Abstract
Polarity formation is central to leaf morphogenesis, and several key genes that function in adaxial-abaxial polarity establishment have been identified and characterized extensively. We previously reported that Arabidopsis thaliana ASYMMERTIC LEAVES1 (AS1) and AS2 are important in promoting leaf adaxial fates. We obtained an as2 enhancer mutant, asymmetric leaves enhancer3 (ae3), which demonstrated pleiotropic plant phenotypes, including a defective adaxial identity in some leaves. The ae3 as2 double mutant displayed severely abaxialized leaves, which were accompanied by elevated levels of leaf abaxial promoting genes FILAMENTOUS FLOWER, YABBY3, KANADI1 (KAN1), and KAN2 and a reduced level of the adaxial promoting gene REVOLUTA. We identified AE3, which encodes a putative 26S proteasome subunit RPN8a. Furthermore, double mutant combinations of as2 with other 26S subunit mutations, including rpt2a, rpt4a, rpt5a, rpn1a, rpn9a, pad1, and pbe1, all displayed comparable phenotypes with those of ae3 as2, albeit with varying phenotypic severity. Since these mutated genes encode subunits that are located in different parts of the 26S proteasome, it is possible that the proteolytic function of the 26S holoenzyme is involved in leaf polarity formation. Together, our findings reveal that posttranslational regulation is essential in proper leaf patterning.
INTRODUCTION
The leaf primordium originates from the peripheral zone of the shoot apical meristem. Once leaf primordia are formed, they require establishment of the proximodistal, mediolateral, and ad/abaxial axes (Hudson, 2000). Among these three, the establishment of the ad/abaxial axis, which is required for subsequent asymmetric leaf growth, may be of primary importance (Bowman et al., 2002). Cell differentiation along this axis results in the two leaf faces being distinct both in structure and in their biological functions (McConnell et al., 2001).
Recent studies have identified several genes that play critical roles in establishing leaf adaxial/abaxial polarity. Genes in the class III HD-ZIP family, including PHABULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV), are important for specifying adaxial identity (McConnell and Barton, 1998; McConnell et al., 2001; Emery et al., 2003). Members in the YABBY (YAB) and KANADI (KAN) gene families contribute to the abaxial pattern formation of the leaf (Chen et al., 1999; Sawa et al., 1999; Siegfried et al., 1999; Eshed et al., 2001, 2004; Kerstetter et al., 2001). In addition, the ASYMMERTIC LEAVES1 (AS1) and AS2 genes have also been demonstrated to play critical roles in specifying leaf adaxial identity (Lin et al., 2003; Xu et al., 2003; Garcia et al., 2006). AS1 encodes a protein that contains an R2-R3 MYB domain (Byrne et al., 2000; Sun et al., 2002), suggesting that it may bind to DNA and directly regulate transcription. The AS2 protein contains a Leu-zipper motif and can associate with AS1 (Iwakawa et al., 2002; Xu et al., 2002, 2003). Both AS1 and AS2 positively regulate the PHB gene and repress some genes in the YAB and KAN families (Lin et al., 2003; Xu et al., 2003). Since all the genes listed above encode putative transcription factors, regulation at the transcriptional level is likely very important during leaf adaxial/abaxial polarity formation.
In addition to the putative transcription factors, components in RNA silencing pathways also play important roles in specifying leaf adaxial polarity. RNA-DEPENDENT RNA POLYMERASE6 (RDR6) was identified to have such a function (Li et al., 2005). The rdr6 single mutant showed only minor phenotypic changes, whereas the rdr6 as1(2) double mutant displayed dramatically enhanced as1(2) phenotypes with severely abaxialized leaves. The subsequent studies also revealed that the SUPPRESSOR OF GENE SILENCING (SGS3), ZIPPY/ARGONAUTE7 (ZIP), and DICER-LIKE4 (DCL4) genes play similar roles to that of RDR6 in leaf patterning (Garcia et al., 2006; Xu et al., 2006). It is possible that RDR6/SGS3/ZIP/DCL4 act in the same pathway and at least partially function in repressing leaf abaxial promoting genes AUXIN RESPONSE FACTOR3 (ARF3) and ARF4 via production of a trans-acting siRNA, tasiR-ARF (Peragine et al., 2004; Yoshikawa et al., 2005; Garcia et al., 2006; Xu et al., 2006). Recent studies also demonstrated that two microRNAs, miR165 and miR166, participate in leaf adaxial/abaxial polarity formation. These two miRNAs differ by only one nucleotide and have complimentary regions of class III HD-ZIP genes, encoding a so-called START domain (Rhoades et al., 2002). Research has shown that changing a single nucleotide within the START-encoding regions of several class III HD-ZIP genes results in dominant mutations that cause adaxialized leaves (McConnell et al., 2001; Emery et al., 2003; Zhong and Ye, 2004). All these results indicate that in addition to transcriptional regulation, proper leaf polarity formation also requires regulation at the posttranscriptional level.
