Abstract
The regulation of gene expression by cellular calcium is crucial for plant defense against biotic and abiotic stresses. However, the number of genes known to respond to specific transient calcium signals is limited, and as yet there is no definition of a calcium-responsive cis element in plants. Here, we generated specific cytosolic calcium transients in intact Arabidopsis thaliana seedlings and linked them to early transcriptome changes, followed by bioinformatic analysis of the responsive genes. A cytosolic calcium transient induced by calmodulin antagonists and blocked by lanthanides was characterized using aequorin-based luminometry and photon imaging. Analysis of transcriptome changes revealed 230 calcium-responsive genes, of which 162 were upregulated and 68 were downregulated. These include known early stress-responsive genes as well as genes of unknown function. Analysis of their upstream regions revealed, exclusively in the upregulated genes, a highly significant occurrence of a consensus sequence (P < 10−13) comprising two abscisic acid–specific cis elements: the abscisic acid–responsive element (ABRE; CACGTG[T/C/G]) and its coupling element ([C/A]ACGCG[T/C/A]). Finally, we show that a tetramer of the ABRE cis element is sufficient to confer transcriptional activation in response to cytosolic Ca2+ transients. Thus, at least for some specific Ca2+ transients and motif combinations, ABREs function as Ca2+-responsive cis elements.
INTRODUCTION
Ca2+ is a key second messenger in both animals and plants (Harper et al., 2004; Hetherington and Brownlee, 2004; Reddy and Reddy, 2004; Hepler, 2005). In plants, Ca2+ transients mediate responses to environmental stresses, including salt, drought, cold, heat, UV light, and touch. The stress triggers cytosolic Ca2+ bursts (Knight, 2000), which are transduced by Ca2+ binding proteins such as calmodulin (CaM), CaM-related proteins (Bouché et al., 2005), Ca2+-dependent protein kinases (Harper et al., 2004), and calicneurin-like proteins (Luan et al., 2002). The Ca2+ signals confer changes in enzyme activity, cell structure, and gene expression, which, collectively, allow plants to cope with the ever-changing environment. In several instances, the Ca2+ signal was shown to be crucial in translating a stress stimulus into the induction of gene expression. Typically, inhibition of Ca2+ transients by Ca2+ channel blockers inhibits the expression of these specific genes (Polisensky and Braam, 1996; Knight et al., 1997). Very few examples are known for a role of Ca2+ in repressing gene expression (Neuhaus et al., 1997). A considerable portion of the stress-induced genes are induced by more than one stress (Seki et al., 2002b), of which the touch-induced genes (TCHs) that also respond to cold and heat are a good example (Braam et al., 1997). Moreover, exposure of Arabidopsis thaliana cells to a sudden increase of external Ca2+, which causes an immediate increase in cytosolic Ca2+ concentration ([Ca2+]cyt), is sufficient to induce the expression of a subset of the TCH genes (Braam, 1992). However, to date, the number of genes whose expression is known to be modulated by Ca2+ transients in plants is limited, and the mechanisms underlying the regulation of gene expression by Ca2+ signaling are largely unknown. In fact, there is yet no consensus for regulatory cis elements mediating the responsiveness to Ca2+ signals in plants.
CaM, a well-known transducer of Ca2+ signals, is a protein containing four EF-hand Ca2+ binding motifs. It is present in all eukaryotes, including animals, yeast, and plants. Unlike animals, plants contain a large family of CaM-related proteins (McCormack and Braam, 2003) with diverse structures, only some of which are highly similar (up to 90% identity in amino acid sequence) to mammalian CaM. CaM has no catalytic activity of its own but is capable of binding diverse target proteins and modulating their activity (Snedden and Fromm, 2001; Reddy et al., 2002; Bouché et al., 2005). One of the important roles CaM plays in both plants and animals is in the regulation of cytosolic Ca2+ levels. In contrast with animals, which contain CaM-stimulated Ca2+-ATPases in the plasma membrane, plants contain CaM-stimulated Ca2+ pumps in both the plasma membrane and endomembranes (Sze et al., 2000). In animals, CaM is capable of modulating several different types of Ca2+ channels. For example, dependent on the particular conditions, animal L-type Ca2+ channels are either inhibited or activated by CaM (Zuhlke et al., 1999). The role of CaM in regulating plant Ca2+ channels is much less understood. Ca2+/CaM has been proposed to activate the slow vacuolar cation channels of barley (Hordeum vulgare) aleurone cells (Bethke and Jones, 1994) and to inhibit Ca2+ fluxes in carrot (Daucus carota) cells (Kurosaki et al., 1994).
A variety of CaM antagonists have been used to study Ca2+/CaM-mediated pathways in animals and plants, among them trifluoperazine (TFP) and N-(6-aminohexyl)-5-chloro-1-naphthelenesulfonamid-hydrochloride (W7). CaM antagonists have been reported to increase [Ca2+]cyt in animal cells (Harper and Daly, 2000; Jan and Tseng, 2000). In some cases, CaM antagonists have been reported to interact directly with and to inhibit cation channel activity independently of CaM. For instance, TFP and calmidazolium chloride inhibit the extent of Ca2+ release from type 1 inositol triphosphate receptor channels (Khan et al., 2001). In plants, Gilroy and colleagues (1987) reported that CaM antagonists induced a slow and steady increase in cytosolic Ca2+ in carrot protoplasts. Furthermore, TFP, W7, and calmidazolium have all been reported to inhibit the slowly activating vacuolar ion channels of broad bean (Vicia faba) and sugar beet (Beta vulgaris) (Schulz-Lessdorf and Hedrich, 1995).
A direct link between Ca2+ transients and gene expression is still restricted to a limited number of genes. In particular, we are not aware of any report attempting to associate induced specific cytosolic Ca2+ transients with a whole genome–based transcriptome analysis. Here, we have characterized the effect of CaM antagonists on plant cytosolic Ca2+, using the Ca2+ sensor aequorin (Knight et al., 1991), with the intention to link cytosolic Ca2+ transients to early changes in the transcriptome using DNA microarrays. Clustering of genes based upon their expression characteristics has revealed a consensus cis regulatory element in the promoters of Ca2+-responsive genes that matches with two abscisic acid (ABA)–related cis elements: the ABA-responsive element (ABRE) and an ABRE coupling element (ABRE-CE). We show that a tetramer of the ABRE cis regulatory element is sufficient to confer transcriptional activation in response to cytosolic Ca2+ transients.
RESULTS
CaM Antagonists Induce a Cytosolic Ca2+ Burst in Plants
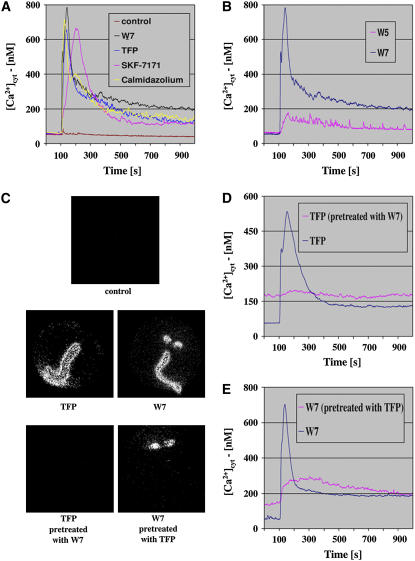
Arabidopsis seedlings expressing apoaequorin in the cytosol (Knight et al., 1991) were used to test the effect of four CaM antagonists, W7, TFP, calmidazolium chloride, and fluphenazine-N-2-chloroethane dihydrochloride (SKF-7171), on [Ca2+]cyt. All tested antagonists triggered a similar Ca2+ response (Figure 1A), an initial burst peaking 30 to 100 s after the addition of antagonists with a peak maximum in the range of 600 to 800 nM [Ca2+]cyt. This peak lasted 5 to 10 min and was followed by steady cytosolic Ca2+ levels that were twofold to threefold higher than the [Ca2+]cyt resting levels. W7 and TFP had similar effects on [Ca2+]cyt in Nicotiana tabacum seedlings expressing aequorin (see Supplemental Figure 1 online). The concentrations needed to trigger maximal [Ca2+]cyt responses were 25, 100, 150, and 600 μM for calmidazolium, SKF-7171, TFP, and W7, respectively. The response to CaM antagonists was concentration-dependent, and the concentrations needed to reach a half-maximal [Ca2+]cyt burst for W7 and TFP were ∼200 and ∼65 μM, respectively.
Figure 1.
Cytosolic Ca2+ in Response to Treatments with Different CaM Antagonists.
Seedlings were treated with different CaM antagonists as indicated 100 s after the luminometer photon counts recording started.
(A) Time course of cytosolic Ca2+ concentrations in response to different CaM antagonists: W7 (600 μM), TFP (150 μM), SKF-7171 (100 μM), and calmidazolium chloride (25 μM). DMSO, the solvent for all antagonists, served as a control.
(B) Comparison of the [Ca2+]cyt response to W7 and W5 at a concentration of 600 μM for both antagonists.
(C) Computer imaging of seedlings treated with CaM antagonists as indicated. Concentrations of CaM antagonists were identical to those in (A). DMSO treatment served as a control.
(D) [Ca2+]cyt plot of seedlings treated with TFP (150 μM) with or without a 15-min pretreatment with W7 (600 μM).
(E) [Ca2+]cyt plot of seedlings treated with W7 (600 μM) with or without a 15-min pretreatment with TFP (150 μM).
W5 [N-(6-aminohexyl)-1-naphthalenesulfoneamide hydrochloride] is commonly used as the negative control for W7-dependent CaM inhibition experiments. We compared the effects of W5 and W7 on the [Ca2+]cyt burst at two concentrations. At 600 μM, at which W7 has a maximal effect on the [Ca2+]cyt response (Figure 1B), W5 triggered a much smaller response than W7, peaking at <200 nM [Ca2+]cyt compared with a peak of 770 nM [Ca2+]cyt for the W7 response. At a lower concentration of 400 μM, at which W7 still triggered a substantial response, W5 had almost no detectable effect (see Supplemental Figure 2 online). These results imply that inhibition of CaM, or a CaM-related protein(s), triggers the Ca2+ response.
