fragment A: 5′-TCCTCCTTTCTATTAGGCATTC-3′ and 5′-CCTGATGACAATCTAACTCCTG-3′; fragment B: 5′-GAATGCCTAATAGAAAGGAGGACCCGGGATCTTTGGTTGCCATTTGTTAACGC-3′ and 5′-AGCTCTAGTGCTATAGCCTTTTCCCGGGGTCTAATGGATACGAGGATGTTGTC-3′; fragment C: 5′-AAAAGGCTATAGCACTAGAGCT-3′ and 5′-ACTGGTTTGCACCACCCTCTTC-3′). The Tn5-Erm transposon was created by cloning the erythromycin resistance gene (ermC) from the pG+host5 shuttle vector (Biswas et al. 1993) into the pMOD-2<MCS> Transposon Construction Vector from Epicentre (KpnI site). The erythromycin gene was PCR amplified with the following primers: 5′-AGGGGTACCAACCCGGTAAGACACGACTTAT-3′ and 5′-CGGGGTACCGGTTATAATGAATCGTTAATAAGC-3′. The Tn5-Kan transposon was obtained from the EZ::TN <oriV/KAN-2> Insertion Kit from Epicentre.
Selective medium contained the following concentrations of antibiotics: chloramphenicol (Cam), 10 μg/mL; spectinomycin (Spc), 300 μg/mL; erythromycin (Erm), 300 μg/mL; kanamycin (Kan), 20 μg/mL; ampicillin (Amp), 100 μg/mL.
Ll.LtrB mobility assay and scoring
The NZ9800 L. lactis strain containing different Ll.LtrB expression plasmids (donors) and the pMNHS recipient plasmid harboring the intron homing site (E1/E2) (Fig. 1A; Cousineau et al. 2000) was grown without shaking at 30°C. Saturated overnight cultures were diluted (1/10 or 1/20) and grown to an optical density at 600 nm (OD600) of 0.5. Ll.LtrB expression was then induced for 3 h by the addition of nisin to the cultures to a final concentration of 10 ng/mL. Cells were recovered by centrifugation, and the plasmid mix was extracted (Fig. 1B). The plasmid mix was transformed in E. coli DH5α by heat shock and plated on LB/Spc plates in order to select cells that acquired either the empty recipient plasmid or a mobility product. For each mobility assay, 100 colonies were patched on an LB/Spc plate, transferred onto a nylon membrane, and hybridized with an intron-specific 32P-labeled probe (5′-CCGTGCTCTGTTCCCGTATCAGC-3′), therefore revealing the percentage of recipient plasmids that were invaded by Ll.LtrB (Fig. 1B).
Tn5 genetic screens
Two independent screens were performed with the Tn5-Erm and Tn5-Kan transposons (Fig. 3). The Tn5-Erm transposon was excised from pMOD<2> by PvuII digestion. The Tn5-Kan double-stranded DNA fragment was taken from the EZ::TN <oriV/KAN-2> Insertion Kit from Epicentre. The excised Tn5-Erm or Tn5-Kan transposons were incubated in a 1:1 molar ratio with the donor plasmid carrying the Ll.LtrB construct expressing LtrA in trans for 2 h at 37°C in the presence of recombinant Tn5 transposase (Epicentre) (Goryshin and Reznikoff 1998). A fraction (10%) of the reaction mixes was electroporated (BioRad MicroPulser) in E. coli DH10β cells, and plated on either LB/Erm or LB/Kan plates to select for Tn5 insertion events. For each Tn5 insertion reaction, plasmids were recovered from a mix of >10,000 resistant colonies (Erm or Kan), creating a bank of random Tn5 insertions in our intron donor plasmid (12.3 kb). Taking into account that Tn5 inserts randomly, and that functional regions of the plasmid cannot sustain Tn5 insertions, we estimate that our banks of Tn5 insertions are saturated, containing one insertion between each nucleotide of Ll.LtrB. The Tn5 insertion banks were then electroporated in L. lactis cells (NZ9800/pMNHS) (GM17/Cam/Spc) to perform mobility assays (Fig. 1A). Again, >10,000 colonies were recuperated, grown at 30°C to an OD600 of 0.5, and induced with 10 ng/mL of nisin for 3 h. The plasmid mixes were recuperated and retransformed in electrocompetent E. coli DH5α cells. These cells were plated on either LB/Spc/Erm or LB/Spc/Kan in order to select for Ll.LtrB mobility products that had received a copy of a marked transposon. The Tn5 insertion sites within Ll.LtrB mobility products were analyzed by sequencing the transposon–intron junctions using primers that are complementary to both extremities of the tranposon (5′-GCCAACGACTACGCACTAGCCAAC-3′; 5′-GAGCCAATATGCGAGAACACCCGAGAA-3′).
