Abstract
Understanding how animal populations have evolved in response to palaeoenvironmental conditions is essential for predicting the impact of future environmental change on current biodiversity. Analyses of ancient DNA provide a unique opportunity to track population responses to prehistoric environments. We explored the effects of palaeoenvironmental change on the colonial tuco-tuco (Ctenomys sociabilis), a highly endemic species of Patagonian rodent that is currently listed as threatened by the IUCN. By combining surveys of modern genetic variation from throughout this species' current geographic range with analyses of DNA samples from fossil material dating back to 10 000 ybp, we demonstrate a striking decline in genetic diversity that is concordant with environmental events in the study region. Our results highlight the importance of non-anthropogenic factors in loss of diversity, including reductions in smaller mammals such as rodents.
Keywords: Ctenomys sociabilis, ancient DNA, phylochronology, climatic change, genetic variability, volcanism
1. Introduction
Tracing genetic lineages through time (phylochronology) bridges the gap between the micro- and macroevolutionary processes that determine faunal diversity (Hadly et al. 2004). When coupled with data on biotic and abiotic environments, such analyses bring us closer to a comprehensive understanding of species' responses to environmental change. Although climatic change and habitat loss are identified as contributing to reductions in faunal diversity (McLaughlin et al. 2002), few studies have examined the impacts of these processes over evolutionary time-scales. Studies of ancient DNA provide an important means of quantifying the effects of prehistoric events on faunal diversity, including reductions in species abundance and genotypic variability (Willerslev & Cooper 2005). Because studies of ancient DNA allow explicit comparisons of current versus historical patterns of genetic variability, they provide a direct means of characterizing temporal changes in population- and species-level diversity.
The colonial tuco-tuco (Ctenomys sociabilis) is a medium-sized, highly endemic subterranean rodent whose range is currently restricted to an area of Neuquén Province, Argentina, of approximately 1500 km2 where it inhabits patches of wet meadow known as ‘mallines’ (Lacey & Wieczorek 2003). Behaviourally, C. sociabilis is the only species of tuco-tuco (n greater than 35 species; Woods 1993) known to be social (Lacey 2000). Group living is associated with limited natal dispersal by members of both sexes (Lacey & Wieczorek 2004), which may be reinforced by the patchy distribution of mallín habitats (Lacey & Wieczorek 2003). These behavioural, demographic and ecological attributes are expected to lead to low levels of genetic variability within and high levels of genetic differentiation among local populations. Accordingly, a survey of 15 microsatellite loci revealed very limited genotypic variability within the population of C. sociabilis that has been the focus of behavioural and ecological studies of this species (Lacey 2001). Preliminary analyses of ancient DNA samples from a site located approximately 6 km southwest of this population (Estancia Nahuel Huapi) revealed the presence of only a single mitochondrial haplotype over the past 1000 years (Hadly et al. 2003), indicating that low levels of genetic variability have persisted in C. sociabilis for at least this many generations.
The geographic range of C. sociabilis is dominated by a steep west-to-east rainfall gradient created by the rainshadow effect of the Andes (Heusser 2003). Due to this gradient, historical region-wide shifts in climate patterns have caused significant changes in the distribution of the local flora and the location of the mesic forest-arid steppe ecotone where C. sociabilis occurs (Markgraf 1983; Pearson & Christie 1985). In addition, this area has a complex history of environmental change associated with volcanism (Heusser 2003), which may have dramatically affected the palaeoenvironment of this species. Collectively, these observations suggest that response to environmental change may be an important component of the evolutionary history of this species, including the low levels of genetic variability and restricted geographic range characteristic of C. sociabilis today.
2. Material and methods
To characterize current haplotypic variability in C. sociabilis, five modern populations were sampled from localities spanning the species' current geographic range: (1) Cerro Monte Redondo (n=13); (2) Rincon Grande (n=9); (3) Altos del Fortin (n=9); (4) Paso Coihue (n=9); (5) Cerro La Lagunita (n=13; figure 1a). Animals were captured and non-destructive tissue samples were collected as described by Lacey (2001). MtDNA was extracted using the Qiagen DNAeasy Kit (Qiagen Inc., Valencia, CA) and cytochrome b (1140 bp) was amplified using primers MVZ05 (Smith 1998) and MVZ108 (E. A. Lacey, unpublished data) under the following conditions: 30–60 μg of DNA was added to 1X PCR buffer with 1.5 mM MgCl2, 0.89 μM dNTP's, 0.22 μM of each primer, and 1.5 U of Taq DNA polymerase in total reaction volume of 27 μl (denature 94 °C for 5 min., 36 cycles of 94 °C 1 min., 48 °C 30 s, 72 °C 1 min., 5 min. extension at 72 °C). PCR products were sequenced in both directions on an ABI 377 automated sequencer.