To elucidate the molecular mechanism for leaf adaxial/abaxial polarity establishment, it seems important as a first step to identify all genetic pathways involved in the regulatory network of leaf development. In these genetic and molecular studies, we show that the genes encoding different 26S proteasome subunits are required for normal adaxial/abaxial leaf patterning, revealing a role for posttranslational regulation in leaf patterning.
The 26S proteasome is a highly conserved protein degradation system in eukaryotes, which contains ∼31 subunits arranged into two protein subcomplexes: the 20S catalytic core particle (CP) and the 19S regulatory particle (RP) (Smalle and Vierstra, 2004). The CP has the capacity to cleave peptide bonds, while the RP assists in recognizing and unfolding target substrates tagged with polyubiquitin chains, removing the chains, and directing the unfolded polypeptides into the CP for degradation (Smalle and Vierstra, 2004). The RP could be further divided into two parts known as the base and the lid (Glickman et al., 1998). The base contains six ATPases (RPT1 to RPT6) and three non-ATPase subunits (RPN1, RPN2, and RPN10), whereas the lid is made up of eight additional RPN subunits (RPN3, RPN5 to RPN9, RPN11, and RPN12) (Voges et al., 1999). In Arabidopsis thaliana, two base subunits, RPN10 and RPT2a (HALTED ROOT [HLR]) were found to have biological functions in the plant's response to abscisic acid signals (Smalle et al., 2003) and in maintaining the meristem integrity both in the shoot and in the root (Ueda et al., 2004). Additionally, one lid subunit, RPN12a, plays a role in controlling the stability of one or more of the factors involved in cytokinin regulation (Smalle et al., 2002). However, functions of most other Arabidopsis 26S proteasome subunits are unknown.
In this article, we report our mutagenesis screen and characterizations of an as1/as2 enhancer, asymmetric leaves enhancer3 (ae3), and show that Arabidopsis AE3 encodes the 26S proteasome lid subunit RPN8a that plays a role in specifying leaf adaxial identity. Moreover, we provide genetic evidence that the proteolytic function of the 26S proteasome is required for the proper leaf adaxial/abaxial polarity establishment.
RESULTS
The as1/as2 Enhancer Mutant ae3-1
We previously reported that AS1 and AS2 are required for establishing leaf adaxial/abaxial polarity (Sun et al., 2002; Xu et al., 2002, 2003). Compared with the wild-type plant (Figure 1A), mutant as2-101 often showed a lotus-leaf structure with petioles growing from underneath the leaf blade (Figure 1B). This structure is believed to be caused by a partial loss of the leaf adaxial identities (Xu et al., 2003). In some extreme cases, the as2-101 plants even produced some abaxialized needle-like structures, though the frequency was very low (Xu et al., 2003; Qi et al., 2004). In the course of completing a mutagenesis screen for as2-101 enhancers, we identified one plant exhibiting apparently increased numbers of needle and lotus leaves (Figure 1C). These types of abnormal leaves usually appear among the first two rosette leaves in as2-101 single mutant plants, whereas they were present in all rosette leaves in the as2 enhancer mutant plants. We demonstrated that phenotypes of the as2 enhancer mutant were caused by as2 and an additional mutation, which was designated as ae3-1 (see Methods).
Figure 1.
ae3 Enhances as2 and as1.
Morphological observations of wild-type and mutant seedlings: the wild type (A), as2-101 (B), ae3-1 as2-101 (C), ae3-1 (D), as1-101 (E), and ae3-1 as1-101 (F). Arrows indicate the lotus leaves, and arrowheads point to the needle-like leaves. Bars = 1 cm.
The ae3-1 single mutant plant was isolated, showing long and narrow rosette leaves (Figure 1D, Table 1). To determine whether the ae3-1 mutation can also enhance as1 phenotypes, we constructed the ae3-1 as1-101 double mutant. The resulting double mutant plants were similar to ae3-1 as2-101 plants, with an increased number of lotus and needle leaves (Figure 1F) compared with the as1-101 single mutant (Figure 1E). These results indicate that AE3 acts synergistically with the AS1/AS2 pathway to regulate leaf development.
Table 1.
ae3-1 Single Mutant Plants Produce Long and Narrow Rosette Leavesa
| Genotype | Length (mm)b | Width (mm)c | Ratio (Length:Width) |
|---|---|---|---|
| Ler (n = 20) | 9.86 ± 0.84 | 4.35 ± 0.26 | 2.27 |
| ae3-1 (n = 30) | 11.55 ± 1.03 | 2.20 ± 0.40 | 5.25 |
First two rosette leaves were analyzed. Values given are average ± se.
Leaves were measured from the petiole end to the blade tip.
Leaves were measured through the central part of blades.