The tissue specificity of the [Ca2+]cyt burst triggered by W7 and TFP was examined with a light-sensitive CCD camera. Both W7 and TFP triggered a response in hypocotyls and roots, whereas W7 also triggered a response in cotyledons (Figure 1C). The additional activity of W7 in cotyledons may explain the larger response to W7 compared with TFP as measured by luminometry (Figure 1A). If the antagonists interact with the same targets and/or if they affect the same Ca2+ pools, then pretreatment with one antagonist might inhibit the response to the other. Indeed, 15 min of pretreatment of seedlings with W7 abolished the Ca2+ response to TFP (Figure 1D). TFP pretreatment for 15 min substantially reduced the Ca2+ response to W7 but did not abolish it completely (Figure 1E). As expected, most of the additional Ca2+ response to W7 after pretreatment with TFP was found in the cotyledons (Figure 1C, bottom).
Specific Ca2+ Channel Blockers Inhibit the Ca2+ Burst
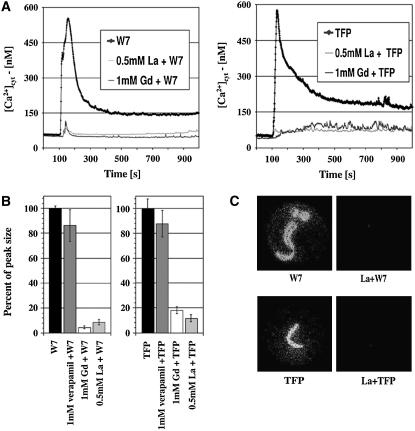
Cytosolic Ca2+ bursts triggered by abiotic stresses such as salt and drought have been shown to be partially inhibited by specific Ca2+ channel blockers (Knight et al., 1997). To identify inhibitors of the [Ca2+]cyt transient detected here and the mechanism by which it was generated, the Ca2+ channel blockers lanthanum (as LaCl3) and gadolinium (as GdCl3) were tested. Pretreatment of seedlings with the trivalent ions of lanthanum or gadolinium (as chloride salts) for 15 min at concentrations of 0.5 or 1 mM, respectively, substantially inhibited the Ca2+ response induced by either TFP or W7 (Figure 2A), indicating that Ca2+-permeable channels mediate the Ca2+ increase. Quantitative analysis (Figure 2B) shows that La3+ and Gd3+ both significantly inhibited the Ca2+ transient peak size by >80%. Moreover, photon imaging clearly shows that LaCl3 at 0.5 mM efficiently blocked the Ca2+ response to TFP and W7 in both roots and cotyledons (Figure 2C). By contrast, verapamil, known as a voltage-gated Ca2+ channel blocker, did not significantly change the Ca2+ response to TFP or W7 (Figure 2B). Furthermore, pretreatment with the general potassium channel blockers tetraethylammonium or CsCl did not significantly affect the Ca2+ burst induced by TFP or W7 (data not shown).
Figure 2.
Effect of Ca2+ Channel Blockers on the [Ca2+]cyt Response to TFP and W7.
(A) [Ca2+]cyt plots of seedlings treated with antagonists with or without pretreatment with the Ca2+ channel blockers lanthanum chloride (La) or gadolinium chloride (Gd). CaM antagonists W7 (left panel) and TFP (right panel) were added 100 s after photon recording started.
(B) Percentage of [Ca2+]cyt peak integral over 5 min after the addition of CaM antagonists, relative to the response to W7 (left panel) or TFP (right panel), in the absence of Ca2+ channel blockers. Error bars indicate se.
(C) Computer imaging of individual seedlings treated with CaM antagonists with or without pretreatment with 0.5 mM lanthanum chloride as indicated.
Transcriptome Changes Are Associated with the Cytosolic Ca2+ Burst
Analysis of gene expression at the whole genome level can potentially reveal gene populations whose expression is associated with a specific stimulus. To assess the role of Ca2+ in mediating gene expression, we designed three sets of microarray experiments using a 12,000 Arabidopsis EST–based chip representing ∼6900 loci (∼25% of the Arabidopsis genome). In experiment A, the effect of W7 was compared with the effect of a control treatment with DMSO (the solvent of W7) to identify the total number of ESTs that are upregulated and downregulated by W7. Experiment B was designed to specifically test the effect of the Ca2+ burst on gene expression. In this set, the effect of W7 was compared with the effect of W7 after pretreatment with 0.35 mM LaCl3, which abolishes the Ca2+ burst (i.e., W7 − Ca). A control experiment (experiment C) was designed to test the effect of W7 on gene expression in the absence of the Ca2+ burst. Here, the effect of W7 after pretreatment with LaCl3 (0.35 mM) was compared with the effect of DMSO after pretreatment with LaCl3 (0.35 mM; i.e., control − Ca).
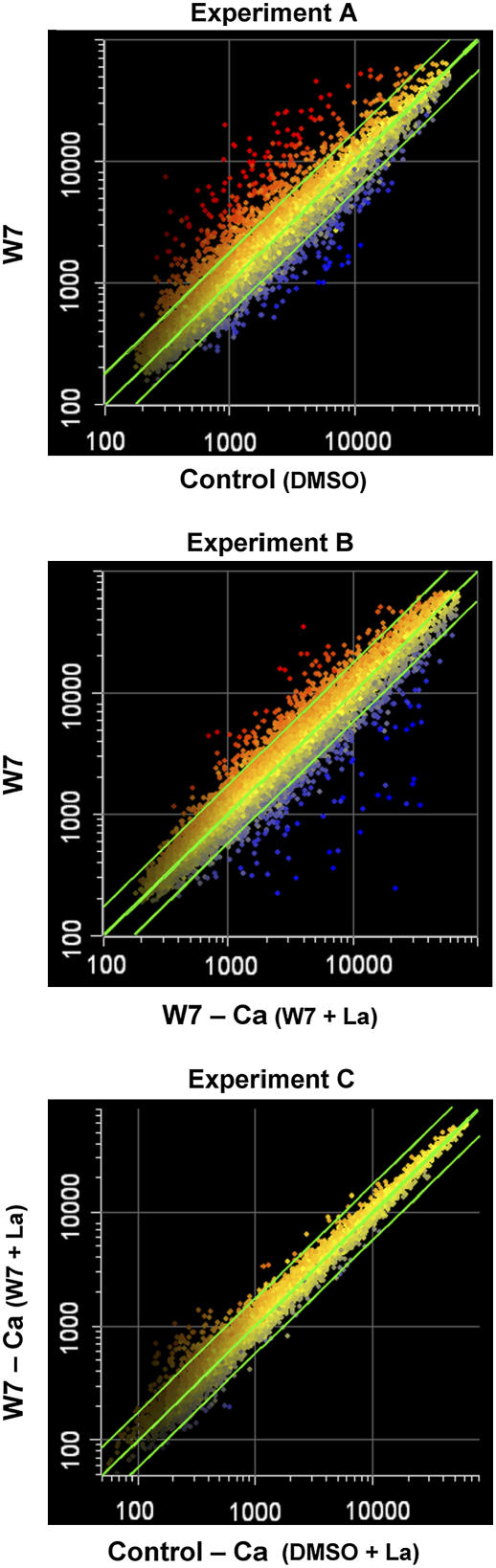
Twelve-day-old Arabidopsis seedlings were treated as indicated above for 1 h, and RNA samples were taken for microarray analysis (see Methods). In all experiments, parallel luminometric Ca2+ measurements were performed as described above (Figures 1 and 2) to confirm the association of gene expression with the Ca2+ burst. Three independent biological repeats were performed for each treatment and subjected to statistical analysis. The averages of the three independent biological repeats were plotted as shown in Figure 3. The median diagonal represents no change in gene expression, whereas the lines parallel to the diagonal represent 1.75-fold change in gene expression; thus, data points exceeding the parallel lines represent transcripts that were activated (above) or repressed (under) by 1.75-fold or more. Apparently, W7 was responsible for a major transcriptome change, both in upregulating and downregulating gene expression by 1.75-fold or more (experiment A; 531 and 317 transcripts, respectively). A similar, though attenuated, picture can be seen in the transcriptome changes in experiment B (363 upregulated and 325 downregulated transcripts), indicating that the major changes in gene expression could be attributed directly to the Ca2+ burst. Importantly, W7 treatment in the absence of the Ca2+ burst (experiment C) had no major effect on the transcriptome, as the expression of very few genes changed by 1.75-fold or more in the absence of the Ca2+ burst (52 upregulated and 2 downregulated transcripts). Therefore, the Ca2+ burst itself is the major factor in mediating gene expression in response to the CaM antagonist.
Figure 3.
Scatterplot Representations of Transcriptome Changes in Seedlings Treated with W7 in the Presence or Absence of a Cytosolic Ca2+ Burst.
Microarray results of W7 versus DMSO (experiment A), W7 versus W7 pretreated with La3+ (W7 − Ca; experiment B), and W7 pretreated with La3+ (W7 − Ca) versus DMSO pretreated with La3+ (Control − Ca; experiment C). The results represent averages of three independent biological repeats. Logarithmic scale values are plotted as indicated, where the top and bottom green linear lines indicate 1.75-fold change in gene expression. Red indicates upregulation of gene expression, yellow indicates constant levels of gene expression, and blue indicates downregulation of gene expression.