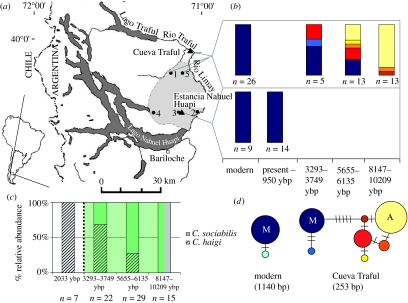
Figure 1.
(a) Map of the study area, including prehistoric fossil sites Estancia Nahuel Huapi (ENH) and Cueva Traful (CT). The geographic distribution of C. sociabilis is shown in light grey. Numbered dots correspond to modern sampling localities: (1) Cerro Monte Redondo; (2) Rincon Grande; (3) Altos del Fortin; (4) Paso Coihue; (5) Cerro la Lagunita. (b) Frequency of haplotypes (based on 253 bp) detected at ENH and CT, presented in time-intervals corresponding to radiocarbon dates for stratigraphic levels. Also shown are haplotype frequencies for the nearest modern collection localities. (c) Relative abundance of C. sociabilis and C. haigi at CT over the last 10 000 years by time-interval. Shading denotes an increase in density of Nothofagus forest at 8500 ybp; dotted line denotes volcanic eruption with ash deposit in CT. (d) Haplotype networks for cytochrome b sequence data from modern and ancient DNA samples. The sizes of the circles indicate the relative abundance of each haplotype; the numbers of tick marks between circles correspond to the number of base pair substitutions that differentiate haplotypes. Colours correspond to those used in (b) to denote haplotype frequencies.
Haplotypic variation in ancient DNA samples was examined using teeth collected from Cueva Traful (40°43′ S, 71°07′ W, elevation 760 m), a Holocene archaeological site and barn owl roost excavated between 1973 and 1978 (Montero et al. 1983). The cave is located in the forest-steppe ecotone approximately 6 km north of the current northern distributional limit for C. sociabilis. Due to the deposition of owl pellets, material excavated from the cave provides a continuous temporal sequence of small mammal bones and teeth dating from approximately 2000 to 10 000 ybp (Pearson & Pearson 1981). For genetic analyses, the protocol of Hadly et al. (2003) was used to extract DNA from 73 teeth recovered from eight stratigraphic levels (ca. 2000–10 000 ybp). For each sample, a total of 253 bp of cytochrome b was amplified and sequenced according to strict ancient DNA standards.
3. Results and discussion
Only a single cytochrome b haplotype was detected in each of the five modern populations of C. sociabilis surveyed (h=0.000, π=0.000, n>9 animals per population). Four of these populations shared the same haplotype, with the haplotype of the fifth differing by only a single third-position transition (0.09% sequence divergence; figure 1d). The predominant haplotype was identical to the comparable portions of the single cytochrome b haplotype recovered at Estancia Nahuel Huapi from 14 specimens dating from the present to 950 ypb (figure 1b; Hadly et al. 2003).
Based on a phylogenetic analysis that included all Ctenomys species found in GenBank (39 species, 129 sequences), we identified 43 out of 73 fossil teeth recovered from Cueva Traful as C. sociabilis (see electronic supplementary material). The remaining 30 teeth were identified as Ctenomys haigi, a non-social tuco-tuco found in the same region of Patagonia. In contrast to the modern populations, eight cytochrome b haplotypes from C. sociabilis were detected among samples from Cueva Traful: the predominant modern haplotype (M) and seven historical variants (n=33 sequences; h=0.71±0.06, π=0.01283±0.00167, calculated using DNAsp v. 4.0.6 (Rozas & Rozas 1999), 4.3% overall sequence divergence; figure 1b). Thus, prior to ca. 3000 ybp, C. sociabilis from within an area of Cueva Traful of roughly 19.6 km2 (mean hunting radius for barn owls=2.5 km; Taberlet 1983) were characterized by greater haplotype diversity than occurs today throughout the species' range (Fisher's exact test, p=0.000, s.e.=0.000; Raymond & Rousset 1995).
The proportion of dental samples from each stratigraphic level that were identified as C. sociabilis indicates that this species declined in relative abundance from approximately 8200 to 3000 ybp (figure 1c). This change in relative abundance, in conjunction with the modern absence of C. sociabilis from the immediate vicinity of Cueva Traful, suggests a numerical decline and possible range contraction. Although it is difficult to obtain direct evidence regarding the cause(s) of this decline and associated loss of genetic diversity, further analysis of the Cueva Traful data suggests that several factors may have contributed to this outcome.