Pleiotropic Phenotypes of the ae3-1 Mutant Plants
To better understand the role that AE3 plays in plant development, we analyzed phenotypes of the ae3-1 single mutant. Compared with the wild type (Figure 2A), ae3-1 was slightly dwarfish (Figure 2B) and exhibited a delayed flowering time with increased numbers of rosette leaves (6.2 ± 0.6 in the wild type versus 9.2 ± 0.7 in the mutant; n = 30). The ae3-1 phyllotaxy was aberrant, with several cauline leaves frequently associated together (Figure 2B, arrowhead). While most other rosette leaves in ae3-1 showed only a long and narrow shape, the first two rosette leaves often exhibited more severe phenotypes. These included (a) very narrow leaves (12/270), (b) lotus leaves with a very long petiole (2/800), (c) horn-like structures (which we refer to as ectopic leaves growing near the leaf tip on the abaxial side) (3/600, arrowhead), and (d) needle-like leaves (10/258) (Figure 2C).
Figure 2.
Pleiotropic Phenotypes of ae3-1.
(A) and (B) The 5-week-old plants of the wild type (A) and ae3-1 mutant (B). Arrowhead in (B) indicates the abnormal phyllotaxy with several cauline leaves associated together.
(C) First-pair rosette leaves of ae3-1 show different shapes: (a) a very narrow rosette leaf, (b) a lotus leaf with a long petiole, (c) a leaf bearing an ectopic structure (arrowhead) at the distal part, and (d) a needle-like leaf.
(D) ae3-1 cauline leaves often produce an ectopic leaf on their abaxial distal parts (arrowheads), and these ectopic structures occasionally develop into a leaflet (arrow and inset).
(E) Siliques of the wild type (left) and ae3-1 (right). ae3-1 siliques usually have a longer pedicel than that of the wild type.
(F) and (G) Trichomes and their support cells (arrowheads) on wild-type (F) and ae3-1 (G) leaf surfaces. ae3-1 support cells protrude from the leaf surface.
Bars = 1 cm in (A), (B), and (E), 5 mm in (C) and (D), and 50 μm in (F) and (G).
Frequently, ae3-1 cauline leaves also formed the ectopic leaves (Figure 2D, arrowheads), and this structure could occasionally further expand, forming a small leaf (Figure 2D, arrow and inset). In addition, although the carpel number in ae3-1 flowers appeared normal, other floral organ numbers were often altered (Table 2). Compared with the wild type (Figure 2E, left), ae3-1 pedicels were longer (Figure 2E, right). Furthermore, although trichomes in the wild type (Figure 2F) and ae3-1 (Figure 2G) were similar in shape, the trichome support cells on ae3-1 leaves were reduced in size and protruded from the leaf surface (arrowheads). The pleiotropic phenotypes of the ae3-1 plant indicate that AE3 has multiple functions in plant development.
Table 2.
Abnormal Floral Organ Numbers in ae3-1a
| Genotype | Sepal | Petal | Stamen | Carpel |
|---|---|---|---|---|
| Ler flowers (n = 40) | 4 ± 0 | 4 ± 0 | 5.9 ± 0.3 | 2 ± 0 |
| ae3-1 flowers (n = 87) | 4.2 ± 0.6 | 3.9 ± 0.9 | 4.8 ± 0.8 | 2 ± 0 |
Values are average ± se.
ae3-1 Produces Abaxialized Leaves
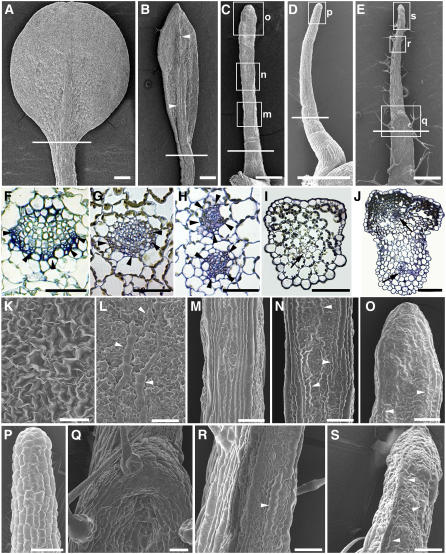
The lotus- and needle-like leaf structures are generally thought to be caused by defective leaf adaxial/abaxial polarity. To determine whether the ae3-1 leaves are defective in adaxial or abaxial identity, we examined the ae3-1 leaf vascular pattern by transverse section through the blade-petiole conjugation region and analyzed the leaf epidermal cell pattern by scanning electron microscopy. In the wild-type leaves (Figure 3A), vascular bundles at this region showed a pattern where xylem develops on the adaxial pole and phloem is located on the abaxial pole (Figure 3F). ae3-1 leaves that were slightly narrow exhibited a vascular pattern similar to that in the wild type. ae3-1 leaves with even narrower blades possessed a thickened midrib (Figure 3B, arrowheads), while the tracheary elements of xylem became thinner (Figure 3G). The very narrow blade leaves (Figure 3C) contained two separated vascular bundles (Figure 3H), each of which exhibited a phloem-surrounding-xylem structure, with the least-developed xylem tissues in the center. There was no apparent vascular bundle in the radially symmetric needle-like leaves (Figure 3D), although the few small cells grouped together might represent a premature vascular bundle (Figure 3I, arrow). We also analyzed an ae3-1 cauline leaf that carried an ectopic leaf (Figure 3E). Similar to the very narrow ae3-1 rosette leaf (Figure 3C), the cauline leaf also contained two vascular bundles (Figure 3J, arrows). The ectopic leaf growing on the cauline leaves contained a premature vascular bundle, similar to that in the needle-like leaves in Figure 3I. The vascular pattern in the ae3-1 leaves suggests that the adaxial identity of some ae3 leaves is defective.