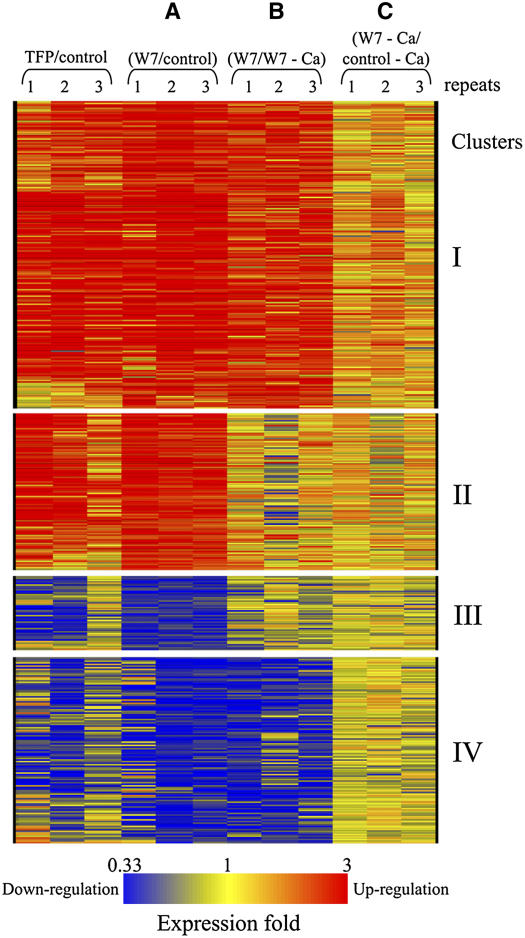
Statistical analysis of the microarray data was performed to find specific genes whose expression is reproducibly modulated by the cytosolic Ca2+ burst (see Methods). Overall, 287 upregulated and 161 downregulated ESTs were identified to be differentially expressed significantly within the set of experiments A, B, and C. k-means clustering (see Methods) produced four clusters (I to IV) as shown in Figure 4. Cluster I included 190 ESTs corresponding to 162 genes that are upregulated in experiments A and B but not in C (i.e., upregulated by W7 in association with the Ca2+ burst; see Supplemental Table 1 online for quantitative details). Cluster II included 97 ESTs representing 81 genes that are upregulated in experiment A but not in B and C (see Supplemental Table 2 online for quantitative details). Cluster III included 46 ESTs representing 38 genes that are downregulated in experiment A but not in B and C (see Supplemental Table 3 online for quantitative details). Cluster IV included 115 ESTs representing 68 genes that are downregulated in experiments A and B but not in C (i.e., downregulated by W7 in association with a Ca2+ burst; see Supplemental Table 4 online for quantitative details). Thus, clusters I and IV represent 230 Arabidopsis genes that are early Ca2+-responsive. However, we note that clusters II and III might represent both Ca2+-dependent and Ca2+-independent genes. As La3+ alone has an effect on gene expression (see, for example, the induction of the EARLY RESPONSIVE TO DEHYDRATION15 (ERD15) gene in Figure 5) (Polisensky and Braam, 1996), we cannot determine the role of the Ca2+ cytosolic transient for the genes of these two clusters.
Figure 4.
Scheme of Gene Clusters of Statistically Significant Differentially Expressed ESTs.
Clusters I to IV (see text) of the statistically significant ESTs are presented. Each column represents one of three independent biological repeats for each of the experiments indicated. Experiments A, B, and C were similar to those presented in Figure 3. TFP treatment was compared with DMSO as a control. Each row represents one EST, and colors represent changes in gene expression levels as indicated at bottom. Quantitative data for each EST of the four clusters appear in Supplemental Tables 1 to 6 online.
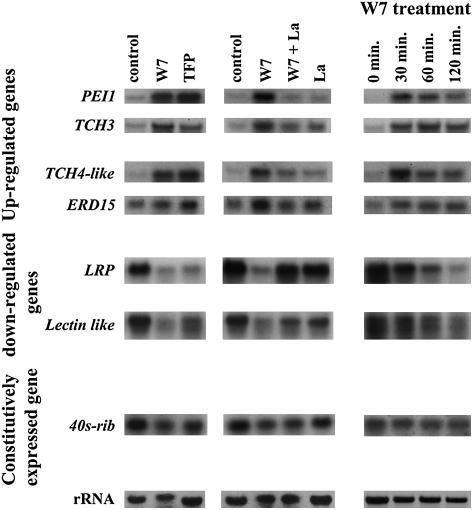
Figure 5.
RNA Gel Blot Expression Analysis of Genes Selected from Microarrays.
Arabidopsis seedlings were treated with DMSO as a control, TFP, W7, W7 + La3+ (W7 − Ca), or DMSO + La3+ (Control − Ca) for 1 h (left and middle panels) or with W7 for the indicated times (right panel). Four upregulated Ca2+-dependent genes, two downregulated Ca2+-dependent genes, and one constitutively expressed gene were probed as indicated. The 25S rRNA band served as an internal standard (bottom).
To further ascertain the specificity of the effect on gene expression mediated by the Ca2+ burst induced by CaM antagonists, W7-induced transcriptome changes were compared with the effect of TFP. Generally, TFP, like W7, had a significant effect on gene expression within 1 h of treatment (data not shown). Moreover, a significant linear correlation (r2 = 0.87) was found between the treatments for genes from clusters I to IV. Nonetheless, although the correlation between treatments was clear and highly significant, for 7.5% of the Ca2+-responsive genes a statistically significant difference was found between the two treatments. However, because W7 triggered a Ca2+ burst in cotyledons and TFP did not (Figure 1C), differences in gene expression between the two treatments were expected. Collectively, the microarray results indicate that a cytosolic Ca2+ burst triggered by CaM antagonists mediates major transcriptome changes of specific sets of upregulated and downregulated genes.
Direct Transcript Analysis Reveals Complex Temporal Changes
To assess the validity of the microarray results, several genes showing significant Ca2+-dependent changes in their expression levels were chosen for RNA gel blot analysis. These included four upregulated genes: PEI1 (At3g55980), an embryo-specific zinc finger protein (Li and Thomas, 1998); TOUCH3 (TCH3) (At2g41090), a touch-responsive CaM-like protein; TCH4-like (At1g65310), similar to a touch-responsive xyloglucan endotransglycosylase (Braam et al., 1997); and ERD15 (At2g41420), an early dehydration-responsive gene. Two downregulated genes were also tested: a lectin-like gene (At3g16420) and a gene encoding a light-regulated protein (LRP; At3g26740). As controls, we also probed the gene encoding a 40S ribosomal protein (At3g02560) that did not respond to the Ca2+ burst in the microarrays. Thus, our gene choice included three known stress-induced genes (TCH3, TCH4-like, and ERD15) and four genes that, to date, have not been associated with stress. We chose genes that were relatively highly induced (PEI1 and TCH3), moderately induced (TCH4-like), and weakly induced (ERD15). In addition, we also studied a gene expression time course after the addition of W7.
RNA gel blot results (Figure 5) confirmed that both TFP and W7 induced the expression of all four upregulated genes within 1 h of treatment. In addition, all tested genes show that the upregulation was Ca2+-dependent, as an apparent difference in the level of expression was found between W7-treated samples and samples pretreated with LaCl3 before W7 treatment (W7 − Ca). No significant differences in the levels of expression were found between LaCl3-pretreated seedlings that were treated with W7 (W7 − Ca) or DMSO (control − Ca). In addition, all of the genes tested responded to W7 within 30 min. However, although some of the genes (ERD15 and TCH3) maintained high levels of mRNA even after 120 min, others, like TCH4-like and PEI1, showed a decline in mRNA levels after reaching a peak high at ∼30 min after treatment. As expected, opposite results were obtained with the two downregulated genes, confirming that repression of gene expression is also dependent on the Ca2+ burst initiated by the CaM antagonists. The expression of the 40S ribosomal protein gene revealed no major changes in any of the treatments. Shorter (30 min) or longer (120 min) time of treatment with W7 did not affect the mRNA levels of the 40S ribosomal gene.
Functional Classification of the Early Ca2+-Responsive Genes
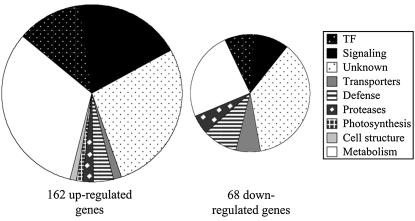
The Ca2+-responsive genes were classified according to their known or predicted physiological functions (see Supplemental Tables 5 and 6 online for details). A scheme of the gene classification is presented in Figure 6. Differences between the clusters of Ca2+-responsive upregulated and downregulated genes were observed. In general, the cluster of upregulated genes contains proportionally more transcription factors/posttranscriptional regulatory proteins and signaling-related genes. Some features are unique to the downregulated genes, including a high proportion of transporters, specifically aquaporins. The group of downregulated genes also includes a high proportion of defense-related genes, particularly peroxidases (for details, see Supplemental Table 6 online). Interestingly, photosynthesis-related proteins were almost absent from the two clusters, and very few transporters or channel genes, other than the aquaporins, were found to respond to Ca2+ within 1 h of treatment.
Figure 6.
Pie Chart Representing the Functional Classification of Ca2+-Responsive Genes.
Upregulated and downregulated Ca2+-responsive genes were classified according to their physiological functions (see Supplemental Tables 5 and 6 online) and represented as a pie chart. TF, transcription factors.
The upregulated genes (see Supplemental Table 1 online) include 19 genes encoding transcription factors and other expression regulators. The transcriptional regulators can be divided into several groups: ethylene-responsive transcription factors (ERFs), basic domain/leucine zipper (bZIP), MYB, B3, and zinc finger proteins. The most prominent groups of transcription factors were the zinc finger proteins, with nine responding to the Ca2+ burst, and ERFs, with three activated by the Ca2+ burst. In addition, the Ca2+ burst upregulated 31 genes encoding signaling proteins. Ca2+ binding proteins are expected to perceive the Ca2+ signal and transduce it to downstream effectors. Indeed, upregulation of five genes encoding CaM-related proteins was observed. CaM-like proteins were not the only Ca2+ binding protein genes to be induced by the Ca2+ burst. The expression of a Ca2+-dependent protein kinase was induced as well. Like CaM, CDPKs can translate Ca2+ signals into downstream effects, including gene expression (Sheen, 1996). The signaling-related genes also include 10 protein kinases and a protein phosphatase 2C. Finally, several other signaling-related proteins, such as RAB-GTPases, hormone-induced proteins, and CaM binding proteins, were activated as well by the Ca2+ signal. Overall, the Ca2+ burst induced a diverse set of signaling molecules. The downregulated genes include a few genes involved in cellular signaling and transcription regulation, including transcription factors from four different families (MYB, bZIP, bHLH, and zinc finger). Similarly, one protein, phosphatase 2C, and one protein kinase were downregulated. Importantly, our analysis revealed 45 upregulated and 21 downregulated genes whose functions are unknown.