RNA isolation and Northern blot hybridization
Total RNA was isolated from NZ9800 L. lactis cells containing different Ll.LtrB expression plasmids grown to an OD600 of 0.5 and induced with nisin (10 ng/mL) for 3 h. The cell pellets were mixed with 500 μL of TRIzol (Invitrogen Life Technologies) and 250 μg of glass beads (Sigma). These mixtures were vortexed for 3 min and incubated for 5 min at 55°C. This treatment was repeated three times. For Northern blot hybridizations, samples containing 2.5 μg of total RNA were run on a 4% polyacrylamide denaturing gel (7 M urea, 1× TBE) and transferred to a nylon membrane (Hybond-N, GE Healthcare Bio-Sciences) by capillarity. The membrane was hybridized with a branch-point-specific probe (5′-ATCTGGGCGCACTAGGGAGGTACCG-3′) labeled at its 5′end with 32P (Fig. 5). This oligodeoxyribonucleotide (40 pmol) was labeled in a final volume of 10 μL containing 13.3 pmol of [γ-32P]ATP (3000 Ci/mmol; GE Healthcare Bio-Sciences) and 5 units of T4 polynucleotide kinase (New England Biolabs) for 1 h at 37°C, and then purified on G-50 sephadex columns. The hybridized membrane was exposed on a phosphor screen and revealed with the Molecular Imager Fx (BioRad).
Poisoned primer extension
Total RNA was first extracted from L. lactis NZ9800 induced cells, expressing different Ll.LtrB variants (3h00; nisin, 10 ng/mL). Five micrograms of total RNA were annealed (7 μL; 60 mM NaCl, 50 mM Tris-HCl [pH 7.5], 10 mM DTT) with 0.5 pmol of a 5′ end-labeled primer (32P, 5′-GCCAGTATAAAGATTCGTAGAAT-3′) complementary to exon II (Fig. 6; Belfort et al. 1990; Watanabe and Lambowitz 2004). After annealing of the primer (3 min, 60°C; 5 min, 4°C), the SuperScript II RT (Invitrogen) (50 units) was added along with nucleotides (3 μL; 125 μM each of dATP, dTTP, and dGTP; 500 μM ddCTP; 1.8 mM MgCl2). The mix was incubated for 30 min at 48°C. The reactions were terminated by the addition of gel loading buffer (10 μL; 97.5% formamide, 10 mM EDTA, 0.3% bromophenol blue and xylene cyanol FF). The extension products were run on an 8% denaturing PAGE gel, exposed on a phosphor screen, and revealed with the Molecular Imager Fx (BioRad). Band intensities were calculated using the Quantity One software.
ACKNOWLEDGMENTS
We thank Dr. S. Moineau and Dr. A.M. Lambowitz for kindly providing the L. lactis subsp. cremoris R1 strain and the Ll.LtrB secondary structure, respectively, used to create Figure 4. We also thank M. Guilmette and J. Wyse for technical assistance and K. Belhocine and K. Yam for providing comments on the manuscript. This work was supported by a CIHR grant to B.C. B.C. was a CIHR New Investigator Scholar and is currently an FRSQ Chercheur-Boursier Junior 2 and a McGill William Dawson Scholar.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.193306.