Figure 3.
Aberrant Adaxial/Abaxial Polarity in ae3-1 Leaves.
(A) A wild-type rosette leaf.
(B) to (D) ae3-1 rosette leaves.
(B) An abaxial view showing a narrow leaf with a thickened midrib (arrowheads).
(C) An adaxial view showing a very narrow leaf with only minimum adaxial/abaxial differentiation.
(D) A radial symmetric needle-like leaf.
(E) A cauline leaf that bears an ectopic leaf.
(F) to (J) Transverse sections. Sections from (F) to (J) are corresponding to the individual leaves from (A) to (E), respectively, with the approximate section regions indicated by the white lines. They are from a wild-type rosette leaf (F), narrow (G) and very narrow (H) ae3-1 rosette leaves, a needle-like rosette leaf (I), and a cauline leaf bearing an ectopic leaf (J). Arrowheads in (F) to (H) indicate phloem, and arrows in (I) and (J) point to vascular bundles.
(K) to (S) Epidermal patterns of leaves.
(K) Adaxial epidermis of wild-type Ler.
(L) Abaxial epidermis of Ler.
(M) to (O) Close-ups of leaf epidermis in (C), corresponding to the boxed regions indicated by m, n, and o, respectively.
(P) Close-up of epidermis of a boxed region in (D).
(Q) to (S) Close-ups of epidermis in (E), corresponding to the boxed regions indicated by q, r, and s, respectively.
Bars = 500 μm in (A) to (E) and 50 μm in (F) to (S).
The scanning electron microscopy analyses showed that the adaxial side of a Landsberg erecta (Ler) leaf contains large and uniformly sized epidermal cells (Figure 3K), while the abaxial surface contains relatively small cells mixed with some long and narrow cells (Figure 3L, arrowheads). On the adaxial side of the very narrow ae3-1 rosette leaf (Figure 3C), although the proximal portion contained cells similar to the petiole surface cells (Figure 3M), the middle and distal portions of the leaf exhibited long and narrow abaxial epidermal cells (Figures 3N and 3O, arrowheads), indicating the leaf is abaxialized. All epidermal cells in the ae3-1 needle-like leaf were rectangular (Figure 3D), which suggests they may not be sufficiently differentiated (Figure 3P). Furthermore, the ectopic leaf on ae3-1 cauline leaves (Figure 3E) also contained abaxial epidermal cells on its adaxial side (Figures 3R and 3S, arrowheads), though the adaxial side of the cauline leaf part (Figure 3Q) still possessed a uniformly adaxial epidermis. All these observations support the possibility that ae3-1 leaves are not demonstrating sufficient adaxial differentiation.
The Severely Defective Adaxial/Abaxial Polarity in the ae3 as2 Leaves
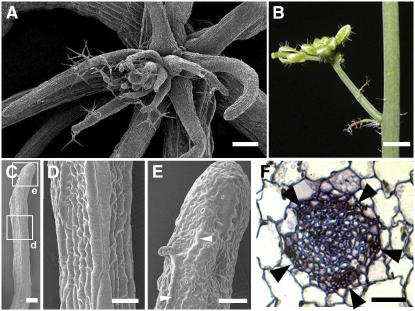
To further understand the roles of AE3 in leaf development, we analyzed leaf phenotypes of the ae3-1 as2-101 double mutant by scanning electron microscopy and sectioning. Although the early-appearing leaves of the double mutant had lotus or needle structures, almost all late-appearing rosette (Figure 4A) and most cauline leaves (Figure 4B) were needle-like. The needle-like leaves in ae3-1 as2-101 can be further divided into two types. One contained abaxial epidermal cells on the adaxial distal portion (Figures 4C and 4E, arrowhead), with the cells on the proximal (data no shown) and middle portions (Figures 4C and 4D) resembling those on wild-type petioles. The other showed rectangular epidermal cells covering almost the entire needle-like structure, similar to those in Figures 3D and 3P. These phenotypes may represent less-differentiated forms. A transverse section through the proximal part of a needle-like leaf (of the first ae3-1 as2-101 type) showed a phloem-surrounding-xylem pattern (Figure 4F). All these results indicate that ae3-1 as2-101 leaves are abaxialized but with much more severe characteristics than those in the ae3-1 and as2-101 single mutant plants.