Specific ABRE-Related cis Regulatory Elements Are Present in the Ca2+-Responsive Upregulated Genes
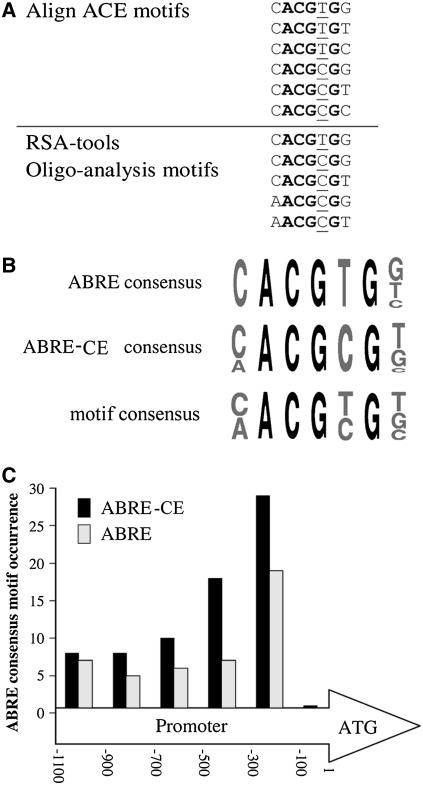
Microarray analysis provided a tool for the identification of several genes responsive to Ca2+. The significance of this can be extended beyond the number of genes identified to reveal regulatory elements, patterns, and gene networks using appropriate bioinformatic analysis. To search for regulatory elements, two unbiased computational analyses were performed. In the first, we examined the putative promoters of 162 of the upregulated genes in the region spanning nucleotides 100 to 600 upstream of their ATG translation initiation codons and searched for any repeating short sequence motifs (nine nucleotides or more) using AlignACE software (see Methods) (Tavazoie et al., 1999). A motif repeating 50 times was found, and the core seven nucleotides of its sequence represent six homologous oligonucleotides (Figure 7A). In an independent approach using the Oligo-analysis of the RSA-tools (see Methods) (van Helden et al., 1998), we searched for motifs that were overrepresented in the upregulated genes compared with their representation in the whole genome. This analysis revealed five homologous motifs (Figure 7A). Combination of the sequences found by the two independent approaches revealed the motif consensus of a cis element [(C/A)ACG(T/C)G(T/G/C)] related to ABRE motif consensus (Figure 7B).
Figure 7.
Potential Regulatory cis Elements in the Upregulated Ca2+-Responsive Genes.
(A) Repeated sequence motifs identified in the upstream regions of 162 Ca2+-responsive upregulated genes, using AlignACE software and Oligo-analysis of the RSA-tools.
(B) Schemes of consensus sequences of the ABRE-related motif derived from AlignACE software and Oligo-analysis.
(C) Distance distribution of the ABRE-related consensus motif along the regions upstream of the ATG initiation codons of the Ca2+-responsive upregulated genes presented as a frequency histogram.
The motif consensus includes both the classical ABRE (Figure 7B; containing the ACGTG core) and the ABRE-CE (Figure 7B; containing the ACGCG core) that were reported in some instances as functional equivalents to classical ABREs (Hobo et al., 1999; Choi et al., 2000; Zhang et al., 2005). The significance of the ABRE-related motifs found in the putative promoters of the Ca2+-responsive genes was assessed by two more computational tests. The spatial distribution of the ABRE-related motifs was tested in the 162 upregulated Ca2+-responsive genes (Figure 7C). This analysis revealed a strong tendency of the ABRE motifs to be localized within 100 to 500 nucleotides upstream of the ATG translation initiation codon and to be almost absent within the 100 nucleotides immediately upstream of the ATG codon. In addition, representation of the motif was compared with its occurrence in the Ca2+-responsive downregulated genes (cluster IV in Figure 4). The results show that overrepresentation of the ABRE motifs occurs exclusively in the Ca2+-responsive upregulated genes with very high statistical significance (Table 1) (P < 10−13). An unrelated cis element of the sequence CACATG, which is important in the early response to cold (ICEI; Chinnusamy et al., 2003) and dehydration (RD22bp1; Abe et al., 1997), was not significantly overrepresented in any of the clusters (Table 1).
Table 1.
Statistical Analysis of the Occurrence of Motifs within 100 to 600 Nucleotides Upstream of the ATG Translation Initiation Codon of Ca2+-Responsive Genes
| Ca2+-Responsive Downregulated Genes (Cluster IV, 66 Genes)
|
Ca2+-Responsive Upregulated Genes (Cluster I, 162 Genes)
|
||||
|---|---|---|---|---|---|
| Motif | Motif Sequence | Motif Occurrence [Observed (Expected)] | P | Motif Occurrence [Observed (Expected)] | P |
| ABRE | CACGTG(T/C/G) | 7 (7.0) | 0.55 | 30 (17.2) | 3.1 × 10−3 |
| ABRE-CE | (C/A)ACGCG(T/C/G) | 5 (5.5) | 0.65 | 52 (13.5) | 1.1 × 10 −15 |
| Consensus | (C/A)ACG(C/T)G(T/C/G) | 18 (19.1) | 0.63 | 109 (46.9) | 5.0 × 10 −13 |
| Rd22bp and ICE1 unrelated cis element | CACATG | 14 (15.4) | 0.70 | 37 (37.9) | 0.62 |
An ABRE cis Element Is Sufficient to Confer Transcriptional Activation in Response to a Cytosolic Ca2+ Transient
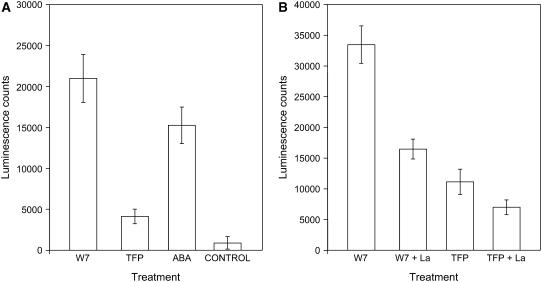
Identification of ABRE-related sequences as potential Ca2+-responsive cis elements prompted us to assess the responsiveness of an ABRE element to cytosolic Ca2+ transients in intact Arabidopsis seedlings. Considering previous observations that the response to ABA requires multiple ABRE-related sequences (multiple ABREs or ABRE with an ABRE-CE) (Hobo et al., 1999; Zhang et al., 2005), we tested a tetramer of the ABRE motif ACACGTGGC fused to the −70 cauliflower mosaic virus (CaMV) 35S promoter to drive the expression of luciferase (see Methods) in transgenic Arabidopsis seedlings. At first, we confirmed that the expression of the reporter responds to ABA (Figure 8A). If the same regulatory element responds to a cytosolic Ca2+ transient, we expected to detect enhanced luciferase expression in response to treatments with W7 and TFP, which would be significantly reduced by La2+ pretreatment. Indeed, both W7 and TFP treatments resulted in significantly enhanced luciferase activity, although the response to W7 was much stronger (Figures 8A and 8B). Importantly, La2+ significantly inhibited the response to W7 and TFP, suggesting that activation of luciferase driven by the ABRE cis element is mediated by a Ca2+ transient. Control transgenic plants with the full-length CaMV 35S promoter upstream of luciferase showed no activation by W7 or TFP (data not shown). These results suggest that a tetramer of the ABRE cis element mediates Ca2+-induced transcription activation.
Figure 8.
Luciferase Assay for Measuring the Ca2+-Responsiveness of the ABRE Motif.
The data represent average luminescence counts emitted over a 6-h period after treatment from each well containing 10 seedlings. Counts were recorded using an intensified CCD camera.
(A) Average luminescence emitted after the addition of 600 μM W-7, 150 μM TFP, 0.3% DMSO (control), or 100 μM ABA. Error bars indicate the se for six replicate well treatments.
(B) Average luminescence emitted after the addition of 600 μM W-7 or 150 μM TFP with and without lanthanum pretreatment. Plants were pretreated with LaCl3 at a concentration of 0.35 mM for 10 min before the addition of the appropriate CaM agonist. Error bars indicate the se for five replicate well treatments.
DISCUSSION
CaM Antagonists Trigger a Cytosolic Ca2+ Burst That Is Specifically Blocked by Lanthanides
In this study, four CaM antagonists representing three chemically unrelated compounds triggered a similar, although not identical, [Ca2+]cyt response in time and shape. Differences in the timing of the maximal peak could be attributed to differences in cell permeability or to some target specificity of the different compounds. Such differences are reflected in the photon-imaging analysis of seedlings treated with W7 compared with TFP, in which only W7 triggered a [Ca2+]cyt response in cotyledons. Nevertheless, in roots and hypocotyls, W7 and TFP seem to deplete the same Ca2+ pools or to affect the same targets, as pretreatment with one completely abolished the effect of the other. Moreover, most of the genes that responded to TFP and W7 are the same. Therefore, overall, it is likely that the various CaM antagonists have similar targets, which produce similar effects. In addition, the CaM antagonists W7 and TFP had similar effects on evoking a [Ca2+]cyt burst in N. tabacum and Arabidopsis seedlings, suggesting that a Ca2+ response triggered by CaM inhibition is common to many plants and that similar targets for the CaM antagonists appear in other plant species. The differences between the effectiveness of W7 and that of its less potent analogue W5 suggest that inhibition of CaM per se, or a CaM-like protein, or a protein containing a CaM-like domain, leads to the Ca2+ burst. We note that the concentrations needed to reach a half-maximal [Ca2+]cyt burst (IC50) for W7 and TFP were approximately fivefold higher than the known IC50 for CaM inhibition in mammalian cell cultures. This could be attributed to the fact that our experiments were performed on an intact organism (seedlings). Probably, the concentrations needed at the cellular level would be substantially lower. Furthermore, as CaM antagonists may interact with different CaM-like proteins and with proteins with CaM-like domains, the precise nature of the target(s) that interact with CaM antagonists to evoke the Ca2+ burst remains unknown. For instance, a putative Ca2+-permeable channel with an EF-hand calcium binding domain (Peiter et al., 2005) is an example of a possible target.
Lanthanum and gadolinium chloride, known as blockers of Ca2+-permeable channels and of nonselective cation channels, completely inhibited the Ca2+ burst mediated by the CaM antagonists W7 and TFP, indicating that the Ca2+ burst is mediated by a Ca2+ flux through Ca2+ channels. By contrast, verapamil, a blocker of voltage-activated Ca2+ channels, had no significant effect on the Ca2+ burst, suggesting that a specific set of plant Ca2+-permeable channels mediates the Ca2+ burst in response to CaM antagonists. In addition, potassium channel blockers did not affect the Ca2+ burst triggered by CaM antagonists. However, it is important to note that although Ca2+ flux through Ca2+-permeable channels is certainly required for the [Ca2+]cyt burst, we cannot exclude a role for Ca2+-ATPases in this process, as suggested by Gilroy and colleagues (1987). Finally, the rapid response to CaM antagonists (within seconds) and the blockage by lanthanides suggest that the modulated Ca2+ channels are in the plasma membrane. However, the involvement of channels in endomembranes cannot be ruled out.