REFERENCES
- Belfort, M., Chandry, P.S., Pedersen-Lane, J. Genetic delineation of functional components of the group I intron in the phage T4 td gene. Cold Spring Harb. Symp. Quant. Biol. 1987;52:181–192. doi: 10.1101/sqb.1987.052.01.023. [DOI] [PubMed] [Google Scholar]
- Belfort, M., Ehrenman, K., Chandry, P.S. Genetic and molecular analysis of RNA splicing in Escherichia coli . Methods Enzymol. 1990;181:521–539. doi: 10.1016/0076-6879(90)81149-o. [DOI] [PubMed] [Google Scholar]
- Belfort, M., Derbyshire, V., Parker, M.M., Cousineau, B., Lambowitz, A.M. Mobile introns: Pathways and proteins. In: Craig N.L., et al., editors. Mobile DNA II. American Society for Microbiology Press; Washington DC: 2002. pp. 761–783. [Google Scholar]
- Belhocine, K., Plante, I., Cousineau, B. Conjugation mediates transfer of the Ll.LtrB group II intron between different bacterial species. Mol. Microbiol. 2004;51:1459–1469. doi: 10.1111/j.1365-2958.2004.03923.x. [DOI] [PubMed] [Google Scholar]
- Belhocine, K., Yam, K.K., Cousineau, B. Conjugative transfer of the Lactococcus lactis chromosomal sex factor promotes dissemination of the Ll.LtrB group II intron. J. Bacteriol. 2005;187:930–939. doi: 10.1128/JB.187.3.930-939.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, I., Gruss, A., Ehrlich, S.D., Maguin, E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 1993;175:3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buist, G., Karsens, H., Nauta, A., van Sinderen, D., Venema, G., Kok, J. Autolysis of Lactococcus lactis caused by induced overproduction of its major autolysin, AcmA. Appl. Environ. Microbiol. 1997;63:2722–2728. doi: 10.1128/aem.63.7.2722-2728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coros, C.J., Landthaler, M., Piazza, C.L., Beauregard, A., Esposito, D., Perutka, J., Lambowitz, A.M., Belfort, M. Retrotransposition strategies of the Lactococcus lactis Ll.LtrB group II intron are dictated by host identity and cellular environment. Mol. Microbiol. 2005;56:509–524. doi: 10.1111/j.1365-2958.2005.04554.x. [DOI] [PubMed] [Google Scholar]
- Cousineau, B., Smith, D., Lawrence-Cavanagh, S., Mueller, J.E., Yang, J., Mills, D., Manias, D., Dunny, G., Lambowitz, A.M., Belfort, M. Retrohoming of a bacterial group II intron: Mobility via complete reverse splicing, independent of homologous DNA recombination. Cell. 1998;94:451–462. doi: 10.1016/s0092-8674(00)81586-x. [DOI] [PubMed] [Google Scholar]
- Cousineau, B., Lawrence, S., Smith, D., Belfort, M. Retrotransposition of a bacterial group II intron. Nature. 2000;404:1018–1021. doi: 10.1038/35010029. [DOI] [PubMed] [Google Scholar]
- Curcio, M.J., Belfort, M. Retrohoming: cDNA-mediated mobility of group II introns requires a catalytic RNA. Cell. 1996;84:9–12. doi: 10.1016/s0092-8674(00)80987-3. [DOI] [PubMed] [Google Scholar]
- de Lencastre, A., Hamill, S., Pyle, A.M. A single active-site region for a group II intron. Nat. Struct. Mol. Biol. 2005;12:626–627. doi: 10.1038/nsmb957. [DOI] [PubMed] [Google Scholar]
- de Ruyter, P.G., Kuipers, O.P., Meijer, W.C., de Vos, W.M. Food-grade controlled lysis of Lactococcus lactis for accelerated cheese ripening. Nat. Biotechnol. 1997;15:976–979. doi: 10.1038/nbt1097-976. [DOI] [PubMed] [Google Scholar]