Figure 4.
Phenotypes of ae3-1 as2-101 Leaves.
(A) Later-appearing ae3-1 as2-101 rosette leaves, which all show a needle-like structure.
(B) An inflorescence branch of ae3-1 as2-101, showing that cauline leaves are needle-like.
(C) to (E) Epidermis of a needle-like leaf in ae3-1 as2-101.
(D) and (E) Close-ups of epidermis in (C), corresponding to the boxed regions indicated by d and e, respectively.
(F) A transverse section, showing a vascular bundle at the proximal part of an ae3-1 as2-101 needle-like rosette leaf with a phloem-surrounding-xylem structure. Arrowheads indicate phloem.
Bars = 150 μm in (A) and (C), 5 mm in (B), and 50 μm in (D) to (F).
Altered Expression of the Leaf Polarity Genes in ae3 and ae3 as2 Leaves
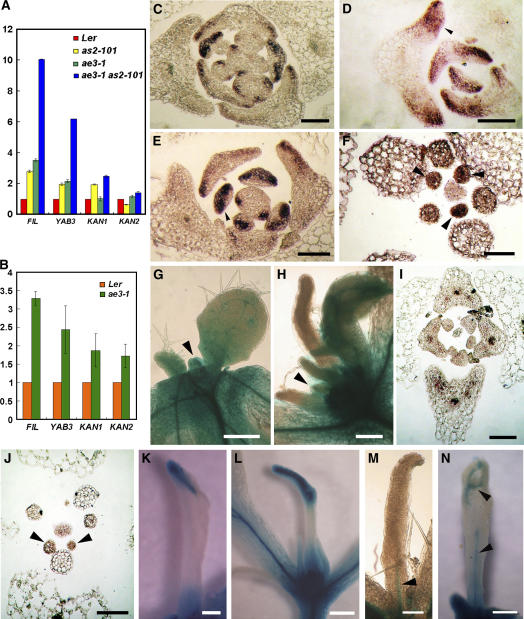
Phenotypic analyses of ae3-1 single and ae3-1 as2-101 double mutants indicate that the AE3 gene is required for specifying the leaf adaxial identity. To obtain molecular evidence for the AE3 function, we analyzed FILAMENTOUS FLOWER (FIL), YAB3, KAN1, and KAN2 mRNA levels by quantitative real-time RT-PCR. Transcript levels of these genes were generally elevated in the single mutant leaves, but they were enhanced even more in the ae3-1 as2-101 double mutant leaves (Figure 5A). Similar to the rosette leaves, the ae3-1 cauline leaves carrying the ectopic leaf also contained higher transcript levels of the four genes than those in the wild-type leaves (Figure 5B).
Figure 5.
Expression of the Leaf Polarity Marker Genes.
(A) and (B) Analyses by real-time RT-PCR for transcript levels of FIL, YAB3, KAN1, and KAN2 in wild-type Ler, as2-101, ae3-1, and ae3-1 as2-101 rosette leaves (A) or in the Ler and ae3-1 cauline leaves (B). Bars show standard error.
(C) to (F) In situ hybridization of FIL expression in Ler (C), as2-101 (D), ae3-1 (E), and ae3-1 as2-101 (F).
(G) and (H) Analyses of REV expression using an rev-9 enhancer trap line. Note that only wild-type and ae3-1 as2-101 mutant plants that are hemizygous for rev-9 were characterized. Shoot apices from 12-d-old wild-type Ler (G) or ae3-1 as2-101 (H) are shown.
(I) and (J) In situ hybridization of REV expression in ae3-1 (I) and ae1-3 as2-101 (J).
(K) to (N) Needle-like leaves of ae3-1 as2-101. Note that these needle-like leaves show varying degrees of adaxial/abaxial differentiation and vascular tissue formation. Arrowheads in (M) and (N) show GUS staining, indicating the insufficiently developed vascular tissues.
Bars = 100 μm in (C) to (F), (I), and (J), 200 μm in (G), (H), and (K) to (M), and 500 μm in (N).
To investigate whether the altered expression levels of the abaxial-promoting genes were also accompanied by changes in the expression pattern, we analyzed one of these genes, FIL, using in situ hybridization. FIL is usually expressed on the abaxial side of leaves (Figure 5C) but appeared to be extended toward the adaxial side in some young leaves in as2-101 (Figure 5D, arrowhead) and ae3-1 (Figure 5E, arrowhead). FIL was expressed throughout the entire primordium of the ae3-1 as2-101 needle-like leaves (Figure 5F, arrowheads). The increased transcript levels of the abaxial-promoting genes and the expanded expression pattern of FIL in ae3 single and ae3 as2 double mutants support the idea that AE3 is required for specifying adaxial identity in leaves.