[Ca2+]cyt Transients Have a Profound Effect on the Plant Transcriptome That Is Associated with Responses to ABA and Environmental Stress
Although the induction of the cytosolic Ca2+ burst was artificial, it allowed us to identify genes that respond to a rapid cytosolic Ca2+ burst in the absence of an applied environmental stress. Moreover, it is not unlikely that Ca2+ transients evoked by CaM antagonists mimic natural mechanisms that regulate CaM function in vivo. For example, posttranslational modifications of CaM (e.g., phosphorylation and ubiquitination) may affect its functions (Parag et al., 1993; Benaim and Villalobo, 2002), leading to Ca2+ entry into the cytosol.
We applied a novel approach to associate a specifically induced [Ca2+]cyt burst with rapid transcriptome changes. In this approach, W7 and TFP treatments resulted in the upregulation or downregulation of several hundred Arabidopsis genes. Results of control transcriptome profiles (±W7, ±TFP, ±La3+ in various combinations) suggest that the activation of the majority of these genes could be traced to the Ca2+ burst per se. The number of Ca2+-responsive genes identified in our study (162 upregulated and 68 downregulated genes) is much larger than the number of genes reported to date to respond to Ca2+ transients in plants and is based on a microarray chip representing ∼25% of the Arabidopsis genome (see Methods). Thus, in the whole genome, there might be ∼650 (∼2.3% of the genome) upregulated genes and 280 (∼1.0% of the genome) downregulated genes responding to a similar cytosolic Ca2+ burst. Interestingly, whole genome–based transcriptome analysis of human T lymphocyte cells (Feske et al., 2001) from patients with defective transmembrane Ca2+ influx compared with normal cells revealed similarities with our findings. The results of the T lymphocyte study showed that Ca2+ is necessary for both the activation and the repression of genes, that more genes were upregulated than downregulated, and that ∼2.1% of the genes were responsive to Ca2+.
A striking feature of the upregulated genes is that they include many known early stress-induced genes (Table 2). Among those are the early touch-induced genes (TCH2, TCH3, and TCH4-like), cold-induced genes (COR47 and KIN2), and dehydration early-responsive genes (ERD1, ERD6, ERD10, ERD13, ERD14, and ERD15). Compared with previously reported stress-responsive genes, mainly by Seki et al. (2002a), who reported on salt-, drought-, cold-, and ABA-responsive genes of Arabidopsis based on microarrays, 25% of the Ca2+-responsive upregulated genes that we identified were induced by at least one of these stresses and/or ABA (Table 2). Twenty-seven of those (∼17% of the total Ca2+-dependent upregulated genes) were reported to be induced by more than one stress (Table 2).
Table 2.
Ca2+-Responsive Upregulated Genes That Are Induced by Stress
| Locus | Description | Drought/Dehydration | Salt | Cold | ABA/Embryo-Specific | Touch |
|---|---|---|---|---|---|---|
| Transcription factors and DNA/RNA-related proteins | ||||||
| At4g17500 | ERF-1, ethylene-responsive element binding factor1 | 1 | 1 | 1 | ||
| At3g15210 | ERF4, ethylene-responsive element binding factor4 | 4 | 4 | 4 | ||
| At1g42990 | bZIP family transcription factor | 1 | 1 | |||
| At1g27730 | ZAT10, salt-induced zinc finger protein | 1 | 1 | 1 | 1 | |
| At1g08930 | ERD6, zinc finger protein ATZF1 | 1, 2 | ||||
| At2g28840 | Putative RING zinc finger ankyrin protein | 1 | ||||
| At2g40140 | CCCH-type zinc finger protein containing an ankyrin repeat | 1 | ||||
| At3g55980 | PEI1, CCCH zinc finger transcription factor | 5 | ||||
| Signaling-related proteins | ||||||
| CaM-related | ||||||
| At2g41110 | CAL4 | 9 | ||||
| At2g41100 | CAL5-TCH3-Ca–related protein | 1 | 1 | 1 | 1, 3 | |
| At5g37770 | CaM-related protein2 (TCH2) | 3 | ||||
| At5g42380 | CaM-related protein | 1 | 1 | |||
| Kinases | ||||||
| At3g45640 | Mitogen-activated protein kinase3 | 6 | 6 | 6 | ||
| At3g59350 | Protein kinase, Pto kinase interactor1 | 1 | 1 | 1 | 1 | |
| Others | ||||||
| At1g09070 | Similar to Gly SRC2 | 8 | ||||
| At5g15970 | Cold-regulated protein COR6.6 (KIN2) | 1 | 1 | 1 | ||
| At1g20440 | COR47-ERD10 | 1, 2 | 1 | 1 | 1 | |
| At2g41430 | ERD15 | 2 | ||||
| At1g35140 | phi1, phosphate-induced protein | 7 | ||||
| At2g17840 | Putative senescence-associated protein12 | 1 | 1 | 1 | 1 | |
| At1g76180 | Dehydrin, putative ERD14 | 2 | ||||
| Cytoskeleton and cell structure | ||||||
| At2g32240 | Myosin heavy chain–related | 1 | ||||
| Defense- and PCD-related genes | ||||||
| At2g40000 | Nematode resistance protein–related | 1 | 1 | 1 | ||
| At5g06320 | NDR1/HIN1-like protein3 (NHL3) | 1 | 1 | |||
| Proteases | ||||||
| At4g02380 | ERD1-SAG21-clp protease | 1, 2 | ||||
| Cellular metabolism and others | ||||||
| At5g42650 | AOS, allene oxide synthase, JA biosynthesis | 1 | ||||
| At1g65310 | Xyloglucan endotransglycosylase, TCH4-like | 3 | 3 | |||
| At1g62660 | Glycosyl hydrolase family 32, identical to β-fructosidase | 1 | ||||
| At3g55430 | Glycosyl hydrolase family 17, β-1,3-glucanase | 1 | 1 | |||
| At5g64310 | AGP1, arabinogalactan protein | 1 | 1 | 1 | ||
| At1g03220 | Extracellular dermal glycoprotein (EDGP) | 1 | ||||
| At1g78850 | Curculin-like (mannose binding) lectin family | 1 | ||||
| At2g22500 | Mitochondrial carrier protein family | 1 | ||||
| At4g24570 | Mitochondrial carrier protein family | 1 | 1 | |||
| At2g30870 | GST-ERD13 | 2 | ||||
| Unknown genes | ||||||
| At2g23120 | Unknown protein | 1 | 1 | 1 | 1 | |
| At1g19180 | Unknown protein | 1 | 1 | |||
| At1g30360 | Unknown protein | 1 | ||||
| At1g69890 | Unknown protein | 1 | 1 | |||
| At2g41640 | Unknown protein | 1 | 1 | 1 | ||
| At4g39670 | Unknown protein | 1 | 1 | |||
| At5g42050 | Unknown protein | 1 | 1 |
Numbers indicate references for induction by specific stress, as follows: 1, Seki et al. (2002a); 2, Taji et al. (1999); 3, Braam et al. (1997); 4, Fujimoto et al. (2000); 5, Li and Thomas (1998); 6, Mizoguchi et al. (1996); 7, Sano and Nagata (2002); 8, Takahashi and Shimosaka (1997); 9, Ito et al. (1995).
Fold induction in experiment B, W7 versus W7 − Ca (W7 + La). If more than one EST on the chip represented one locus, a representative result for one of the ESTs is presented.
ABREs as Ca2+-Responsive cis Elements
Our in vivo experiments with Arabidopsis seedlings indicate that at least the classical ABRE motif in a tetrameric configuration may function as a Ca2+-responsive cis element. Computational analysis of upstream regions of the Ca2+-responsive upregulated genes revealed both the ABRE motif and the related ABRE-CE motif. Interestingly, one ABRE by itself is not sufficient to confer ABA-responsiveness. On the other hand, two or more ABRE-related motifs within the same promoter or one ABRE together with an ABRE-CE appear to form active ABA-responsive units (Hobo et al., 1999; Zhang et al., 2005). Indeed, many of the Ca2+-responsive upregulated genes found in our study contained more than one copy of ABRE-CE, classical ABRE, or a combination of both (Table 3). Of the 75 genes containing these elements, 15 contain the ABRE coupling motif at least twice, 9 contain the classical ABRE motif at least twice, and 13 contain both the ABRE-CE and classical ABRE motifs. A total of 29 (38.7%) of the ABRE-containing genes have at least two copies of ABRE motifs. As the activation of genes by ABRE motifs requires more than one motif in each promoter, the sizable multiple elements in our data are another indication of their functionality. Interestingly, at the whole genome level, we found ∼2400 genes containing multiple ABREs (classical ABRE or ABRE-CE). Zhang et al. (2005) have shown that two ABRE-related motifs in the same promoter can correctly predict ∼40% of ABA- or abiotic stress–responsive genes. Hence, we predict that at the whole genome level of Arabidopsis, there are ∼1000 genes that are Ca2+-responsive.
Table 3.
Ca2+-Responsive Upregulated Genes Containing Multiple ABRE or ABRE-CE Motifs in Their Promoters
| Locus | Description | ABRE Motifs Found within Promotersa | ABRE-CE Motif Found within Promotersa |
|---|---|---|---|
| At5g37770 | CALMODULIN-RELATED PROTEIN2 (TCH2) | 124, 143 | |
| At3g56880 | Unknown protein | 204, 234 | |
| At1g03730 | Unknown protein | 213, 257 | |
| At5g64310 | AGP1, arabinogalactan protein | 252, 328, 347, 443 | |
| At1g59820 | Similar to potential phospholipid-transporting ATPase | 338, 374, 392, 406 | |
| At5g11740 | AGP15, ARABINOGALACTAN PROTEIN | 360, 410 | |
| At1g62870 | Unknown protein | 375, 359 | |
| At3g51550 | Receptor-protein kinase | 382, 625 | |
| At1g52890 | Unknown protein | 795, 858 | |
| At2g45820 | Remorin, a nonspecific DNA binding protein | 240 | 140, 742, 771 |
| At5g03210 | Unknown protein | 351 | 175, 539 |
| At1g59870 | ABC transporter family protein | 495 | 359, 375 |
| At1g70090 | Glycosyltransferase family 8 | 546 | 15, 201, 219, 935 |
| At1g73540 | Similar to diphosphoinositol polyphosphate phosphohydrolase | 641 | 176, 504, 549 |
| At1g19180 | Unknown protein | 284, 342 | 666, 816 |
| At3g07180 | Unknown protein | 171 | 155 |
| At2g45300 | 5-Enolpyruvylshikimate-3-phosphate (EPSP) synthase | 263 | 950 |
| At2g23810 | Unknown protein | 525 | 387 |
| At2g41430 | ERD15 | 602 | 611 |
| At4g25030 | Unknown protein | 676 | 793 |
| At1g47380 | Protein phosphatase 2C (PPTC) | 919 | 720 |
| At1g42990 | bZIP family transcription factor | 317, 158 | 174 |
| At5g59550 | COP1-interacting protein CIP8 | 107, 171, 251 | |
| At5g15970 | Cold-regulated protein COR6.6 (KIN2) | 128, 146, 219, 440 | |
| At5g65630 | Unknown protein | 162, 431 | |
| At1g74950 | Unknown protein | 255, 860 | |
| At1g76180 | Dehydrin, putative, ERD14 | 275, 845 | |
| At2g05710 | Cytoplasmic aconitate hydratase | 647, 932 | |
| At1g72450 | Unknown protein | 904, 941 |
Numbers indicate distance in nucleotides upstream of ATG.