First, consistent changes in vegetation may have altered the habitat available to C. sociabilis. Pollen core analyses from this region indicate a sustained increase in the prevalence of Nothofagus forest from approximately 8500 to 3000 ybp, which resulted in a local decline in the abundance of the open grassland habitat containing the mallines in which C. sociabilis occurs (Markgraf 1983).
Second, a volcanic eruption approximately 3000 ybp (Montero & Silveira 1983) may have exacerbated the decline of C. sociabilis. Volcanic eruptions have been shown to reduce abundance and genetic diversity in other vertebrate species, including other Ctenomys in the Andean region (Gallardo & Köhler 1994). C. sociabilis disappears from Cueva Traful in the stratigraphic level immediately postdating the tephra layer.
Finally, competition with C. haigi, the other local species of ctenomyid, may have contributed to the reduction in C. sociabilis. Although the relative abundance of C. haigi is low during the early Holocene, it increases steadily throughout the temporal period covered by the Cueva Traful deposit. Currently, C. haigi occupies an extensive area to the north, south, and east of the geographic range of C. sociabilis, and is the extant tuco species that occurs in closest proximity to Cueva Traful.
Although these historical hypotheses require testing, it is clear that C. sociabilis experienced a marked reduction in genetic variation during the past 10 000 years. While low levels of genetic variation are often attributed to historical bottlenecks (Amos & Harwood 1998), few studies have tracked directly the genetic changes associated with such events, particularly over a millennial time-scale (but see Shapiro et al. 2004). The reduction in genetic variation we have documented is particularly unusual given that (i) C. sociabilis is a rodent, rather than a megafaunal mammal, and (ii) no single environmental event (e.g. deglaciation or human over-kill) is clearly associated with this loss of diversity. Absence of genetic diversity has important implications for evolutionary and conservation biology and is particularly important in the context of future climatic modifications, since animal populations may require a repertoire of potential responses to adapt to varying conditions (Lande & Shannon 1996).
How has a species with high prehistoric levels of genetic diversity persisted for the past several thousand years with so little variation? Detailed analyses of additional palaeoecological sites in Neuquén Province should provide a better understanding of the temporal and spatial dynamics of the decrease in relative abundance and genetic diversity detected for C. sociabilis at Cueva Traful. At the same time, comparative analyses of other, non-social tuco-tuco species from this region may yield insights into the role that the distinctive behavioral and demographic attributes of C. sociabilis have played in this decline. As our study exemplifies, the ability to track genetic variation through time provides a powerful tool for identifying and interpreting historical relationships between species and their environments.
Acknowledgments
We thank J. Mountain, K. O'Keefe, S. Palumbi, U. Ramakrishnan, P. Spaeth and M. van Tuinen for their comments. For permission to conduct fieldwork, we thank C. Chehebar and E. Ramilio of the Delegacion Tecnica, Parques Nacionales Argentinas, A. Thouyaret of Estancia Rincon Grande and the Jones family of Estancia Nahuel Huapi. For assistance in the field, we thank A. Barnosky, C. Bailey, M. Christie and, especially, A. Pearson. E.A.L would especially like to thank J. Wieczorek. Funding was provided by grants from the National Science Foundation to E.A.H (DEB-0108541, EPS-9640667) and E.A.L (DEB-9704462, DEB-0128857) and by an EPA STAR Fellowship to Y.L.C.