To better understand the adaxial/abaxial polarity formation in ae3 as2 leaves, we also analyzed expression patterns of the adaxial-promoting gene REV by in situ hybridization. In addition, we also crossed ae3-1 as2-101 to rev-9, a T-DNA enhancer trap line, in which β-glucuronidase (GUS) staining can indicate the REV expression (Emery et al., 2003; Hawker and Bowman, 2004). Although REV was expressed in leaf primordia in both rev-9/+ (Figure 5G, arrowhead) and rev-9/+ ae3-1 as2-101 (Figure 5H, arrowhead) plants, GUS activity was diminished in the more developed primordia in rev-9/+ ae3-1 as2-101 (Figure 5H). In wild-type plants, REV is usually expressed in the adaxial domain of leaf primordia (Emery et al., 2003; Li et al., 2005) and in vascular tissues of the developing leaves (Zhong and Ye, 1999). In comparison with the expanded ae3-1 leaves that showed a polar expression of REV (Figure 5I) similar to that in the wild type, the REV transcripts were detected throughout the very young leaf primordia of ae3-1 as2-101 and diminished rapidly upon further leaf development (Figure 5J), consistent with the GUS staining results in Figure 5H. The adaxial/abaxial polarity formation in some needle-like leaves of rev-9/+ ae3-1 as2-101 appeared to be delayed, and only some of these leaves with partial adaxial/abaxial differentiation at the distal portion exhibited GUS staining on the adaxial side (Figures 5K and 5L). For those that did not show leaf adaxial/abaxial differentiation, GUS staining was observed only in the vascular tissues (Figure 5M, arrowhead). GUS staining in mature needle-like leaves was concentrated only in the vascular tissues (Figure 5N, arrowheads). These results suggest that the AE3 and AS2 roles in normal leaf polarity formation may be via regulation of the leaf polarity controlling genes.
Double Mutants of ae3-1 rev-9 and ae3-1 rdr6-3
To understand the genetic interaction between AE3 and other polarity-controlling pathways in leaf patterning, we characterized ae3-1 rev-9 and ae3-1 rdr6-3 double mutants. The recessive rev-9 mutant (Ler) has normal leaf phenotypes, although the axillary inflorescence number was reduced (Emery et al., 2003) (Figure 6A). Compared with the rev-9 mutant plant, ae3-1 rev-9 exhibited narrow early-appearing rosette leaves (Figure 6B) similar to those of the ae3-1 single mutant. However, the ae3-1 rev-9 seedling was aberrant with late-appearing leaves, which were arrested at early developmental stages (Figure 6B). Scanning electron microscopy showed that these late-appearing needle-like leaves were covered with long and straight cells (Figures 6C and 6D), which mimicked those of petiole epidermal cells and failed to demonstrate additional differentiation. The ae3-1 rev-9 phenotypes suggest that AE3 genetically interacts with REV to specify leaf adaxial polarity.
Figure 6.
Phenotypes of the ae3-1 rev-9 and ae3-1 rdr6-3 Double Mutants.
(A) A rev-9 mutant, with arrowheads indicating the nodes with no axillary inflorescence.
(B) An ae3-1 rev-9 mutant, showing the arrested leaves.
(C) and (D) Close-ups of the ae3-1 rev-9 leaves.
(E) An ae3-1 rdr6-3 rosette leaf carrying the ectopic leaf.
(F) An ae3-1 rdr6-3 rosette leaf showing a dramatically thickened midrib (arrowheads), which is associated with the leaf petiole (arrow).
(G) Close-up of (F).
Bars = 1 cm in (A) and (B), 1 mm in (C), (E), and (F), and 100 μm in (D) and (G).
The rdr6-3 mutant revealed only minor changes in the phenotypes (Li et al., 2005), whereas ae3-1 rdr6-3 exhibited apparently enhanced ae3-1 phenotypes. The frequency of ectopic leaves on the ae3-1 rdr6-3 rosette leaves was higher than that on the ae3-1 rosette leaves, and the size of the ae3-1 rdr6-3 ectopic leaves became bigger (Figure 6E; for comparison, see Figure 2C). In addition, the frequency of lotus- and needle-like rosette leaves in the double mutant increased markedly (3.3% in ae3-1, n = 122, versus 34% in ae3-1 rdr6-3, n = 178). Furthermore, petioles of some leaves in the double mutants formed distinct adaxial and abaxial parts. The adaxial part was equivalent to a normal-sized petiole (Figures 6F and 6G, arrows), while the abaxial part was broader (Figures 6F and 6G, arrowheads), which connected with the dramatically thickened midrib. These results suggest that AE3 genetically interacts with the RNA silencing pathway to regulate leaf patterning.