However, further investigations are required to elucidate the relevance of the association of ABREs with ABRE-CE sequences with respect to responsiveness to Ca2+. Moreover, based on our investigation, we cannot suggest that every combination of cis elements that responds to ABA also responds to Ca2+. In addition, because our transcriptome analysis was based on a cytosolic Ca2+ burst evoked by a specific CaM antagonist, there are likely many other genes that respond to different patterns of cellular Ca2+ transients, which may be transduced to the transcription machinery via other types of Ca2+-responsive cis elements. We also note that the ABRE and ABRE-CE may respond to various signals in a Ca2+-independent manner by interaction with different transcription factors.
Ca2+ Signal Transducers and Ca2+-Responsive Transcription Factors
Calcium binding proteins are expected to transduce the transient cytosolic Ca2+ burst to downstream effectors, including kinases and transcription factors. One of the intriguing puzzles is the mechanism underlying stimulus–response specificity. Our study focused on genes whose expression was enhanced within 1 h of the cytosolic Ca2+ transient. Moreover, kinetic studies of representative Ca2+-responsive genes (Figure 5) showed that some of the Ca2+-responsive genes reached their maximal expression levels within 30 min (or less) after stimulus (e.g., the PEI1 gene). Therefore, we expect that some Ca2+-responsive proteins, which are already present at the time of the Ca2+ pulse, are responsible for mediating the apparent rapid gene activation. The major known families of Ca2+ transducers in plant cells are (1) calcineurin-B–like proteins (CBLs) (Kim et al., 2000; Luan et al., 2002) together with their interacting kinases (CIPKs; CBL-interacting kinases) (Liu and Zhu, 1998; Luan et al., 2002), (2) CaMs (Bouché et al., 2005), and (3) CDPKs (Harper et al., 2004). However, none of the CBLs represented on the chip were upregulated in our study. Moreover, induction of CBL1 by cold and salt requires at least 2 h (Kudla et al., 1999). CIPKs play an important role in ABA stress and Ca2+-signaling and are themselves induced by ABA and stress signals (Kim et al., 2003). CIPK3, for example, is induced by cold, salt, ABA, drought, and wounding (Kim et al., 2003) and modulates the expression of genes that are induced by salt and cold but not by drought (Kim et al., 2003). However, induction of CIPK3 in response to salt and cold occurs within 3 h (Kim et al., 2003). Interestingly, five CaM-related genes were rapidly induced by the calcium burst in our study. Among them were TCH2, TCH3, and CAL4 (Table 2), all of which were previously found to be rapidly induced by various stresses (Ito et al., 1995; Polisensky and Braam, 1996). In addition, one CDPK was also upregulated by the Ca2+ burst. Therefore, we speculate that more than one mechanism is involved in the gene expression responses observed in our study, which is consistent with the different kinetics of gene activation we observed by RNA gel blot analysis (Figure 5).
Another open question concerns the identity of the transcription factors that respond to the rapid transient Ca2+ signals and that subsequently activate gene expression through ABRE and ABRE-CE cis elements. Some bZIP transcription factors are known to bind with high affinity to the G-box ABRE motifs (ABFs) and with lower affinity to motifs containing the ABRE-CE sequence (e.g., ABF1 [Choi et al., 2000] and TRAB1 [Hobo et al., 1999]). Consequently, Choi et al. (2000) speculated that factors other than ABF must bind to the ABRE-CE motif with higher affinity. Interestingly, the ABRE-CE core sequence ACGCG(G/T/C) matches the major consensus binding site of a recently discovered family of calcium-dependent CaM binding transcription activators designated CAMTAs (or SRs) ( Bouché et al., 2002; Yang and Poovaiah, 2002; Choi et al., 2005). Furthermore, Mitsuda et al. (2003) showed that a single ABRE-CE sequence can mediate gene activation by CAMTA in plant cells. A rice (Oryza sativa) CAMTA (CBT) binds not only to the ABRE-CE motif but also to the ABRE G-box sequence (Choi et al., 2005), and although the rice CAMTA functions as a transcription activator in vivo, coexpression of CaM negatively regulates transcription by CAMTA (Choi et al., 2005). Thus, CAMTAs bind in vitro to ABRE-CE and ABRE sequence motifs and activate or repress gene expression in vivo through these cis elements. However, the role of CAMTAs in stress responses and in ABA and Ca2+ signaling remains to be elucidated. Finally, one should also consider the possible occurrence of transcription factors that directly respond to Ca2+ signals, such as the mammalian DREAM protein (Carrion et al., 1999). In this case, Ca2+ binding to an EF-hand motif of the transcription factor causes its release from its DNA binding site, thus alleviating the suppression of gene expression (Ikura et al., 2002). However, transcription factors of this nature have not yet been characterized in plants. All in all, there is a plethora of calcium-responsive proteins in plants that could potentially be involved in the transduction of cytosolic calcium signals to the transcription machinery (also see Ikura et al., 2002; Reddy and Reddy, 2004), and further multidisciplinary studies are required to elucidate the mechanisms underlying stimulus–response specificity.
In summary, by simple and specific modulation of cytosolic Ca2+ using CaM antagonists and Ca2+ channel blockers, we were able to identify early Ca2+-responsive upregulated genes, which include many stress- and signaling-related genes. In addition, we show that this Ca2+ burst is sufficient to induce either the activation or the repression of plant genes, allowing us to identify many new genes that are responsive to cytosolic Ca2+ transients. Our bioinformatic analysis identified an ABRE-related motif as a potential Ca2+-responsive cis element, which was confirmed experimentally in intact Arabidopsis seedlings. In conclusion, the results presented here should facilitate the understanding of the mechanisms that link Ca2+ signaling with the transcriptional machinery in plants.
METHODS
Plant Materials, Growth Conditions, and Transgenic Plant Construction
Arabidopsis thaliana plants (Wassilewskija ecotype), expressing the aequorin reporter gene (Knight et al., 1991), were used for all experiments. Aequorin cDNA was cloned into a Ti plasmid, under the transcriptional regulation of the CaMV 35S promoter, with a CaMV-35S transcription terminator sequence and containing a hph gene for hygromycin resistance in plants. Plants were transformed with Agrobacterium tumefaciens strain GV3101-PM90RK essentially as described (Clough and Bent, 1998). Transgenic plants were selected on agar plates supplemented with 8 mg/L hygromycin. Seeds were sown on 0.8% agar (purchased from Duchefa) plates, and plants were grown under white light in a 16-h-light/8-h-dark cycle at 21°C.
Microarray Analysis
Total RNA was extracted using the RNeasy Midi kit (Qiagen), and 100 μg of RNA was subjected to reverse transcription reaction. cDNA products were then labeled with Cy3 and Cy5 by the indirect amino-allyl method. Hybridization was performed on slides containing 12,000 Arabidopsis ESTs (Keck Biotechnology Resource Laboratory, Yale University) (Zik and Irish, 2003). Three biological repeats were performed for each experiment, and hybridization with swapped dye-labeling was performed for one sample per experiment. Separate images were acquired using ScanArray 4000 software (Packard BioScience) for each fluorescence level at a resolution of 10 μm per pixel, adjusting the photomultiplier and laser power to achieve an optimal distribution of signals without minimal saturation. Image analysis was performed using QuantArray version 3 software (Perkin-Elmer Life Sciences). Spots were quantified using the fixed-circle method, measuring the mean of pixels encompassing the spot and subtracting the local background areas.
Clustering and Statistical Analysis of Microarray Results
Data analysis was performed with GeneSpring 5.0 software (Silicon Genetics) applying per spot and per chip: intensity dependent (lowess) normalization. Average fold induction values were calculated and are presented with se values in the supplemental tables online. The following clustering procedure was done for screening purposes. To filter relevant clones, ESTs that did not respond to W7 (experiment A fold change < 1.5) were omitted. ESTs with P < 0.05 based on the deviation from 1 error model and with upregulation or downregulation > 1.75-fold in one of the experiments (A, B, or C) were taken for further analysis. One-way analysis of variance was then applied, and ESTs with P < 0.05 were clustered into two steps by k-mean clustering using smooth correlation from the GeneSpring software. The first step involved clustering into two groups of upregulated and downregulated ESTs followed by subclustering of the latter two groups into two more groups to get the four clusters presented.
Chemical Treatment of Plants
W7, W5, LaCl3, GdCl3, and CsCl were purchased from Sigma-Aldrich. All other inhibitors or pharmacological agents were purchased from Calbiochem. Unless indicated otherwise, seedlings were placed in a basic macroelements medium [100 μM (NH4)2HPO4 600 μM KNO3, 400 μM Ca(NO3)2, and 200 μM MgSO4]. For aequorin measurements, pretreatment of all pharmacological agents was done 15 min before the addition of CaM inhibitors. Final concentrations, unless stated otherwise, were as follow: LaCl3, 0.5 mM; GdCl3, 1 mM; verapamil chloride, 1 mM; CsCl, 2 mM; tetraethylammonium, 5 mM. For calmidazolium chloride and SKF-7171 treatments, the basic macroelements medium was replaced by distilled water to prevent precipitation. In all experiments, CaM antagonists were dissolved in 0.1% (v/v) DMSO. Other chemicals were dissolved in water.