- Frazier, C.L., San Filippo, J., Lambowitz, A.M., Mills, D.A. Genetic manipulations of Lactococcus lactis by using targeted group II introns: Generation of stable insertions without selection. Appl. Environ. Microbiol. 2003;69:1121–1128. doi: 10.1128/AEM.69.2.1121-1128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goryshin, I.Y., Reznikoff, W.S. Tn5 in vitro transposition. J. Biol. Chem. 1998;273:7367–7374. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- Guo, H., Karberg, M., Long, M., Jones, J.P., III, Sullenger, B., Lambowitz, A.M. Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science. 2000;289:452–457. doi: 10.1126/science.289.5478.452. [DOI] [PubMed] [Google Scholar]
- Ichiyanagi, K., Beauregard, A., Lawrence, S., Smith, D., Cousineau, B., Belfort, M. Retrotransposition of the Ll.LtrB group II intron procedes predominantly via reverse splicing into DNA targets. Mol. Microbiol. 2002;46:1259–1272. doi: 10.1046/j.1365-2958.2002.03226.x. [DOI] [PubMed] [Google Scholar]
- Ichiyanagi, K., Beauregard, A., Belfort, M. A bacterial group II intron favors retrotransposition into plasmid targets. Proc. Natl. Acad. Sci. 2003;100:15742–15747. doi: 10.1073/pnas.2536659100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.P., 3rd, Kierlin, M.N., Coon, R.G., Perutka, J., Lambowitz, A.M., Sullenger, B.A. Retargeting mobile group II introns to repair mutant genes. Mol. Ther. 2005;11:687–694. doi: 10.1016/j.ymthe.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Kennel, J.C., Moran, J.V., Perlman, P.S., Butow, R.A., Lambowitz, A.M. Reverse transcriptase activity associated with maturase-encoding group II introns in yeast mitochondria. Cell. 1993;73:133–146. doi: 10.1016/0092-8674(93)90166-n. [DOI] [PubMed] [Google Scholar]
- Kuipers, O.P., Beerthuyzen, M.M., Siezen, R.J., De Vos, W.M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- Lambowitz, A.M., Zimmerly, S. Mobile group II introns. Annu. Rev. Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- Madsen, S.M., Arnau, J., Vrang, A., Givskov, M., Israelsen, H. Molecular characterization of the pH-inducible and growth phase-dependent promoter P170 of Lactococcus lactis . Mol. Microbiol. 1999;32:75–87. doi: 10.1046/j.1365-2958.1999.01326.x. [DOI] [PubMed] [Google Scholar]
- Matsuura, M., Saldanha, R., Ma, H., Wank, H., Yang, J., Mohr, G., Cavanagh, S., Dunny, G., Belfort, M., Lambowitz, A.M. A bacterial group II intron encoding reverse transcriptase, maturase, and endonuclease activities: Biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes & Dev. 1997;11:2910–2924. doi: 10.1101/gad.11.21.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, D.A., McKay, L.L., Dunny, G.M. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J. Bacteriol. 1996;178:3531–3538. doi: 10.1128/jb.178.12.3531-3538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, D.A., Manias, D.A., McKay, L.L., Dunny, G.M. Homing of a group II intron from Lactococcus lactis subsp. lactis ML3. J. Bacteriol. 1997;179:6107–6111. doi: 10.1128/jb.179.19.6107-6111.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr, G., Smith, D., Belfort, M., Lambowitz, A.M. Rules for DNA target-site recognition by a lactococcal group II intron enable retargeting of the intron to specific DNA sequences. Genes & Dev. 2000;14:559–573. [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan, D., Ross, R.P., Twomey, D.P., Fitzgerald, G.F., Hill, C., Coffey, A. Naturally occurring lactococcal plasmid pAH90 links bacteriophage resistance and mobility functions to a food-grade selectable marker. Appl. Environ. Microbiol. 2001;67:929–937. doi: 10.1128/AEM.67.2.929-937.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutka, J., Wang, W., Goerlitz, D., Lambowitz, A.M. Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J. Mol. Biol. 2004;336:421–439. doi: 10.1016/j.jmb.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Saldanha, R., Chen, B., Wank, H., Matsuura, M., Edwards, J., Lambowitz, A.M. RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry. 1999;38:9069–9083. doi: 10.1021/bi982799l. [DOI] [PubMed] [Google Scholar]