Footnotes
Deceased 4 March 2003
Supplementary Material
References
- Amos W, Harwood J. Factors affecting levels of genetic diversity in natural populations. Phil. Trans. R. Soc. B. 1998;353:177–186. doi: 10.1098/rstb.1998.0200. doi:10.1098/rstb.1998.0200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo M.H, Köhler N. Demographic changes and genetic losses in populations of a subterranean rodent (Ctenomys maulinus brunneus) affected by a natural catastrophe. Zeitschrift für Säugetierkunde. 1994;59:358–365. [Google Scholar]
- Hadly E.A, Van Tuinen M, Chan Y, Heiman K. Ancient DNA evidence of prolonged population persistence with negligible genetic diversity in an endemic tuco-tuco (Ctenomys sociabilis) J. Mammal. 2003;84:403–417. doi:10.1644/1545-1542(2003)084<0403:ADEOPP>2.0.CO;2 [Google Scholar]
- Hadly E.A, Ramakrishnan U, Chan Y.L, Van Tuinen M, O'Keefe K, Spaeth P.A, Conroy C.J. Genetic response to climatic change: insights from ancient DNA and phylochronology. Public Lib. Sci. Biol. 2004;2:1600–1609. doi: 10.1371/journal.pbio.0020290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusser C.J. Ice age southern Andes: a chronicle of palaeoecological events. In: Rose J, editor. Developments in quaternary science. vol. 3. Elsevier; New York: 2003. pp. 40–43. [Google Scholar]
- Lacey E.A. Spatial and social systems of subterranean rodents. In: Lacey E.A, Patton J.L, Cameron G.N, editors. Life underground: the biology of subterranean rodents. University of Chicago Press; Chicago, IL: 2000. pp. 257–296. [Google Scholar]
- Lacey E.A. Microsatellite variation in solitary and social tuco-tucos: molecular properties and population dynamics. Heredity. 2001;86:628–637. doi: 10.1046/j.1365-2540.2001.00881.x. doi:10.1046/j.1365-2540.2001.00881.x [DOI] [PubMed] [Google Scholar]
- Lacey E.A, Wieczorek J.R. Ecology of sociality in rodents: a ctenomyid perspective. J. Mammal. 2003;84:1198–1211. doi:10.1644/BLe-014 [Google Scholar]
- Lacey E.A, Wieczorek J.R. Kinship in colonial tuco-tucos: evidence from group composition and population structure. Behav. Ecol. 2004;15:988–996. doi:10.1093/beheco/arh104 [Google Scholar]
- Lande R, Shannon S. The role of genetic variation in adaptation and population persistence in a changing environment. Evolution. 1996;50:434–437. doi: 10.1111/j.1558-5646.1996.tb04504.x. [DOI] [PubMed] [Google Scholar]
- Markgraf V. Late and postglacial vegetational and paleoclimatic changes in subantarctic, temperate, and arid environments in Argentina. Palynology. 1983;7:43–70. [Google Scholar]
- McLaughlin J.F, Hellmann J.J, Boggs C.L, Ehrlich P.R. Climate change hastens population extinctions. Proc. Natl Acad. Sci. USA. 2002;99:6070–6074. doi: 10.1073/pnas.052131199. doi:10.1073/pnas.052131199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero E.A.C, Silveira M.J. Radiocarbon chronology of a tephra layer in Rio Traful Valley, Province of Neuquén, Argentina. Quaternary of South America and Antarctic Peninsula. 1983;1:135–150. [Google Scholar]
- Montero E.A.C, Curzio D.E, Silveira M.J. La estratigrafia de la Cueva Traful I (Provincia del Neuquén) Praehistoria. 1983;1:9–160. [Google Scholar]
- Pearson O.P, Christie M.I. Los tuco-tucos (genero Ctenomys) de los parques nacionales Lanin y Nahuel Huapi, Argentina. Hist. Nat. 1985;5:337–343. [Google Scholar]
- Pearson O.P, Pearson A.K. La fauna mamiferos pequenos cerca de Cueva Traful, Argentina: pasado y presente. Praehistoria. 1981:1. [Google Scholar]
- Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DNAsp, version 3: an integrated program for molecular population genetics and moleculat evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. doi:10.1093/bioinformatics/15.2.174 [DOI] [PubMed] [Google Scholar]
- Shapiro B, et al. Rise and fall of the Beringian steppe bison. Science. 2004;306:1561–1565. doi: 10.1126/science.1101074. doi:10.1126/science.1101074 [DOI] [PubMed] [Google Scholar]
- Smith M.F. Phylogenetic relationships and geographic structure in pocket gophers in the genus Thomomys. Mol. Phylogenet. Evol. 1998;9:1–14. doi: 10.1006/mpev.1997.0459. doi:10.1006/mpev.1997.0459 [DOI] [PubMed] [Google Scholar]
- Taberlet P. An estimation of the average foraging radius of the barn owl based upon rejection pellets analysis. Révue D'Ecologie—la Terre et la Vie. 1983;38:171–177. [Google Scholar]
- Willerslev E, Cooper A. Ancient DNA. Proc. R. Soc. B. 2005;272:3–16. doi: 10.1098/rspb.2004.2813. doi:10.1098/rspb.2004.2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods C.A. Suborder Hystricognathi. In: Wilson D.E, Reeder D.M, editors. Mammal species of the world: a taxonomic and geographic reference. Smithsonian Institution Press; Washington, DC: 1993. pp. 771–806. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.