In Vivo Reconstitution of Aequorin, and Ca2+-Dependent Luminescence Measurements
Luminescence measurements were performed using a luminometer consisting of a 9829A photomultiplier tube with a 1.5-kV potential from a FACT50 air-cooled thermoelectric housing and an AD2 amplifier/discriminator (all from Thorn EMI) to produce a numerical output that was stored on a personal computer. Reconstitution of aequorin in vivo was performed overnight with 12-d-old Arabidopsis seedlings placed in basic macroelement medium supplied with 10 μM coelenterazine. Eight washed seedlings were used for each luminometric experiment. Real-time measurements of aequorin luminescence were taken throughout the experiment every 1 s. To convert luminescence into Ca2+ concentrations, 2 M CaCl2 and 30% ethanol were added to discharge the remaining aequorin. Calculations of Ca2+ concentrations were performed according to Knight and Knight (2000). For Ca2+ imaging, reconstitution of aequorin was performed in vivo essentially as described previously by floating seedlings on water containing 10 μM coelenterazine in the dark overnight at 20°C. Aequorin imaging was performed using an intensified CCD camera (model EDC-02) (Campbell et al., 1996), with a camera control unit (HRPCS-2) and image acquisition and processing software (IFS216), all from Photek, as described previously (Knight and Knight, 2000).
RNA Gel Blot Analysis
Total RNA was extracted using the RNeasy Midi kit (Qiagen). Ten micrograms of total RNA was separated by gel electrophoresis and blotted onto a Hybond-XL (Amersham) nylon membrane. Probe labeling with [32P]dCTP was done using Rediprime II (Amersham), and prehybridization and hybridization were done with Rapid-Hyb (Amersham) buffer according to the manufacturer's protocol. Products of RT-PCR amplification on Arabidopsis RNA using specific primers were used as probes, as follows: TCH3 (At2g41100), forward (5′-TTGTACCTAATGGCTAAGAATCAAGGTCA-3′) and reverse (5′-AAACCTATCTTTCATGAAGTCGGAAATTG-3′); ERD15 (At2g41430), forward (5′-CGATGGTATCAGGAAGACGATCTACTCTA-3′) and reverse (5′-CACTTGATTTCTTAACCATCTCACCATTC-3′); TCH4 (At1g65310), forward (5′-ACTAATGTTTACACAAAAGGCACAGGAGA-3′) and reverse (5′-CCTTCTTTTATCGGTGCAATAGTTGTAGA-3′); PEI1 (At3g55980), forward (5′-GAAGGGGTTTATGGAAGTGATGAGTTTAG-3′) and reverse (5′-CCTGTAGTCTCTAGCTGCGTTATTGTTGT-3′); LRP (At3g26740), forward (5′-GGGAGCTCTGTTTATCAAACCAACTATTC-3′) and reverse (5′-TTTTGGGGCTGTTATACTCTTCGTATTCT-3′); Lectin-like (At3g15420), forward (5′-CAATCTTAATTCTCTTGGGGCTTACTTTG-3′) and reverse (5′-GGAGACGTTTGCTTATTAGTCTTGAACCT-3′); 40S-rRNA (At3g02560), forward (5′-AGCTTGAGAAGAAGTTCAGTGGAAAAGAT-3′) and reverse (5′-AGCTTCTATGACTGGGTACTCGAAAACT-3′).
Promoter Analysis
To retrieve upstream sequences of promoters, we used the RSA-tools Web interface (http://rsat.ulb.ac.be/rsat/). A Linux version of the AlignACE package (Tavazoie et al., 1999) was used to search for short sequence motifs in the defined upstream regions (100 to 600 nucleotides upstream of ATG translation initiation codon). The GC content value was set to 0.32, matching the GC content in the nontranslated regions of Arabidopsis. The minimal motif length was set to nine, and oligonucleotides representing the seven-nucleotide core of the motif found are presented. The same upstream regions were searched online with the Oligo-analysis software (http://rsat.ulb.ac.be/rsat/) of RSA-tools (van Helden et al., 1998) for overrepresentation of oligonucleotides that are six nucleotides long. Computation of occurrence P values (P ≥ n occurrences) was performed according to van Helden et al. (1998) with a reference database representing 100 to 600 nucleotides upstream of the ATG codons of the Arabidopsis genes in the whole genome.
Luciferase Assay
An [ABRE]4:LUC chimeric gene was produced, consisting of four copies of the ABRE motif fused to a minimal −70 CaMV 35S promoter to drive a modified firefly luciferase coding sequence (Mankin et al., 1997) and ending with a nopaline synthase transcriptional terminator sequence. A pair of oligonucleotide primers, 5′-GCGCAAGCTTACACGTGGCACACGTGGCACACGTGGCACACGTGGCGATATCTCCACTGACGTAAG-3′ and 5′-GCTGCAGGAATTCCCGATCT-3′, was used to amplify this construct from the plasmid pLuk07 (Mankin et al., 1997) by polymerase chain reaction and to introduce HindIII and EcoRI restriction sites at the 5′ and 3′ ends of the gene, respectively. The whole chimeric construct was then transferred to the Agrobacterium binary vector pBIN19 (Bevan, 1984) as a HindIII-EcoRI insert. The resulting plasmid was purified from Escherichia coli and then transferred to Agrobacterium, and plants were transformed exactly as described previously (Knight et al., 2004). Expressing lines were identified using in vitro assays of luciferase activity and the luciferase assay system (Promega), and ABA inducibility was confirmed. Wild-type Columbia plants expressing the [ABRE]4:LUC construct were grown as described previously (Knight et al., 1996) on full-strength Murashige and Skoog (1962) medium containing 100 mg/L kanamycin sulfate and 0.8% (w/v) agar, under a 16-h-light/8-h-dark cycle, and maintained at a constant temperature of 20°C. Eight-day-old seedlings were reconstituted overnight in 1 mM luciferin (monopotassium salt; Melford Laboratories) in aqueous solution, floating in 24-well bored metal plates. Each well contained 10 seedlings, floating on 1 mL of luciferin solution. Plates were covered with a sealed Perspex lid and wrapped in foil to prevent the entry of light or the loss of liquid by evaporation. Dishes were incubated at 20°C overnight. After this overnight incubation, 0.5 mL of CaM agonist or control solution was added to each 1-mL well with mixing, to give final concentrations of 600 μM W-7, 150 μM TFP, and 100 μM ABA. The original stock solutions used were 200 mM W-7 and 50 mM TFP in DMSO. Dilution of these stock solutions to the degree described above resulted in a final concentration of 0.3% DMSO; therefore, this concentration was used for the DMSO control treatment. An ABA stock solution of 100 mM in ethanol was diluted to give a final concentration of 100 μM ABA. A separate experiment confirmed that this concentration of ethanol (0.1% ethanol) had no effect on luciferase counts (data not shown). Experiments were performed on five or six replicate wells per treatment and repeated twice.
The multiwell dish was placed immediately under an intensified CCD camera (Photek) (Knight et al., 1999), and luciferase counts were recorded for 6 h. In the experiments in which a lanthanum pretreatment was used, 100 μL of freshly made lanthanum chloride solution (3.5 mM) was added to seedlings in 900 μL of luciferin solution and incubated at room temperature for 10 min. After this, CaM agonists were added as described above and imaging commenced immediately. A separate experiment confirmed that the agonists had no effect on luciferase emission from 35S:LUC-expressing plants (data not shown).
Accession Numbers
The complete expression data set is available as accession numbers GSM127605, GSM128426 to GSM128436, and GSE5517 in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. W7-Induced Calcium-Responsive Upregulated ESTs.
Supplemental Table 2. W7-Induced Calcium-Independent Upregulated ESTs.
Supplemental Table 3. W7-Repressed Calcium-Independent Downregulated ESTs.
Supplemental Table 4. W7-Repressed Calcium-Responsive Downregulated ESTs.
Supplemental Table 5. Classification of Calcium-Responsive Upregulated Genes.
Supplemental Table 6. Classification of Calcium-Responsive Downregulated Genes.
Supplemental Figure 1. Measurements of Plant [Ca2+]cyt in Response to Treatments with Different Calmodulin Antagonists.
Supplemental Figure 2. Comparison of the Arabidopsis [Ca2+]cyt Burst in Response to W7 and Its Weaker CaM Antagonist Analog W5.
Supplementary Material
Acknowledgments
We thank Tony Trewavas for kindly providing the aequorin cDNA and Gideon Baum for advice on establishing the aequorin-based Ca2+ monitoring system. We thank Nava Moran and Aliza Finkler for fruitful discussion and critical review and Ron Ofir for assisting in analyzing microarray results. This research was supported by BARD (the U.S.–Israel Binational Agricultural Research and Development Fund) Grant IS-2971-98 and an Indian–Israeli Scientific Research Cooperation in Biotechnology (Ministry of Science and Technology) grant to H.F. and by MINERVA Foundation and Weizmann-Argentina Fundacion Antorchas grants to R.F.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hillel Fromm (hillelf@post.tau.ac.il).
Online version contains Web-only data.