Molecular Cloning of the AE3 Gene
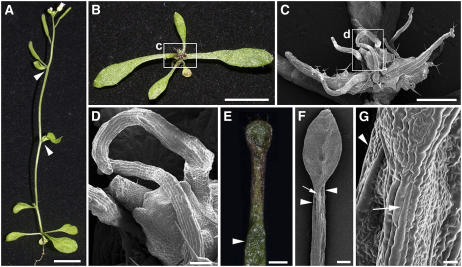
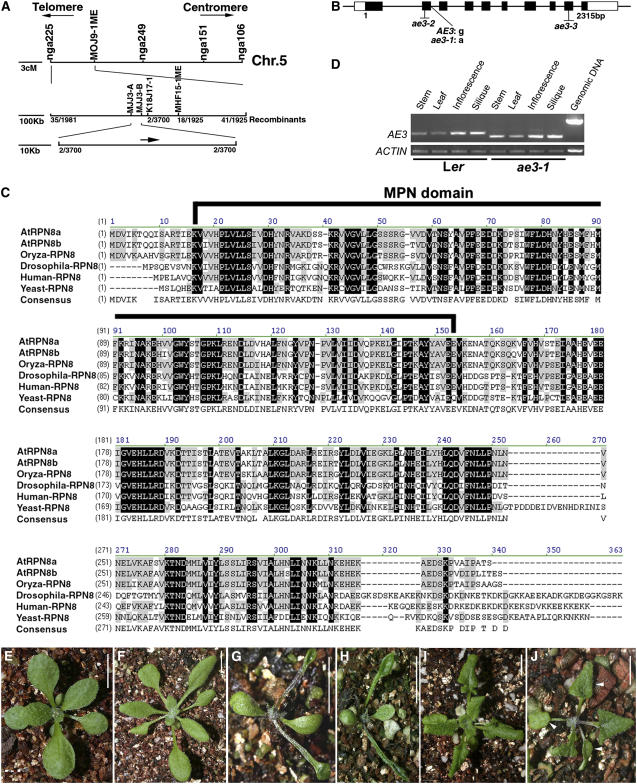
Using ∼3700 recombinant chromosomes, we mapped the AE3 gene on the upper arm of chromosome 5 in a 43-kb region between markers MJJ3-A and MJJ3-B, which contains 13 predicted genes (Figure 7A). Sequencing of these candidate genes revealed that ae3-1 carried a G-to-A nucleotide substitution at the first nucleotide of the second intron in the gene At5g05780 (Figure 7B), which is predicted to encode RPN8a, a 26S proteasome RP subunit (Yang et al., 2004). This nucleotide substitution resulted in AE3 mRNA mis-splicing through elimination of the second exon (sequencing verified; Figures 7B and 7D).
Figure 7.
Molecular Identification of AE3.
(A) Fine structure mapping to localize the AE3 gene on chromosome 5 between markers MJJ3A and MJJ3B.
(B) Structure of the AE3 gene. Black and white boxes indicate the protein coding regions and untranslated regions (UTRs), respectively.
(C) Alignment of the deduced amino acid sequence of AE3 with selected homologous proteins. Residues that are highlighted in black show identity, and conserved residues are highlighted in gray. The MPN domain was indicated by a black line.
(D) Expression pattern of AE3. Note that the PCR products in ae3-1 are smaller in size than those in the wild type due to improper intron splicing.
(E) A T1 transgenic plant carrying a 5-kb complementation fragment is wild-type-like.
(F) An ae3-2 seedling (Col).
(G) An ae3-2 as2-101 double mutant plant.
(H) An ae3-3 as2-101 double mutant plant. Note that the ae3-2 as2-101 and ae3-3 as2-101 phenotypes are very similar to those of ae3-1 as2-101.
(I) An as2-1 seedling (Col).
(J) An ae3-2 as2-1 double mutant seedling showing lotus leaves (arrowheads), which are not observed in the as2-1 single mutant.
Bars = 1 cm in (E) to (J).
The RPN8a protein contains an MPN domain (for Mov34 and Pad1 N-terminal domain), which is conserved in different species (Figure 7C) (Rinaldi et al., 2004; Yang et al., 2004). In addition to RPN8a, Arabidopsis contains another RPN8 homolog, RPN8b; the RPN8a and b amino acid sequences are very similar (Figure 7C). The rpn8b mutant (SALK _128568) did not show obvious phenotypic changes, and the rpn8b as2-101 double mutant only exhibited the as2-101 phenotypes. These results suggest that RPN8a, but not RPN8b, plays a significant role in determination of leaf polarity.