- Sanders, J.W., Venema, G., Kok, J. A chloride-inducible gene expression cassette and its use in induced lysis of Lactococcus lactis . Appl. Environ. Microbiol. 1997;63:4877–4882. doi: 10.1128/aem.63.12.4877-4882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman, C., Godon, J.J., Gasson, M. Splicing of a group II intron in a functional transfer gene of Lactococcus lactis . Mol. Microbiol. 1996;21:45–53. doi: 10.1046/j.1365-2958.1996.00610.x. [DOI] [PubMed] [Google Scholar]
- Singh, R., Lambowitz, A.M. Interaction of a group II intron ribonucleoprotein endonuclease with its DNA target site investigated by DNA footprinting and modification interference. J. Mol. Biol. 2001;309:361–386. doi: 10.1006/jmbi.2001.4658. [DOI] [PubMed] [Google Scholar]
- Singh, R.N., Saldanha, R.J., D'Souza, L.M., Lambowitz, A.M. Binding of a group II intron-encoded reverse transcriptase/maturase to its high affinity intron RNA binding site involves sequence-specific recognition and autoregulates translation. J. Mol. Biol. 2002;318:287–303. doi: 10.1016/S0022-2836(02)00054-2. [DOI] [PubMed] [Google Scholar]
- Smith, D., Zhong, J., Matsuura, M., Lambowitz, A.M., Belfort, M. Recruitment of host functions suggests a repair pathway for late steps in group II intron retrohoming. Genes & Dev. 2005;19:2477–2487. doi: 10.1101/gad.1345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor, N., Hausner, G., Zimmerly, S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA. 2001;7:1142–1152. doi: 10.1017/s1355838201010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sinderen, D., Karsens, H., Kok, J., Terpstra, P., Ruiters, M.H., Venema, G., Nauta, A. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Mol. Microbiol. 1996;19:1343–1355. doi: 10.1111/j.1365-2958.1996.tb02478.x. [DOI] [PubMed] [Google Scholar]
- Wank, H., San Filippo, J., Singh, R.N., Matsuura, M., Lambowitz, A.M. A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol. Cell. 1999;4:239–250. doi: 10.1016/s1097-2765(00)80371-8. [DOI] [PubMed] [Google Scholar]
- Watanabe, K., Lambowitz, A.M. High-affinity binding site for a group II intron-encoded reverse transcriptase/maturase within a stem-loop structure in the intron RNA. RNA. 2004;10:1433–1443. doi: 10.1261/rna.7730104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, J., Zhong, J., Lambowitz, A.M. Gene targeting using randomly inserted group II introns (targetrons) recovered from an Escherichia coli gene disruption library. Nucleic Acids Res. 2005;33:3351–3362. doi: 10.1093/nar/gki649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, J., Zhong, J., Fang, Y., Geisinger, E., Novick, R.P., Lambowitz, A.M. Use of targetrons to disrupt essential and nonessential genes in Staphylococcus aureus reveals temperature sensitivity of Ll.LtrB group II intron splicing. RNA. 2006;12:1271–1281. doi: 10.1261/rna.68706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, J., Karberg, M., Lambowitz, A.M. Targeted and random bacterial gene disruption using a group II intron (targetron) vector containing a retrotransposition-activated selectable marker. Nucleic Acids Res. 2003;31:1656–1664. doi: 10.1093/nar/gkg248. [DOI] [PMC free article] [PubMed] [Google Scholar]