References
- Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaim, G., and Villalobo, A. (2002). Phosphorylation of calmodulin. Eur. J. Biochem. 269 3619–3631. [DOI] [PubMed] [Google Scholar]
- Bethke, P.C., and Jones, R.L. (1994). Ca2+-calmodulin modulates ion channel activity in storage protein vacuoles of barley aleurone cells. Plant Cell 6 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan, M. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché, N., Scharlat, A., Snedden, W., Bouchez, D., and Fromm, H. (2002). A novel family of calmodulin-binding transcription activators in multicellular organisms. J. Biol. Chem. 277 21851–21861. [DOI] [PubMed] [Google Scholar]
- Bouché, N., Yellin, A., Snedden, W.A., and Fromm, H. (2005). Plant-specific calmodulin-binding proteins. Annu. Rev. Plant Biol. 56 435–466. [DOI] [PubMed] [Google Scholar]
- Braam, J. (1992). Regulated expression of the calmodulin-related TCH genes in cultured Arabidopsis cells: Induction by calcium and heat shock. Proc. Natl. Acad. Sci. USA 89 3213–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam, J., Sistrunk, M.L., Polisensky, D.H., Xu, W., Purugganan, M.M., Antosiewicz, D.M., Campbell, P., and Johnson, K.A. (1997). Plant responses to environmental stress: Regulation and functions of the Arabidopsis TCH genes. Planta 203 (suppl.), S35–S41. [DOI] [PubMed] [Google Scholar]
- Campbell, A.K., Trewavas, A.J., and Knight, M.R. (1996). Calcium imaging shows differential sensitivity to cooling and communication in luminous transgenic plants. Cell Calcium 19 211–218. [DOI] [PubMed] [Google Scholar]
- Carrion, A.M., Link, W.A., Ledo, F., Mellstrom, B., and Naranjo, J.R. (1999). DREAM is a Ca2+-regulated transcriptional repressor. Nature 398 80–84. [DOI] [PubMed] [Google Scholar]
- Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B.-h., Hong, X., Agarwal, M., and Zhu, J.-K. (2003). ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, H.-i., Hong, J.-h., Ha, J.-o., Kang, J.-y., and Kim, S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275 1723–1730. [DOI] [PubMed] [Google Scholar]
- Choi, M.S., et al. (2005). Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.). J. Biol. Chem. 280 40820–40831. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Feske, S., Giltnane, J., Dolmetsch, R., Staudt, L.M., and Rao, A. (2001). Gene regulation mediated by calcium signals in T lymphocytes. Nat. Immunol. 2 316–324. [DOI] [PubMed] [Google Scholar]
- Fujimoto, S.Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy, S., Hughes, W.A., and Trewavas, A.J. (1987). Calmodulin antagonists increase free cytosolic calcium levels in plant protoplasts in vivo. FEBS Lett. 212 133–137. [Google Scholar]
- Harper, J.F., Breton, G., and Harmon, A. (2004). Decoding Ca(2+) signals through plant protein kinases. Annu. Rev. Plant Biol. 55 263–288. [DOI] [PubMed] [Google Scholar]
- Harper, J.L., and Daly, J.W. (2000). Effect of calmidazolium analogs on calcium influx in HL-60 cells. Biochem. Pharmacol. 60 317–324. [DOI] [PubMed] [Google Scholar]
- Hepler, P.K. (2005). Calcium: A central regulator of plant growth and development. Plant Cell 17 2142–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington, A.M., and Brownlee, C. (2004). The generation of Ca(2+) signals in plants. Annu. Rev. Plant Biol. 55 401–427. [DOI] [PubMed] [Google Scholar]
- Hobo, T., Asada, M., Kowyama, Y., and Hattori, T. (1999). ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 19 679–689. [DOI] [PubMed] [Google Scholar]
- Ikura, M., Osawa, M., and Ames, J.B. (2002). The role of calcium-binding proteins in the control of transcription: Structure and function. Bioessays 24 625–636. [DOI] [PubMed] [Google Scholar]
- Ito, T., Hirano, M., Akama, K., Shimura, Y., and Okada, K. (1995). Touch-inducible genes for calmodulin and a calmodulin-related protein are located in tandem on a chromosome of Arabidopsis thaliana. Plant Cell Physiol. 36 1369–1373. [PubMed] [Google Scholar]
- Jan, C.-R., and Tseng, C.-J. (2000). W-7 induces [Ca2+]i increases in Madin-Darby canine kidney (MDCK) cells. J. Pharmacol. Exp. Ther. 292 358–365. [PubMed] [Google Scholar]
- Khan, S.Z., Dyer, J.L., and Michelangeli, F. (2001). Inhibition of the type 1 inositol 1,4,5-trisphosphate-sensitive Ca2+ channel by calmodulin antagonists. Cell. Signal. 13 57–63. [DOI] [PubMed] [Google Scholar]
- Kim, K.N., Cheong, Y.H., Grant, J.J., Pandey, G.K., and Luan, S. (2003). CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.N., Cheong, Y.H., Gupta, R., and Luan, S. (2000). Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases. Plant Physiol. 124 1844–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, H. (2000). Calcium signaling during abiotic stress in plants. Int. Rev. Cytol. 195 269–324. [DOI] [PubMed] [Google Scholar]
- Knight, H., and Knight, M.R. (2000). Imaging spatial and cellular characteristics of low temperature calcium signature after cold acclimation in Arabidopsis. J. Exp. Bot. 51 1679–1686. [DOI] [PubMed] [Google Scholar]
- Knight, H., Trewavas, A.J., and Knight, M.R. (1996). Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8 489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, H., Trewavas, A.J., and Knight, M.R. (1997). Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 12 1067–1078. [DOI] [PubMed] [Google Scholar]
- Knight, H., Veale, E., Warren, G.J., and Knight, M.R. (1999). The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell 11 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, H., Zarka, D.G., Okamoto, H., Thomashow, M.F., and Knight, M.R. (2004). Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via the CRT promoter element. Plant Physiol. 135 1710–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, M.R., Campbell, A.K., Smith, S.M., and Trewavas, A.J. (1991). Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352 524–526. [DOI] [PubMed] [Google Scholar]
- Kudla, J., Xu, Q., Harter, K., Gruissem, W., and Luan, S. (1999). Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. USA 96 4718–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki, F., Kaburaki, H., and Nishi, A. (1994). Involvement of plasma membrane-located calmodulin in the response decay of cyclic nucleotide-gated cation channel of cultured carrot cells. FEBS Lett. 340 193–196. [DOI] [PubMed] [Google Scholar]
- Li, Z., and Thomas, T.L. (1998). PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell 10 383–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., and Zhu, J.K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280 1943–1945. [DOI] [PubMed] [Google Scholar]
- Luan, S., Kudla, J., Rodriguez-Concepcion, M., Yalovsky, S., and Gruissem, W. (2002). Calmodulins and calcineurin B-like proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell 14 (suppl.), S389–S400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankin, S.L., Allen, G.C., and Thompson, W.F. (1997). Introduction of a plant intron into the luciferase gene of Photinus pyralis. Plant Mol. Biol. Rep. 15 186–196. [Google Scholar]
- McCormack, E., and Braam, J. (2003). Calmodulins and related potential calcium sensors in Arabidopsis. New Phytol. 159 585–598. [DOI] [PubMed] [Google Scholar]
- Mitsuda, N., Isono, T., and Sato, M.H. (2003). Arabidopsis CAMTA family proteins enhance V-PPase expression in Pollen. Plant Cell Physiol. 44 975–981. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Irie, K., Hirayama, T., Hayashida, N., Yamaguchi-Shinozaki, K., Matsumoto, K., and Shinozaki, K. (1996). A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol. 15 473–497. [Google Scholar]
- Neuhaus, G., Bowler, C., Hiratsuka, K., Yamagata, H., and Chua, N.-H. (1997). Phytochrome-regulated repression of gene expression requires calcium and cGMP. EMBO J. 16 2554–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parag, H.A., Dimitrovsky, D., Raboy, B., and Gulka, R.G. (1993). Selective ubiquitination of calmodulin by UBC4 and a putative ubiquitin protein ligase (E3) from Saccharomyces cerevisiae. FEBS Lett. 325 242–246. [DOI] [PubMed] [Google Scholar]
- Peiter, E., Maathuis, F.J., Mills, L.N., Knight, H., Pelloux, J., Hetherington, A.M., and Sanders, D. (2005). The vacuolar Ca2+-activated channel TPC1 regulates germination and stomatal movement. Nature 434 404–408. [DOI] [PubMed] [Google Scholar]
- Polisensky, D.H., and Braam, J. (1996). Cold-shock regulation of the Arabidopsis TCH genes and the effects of modulating intracellular calcium levels. Plant Physiol. 111 1271–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, V.S., Ali, G.S., and Reddy, A.S.N. (2002). Genes encoding calmodulin-binding proteins in the Arabidopsis genome. J. Biol. Chem. 277 9840–9852. [DOI] [PubMed] [Google Scholar]
- Reddy, V.S., and Reddy, A.S.N. (2004). Proteomics of calcium-signaling components in plants. Phytochemistry 65 1745–1776. [DOI] [PubMed] [Google Scholar]
- Sano, T., and Nagata, T. (2002). The possible involvement of a phosphate-induced transcription factor encoded by Phi-2 gene from tobacco in ABA-signaling pathways. Plant Cell Physiol. 43 12–20. [DOI] [PubMed] [Google Scholar]
- Schulz-Lessdorf, B., and Hedrich, R. (1995). Protons and calcium modulate SV-type channels in the vacuolar-lysosomal compartment-channel interaction with calmodulin inhibitors. Planta 197 655–671. [Google Scholar]
- Seki, M., et al. (2002. a). Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct. Integr. Genomics 2 282–291. [DOI] [PubMed] [Google Scholar]
- Seki, M., et al. (2002. b). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31 279–292. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (1996). Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274 1900–1902. [DOI] [PubMed] [Google Scholar]
- Snedden, W.A., and Fromm, H. (2001). Calmodulin as a versatile calcium signal transducer in plants. New Phytol. 151 35–66. [DOI] [PubMed] [Google Scholar]
- Sze, H., Liang, F., Hwang, I., Curran, A.C., and Harper, J.F. (2000). Diversity and regulation of plant Ca2+ pumps: Insights from expression in yeast. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 433–462. [DOI] [PubMed] [Google Scholar]
- Taji, T., Seki, M., Yamaguchi-Shinozaki, K., Kamada, H., Giraudat, J., and Shinozaki, K. (1999). Mapping of 25 drought-inducible genes, RD and ERD, in Arabidopsis thaliana. Plant Cell Physiol. 40 119–123. [DOI] [PubMed] [Google Scholar]
- Takahashi, R., and Shimosaka, E. (1997). cDNA sequence analysis and expression of two cold-regulated genes in soybean. Plant Sci. 123 93–104. [Google Scholar]
- Tavazoie, S., Hughes, J.D., Campbell, M.J., Cho, R.J., and Church, G.M. (1999). Systematic determination of genetic network architecture. Nat. Genet. 22 281–285. [DOI] [PubMed] [Google Scholar]
- van Helden, J., Andre, B., and Collado-Vides, J. (1998). Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 281 827–842. [DOI] [PubMed] [Google Scholar]
- Yang, T., and Poovaiah, B.W. (2002). A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 277 45049–45058. [DOI] [PubMed] [Google Scholar]
- Zhang, W., Ruan, J., Ho, T.H., You, Y., Yu, T., and Quatrano, R.S. (2005). Cis-regulatory element based targeted gene finding: Genome-wide identification of abscisic acid- and abiotic stress-responsive genes in Arabidopsis thaliana. Bioinformatics 21 3074–3081. [DOI] [PubMed] [Google Scholar]
- Zik, M., and Irish, V.F. (2003). Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell 15 207–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuhlke, R.D., Pitt, G.S., Deisseroth, K., Tsien, R.W., and Reuter, H. (1999). Calmodulin supports both inactivation and facilitation of L-type calcium channels. Nature 399 159–162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.