To further confirm that we had identified the correct gene, we transformed a 5-kb genomic fragment containing the putative AE3 coding region plus a 1-kb 5′ region and a 1.7-kb 3′ region into the ae3-1 mutant. This construct complemented almost all phenotypes of the mutant (Figure 7E). In addition, we crossed as2-101 to two additional ae3 alleles, SALK_143140 (Figure 7F) and SALK _151595, which are in the Columbia (Col) genetic background and referred to here as ae3-2 and ae3-3, respectively (Figure 7B). Although the morphology of ae3-2 and ae3-3 plants appeared normal, plants with severe phenotypes similar to those of ae3-1 as2-101 were identified from the F2 progeny of the crosses (Figures 7G and 7H). We also crossed as2-1 (Col) (Figure 7I) to ae3-2. Plants carrying as2 mutations in Col, rather than in Ler, usually do not contain lotus- or needle-like rosette leaves (Xu et al., 2003). However, ae3-2 as2-1 double mutant plants showed the enhanced as2 phenotypes with lotus rosette leaves (Figure 7J, arrowheads) and needle-like cauline leaves. These results indicate that although genetic backgrounds may affect the severity of ae3 as2 phenotypes, the defects in different genetic backgrounds are consistent. In addition, the data also confirm that the defective RPN8a corresponds to the ae3-1 phenotypes.
Phenotypes of the Double Mutants with as2 and Several Other 26S Subunit Mutations
The previously reported Arabidopsis mutants of the 26S proteasome subunit genes each have their unique plant phenotypes (Smalle et al., 2002, 2003; Ueda et al., 2004). In addition, the corresponding subunits of these mutants are all located in the 19S RP, either in the base or in the lid subcomplex. It was therefore proposed that each subunit in the 19S RP is able to distinguish a specific set of substrates for degradation (Smalle et al., 2002, 2003) To determine whether the 19S RP-located RPN8a is the only subunit involved in leaf polarity formation, we constructed double mutants by combining as2-101 with other subunit mutants of the 26S proteasome. These included the 19S RP base mutants hlr-2(rpt2a), rpt4a, rpt5a, and rpn1a, the 19S RP lid mutant rpn9a, and the 20S CP mutants pad1 and pbe1.
Phenotypes of all the single mutants listed above appeared normal except hlr-2(rpt2a) (Ueda et al., 2004) and rpt5a, both of which produced slightly long and narrow leaves. However, plants with the ae3-1 as2-101 abnormalities were observed in each F2 population of crosses between as2-101 and the above single mutants, although the phenotypic severity of these plants was different among the populations. Since all of the 26S subunit mutants are from T-DNA insertion and in the Col ecotype, the progeny are thus in the mixed Col/Ler genetic background. To eliminate the background effect on plant phenotypes, we genotyped using PCR and phenotypically analyzed individual plants that had altered leaf phenotypes (>100 in each F2 population).
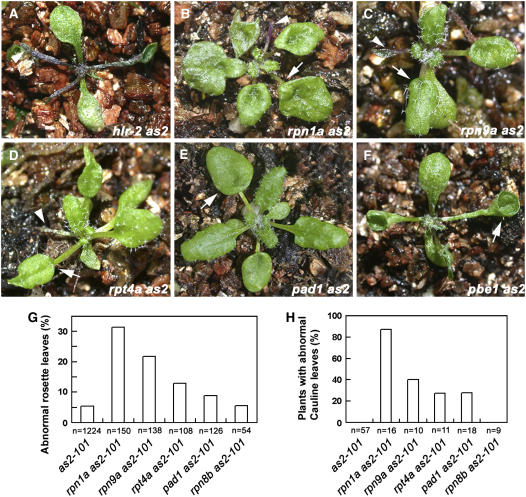
Among the double mutants we identified, leaf phenotypes of hlr-2(rpt2a) as2-101 (Figure 8A), pbe1 as2-101 (Figure 8F), and rpt5a as2-101 (see Supplemental Figure 1 online) appeared more severe than others, with almost all rosette leaves being unexpanded similar to those in the ae3 as2 double mutant. By contrast, rpn1a as2-101 (Figure 8B), rpn9a as2-101 (Figure 8C), rpt4a as2-101 (Figure 8D), and pad1 as2-101 (Figure 8E) produced lotus- and needle-like rosette leaves with a frequency apparently higher than that in the as2-101 single mutant (Figure 8G). The adaxial/abaxial polarity defects are not usually observed in the as2-101 cauline leaves, and the as2 single mutant does not contain ectopic leaves. However, lotus and needle leaves and the ectopic leaves were all found in the less-strong double mutants (Figure 8H). To further rule out the background effects on the double mutant phenotypes, we also analyzed leaf phenotypes of rpn8b as2-101, which is also in the mixed Col/Ler background. Frequency of the abnormal rosette leaves in rpn8b as2-101 was similar to that in as2 (Figure 8G), and the defective cauline leaves were not observed in the double mutant (Figure 8H). Since the aberrant subunits caused by T-DNA insertions are located in different parts of the 26S proteasome, our results strongly suggest that the 26S holoenzyme plays a role in regulating leaf patterning.
Figure 8.
Double Mutants Carrying as2-101 and Several 26S Proteasome Subunit Mutations Displayed Enhanced as2 Abnormalities.
(A) A hlr-2(rpt2a) as2-101 seedling.
(B) An rpn1a as2-101 seedling.
(C) An rpn9a as2-101 seedling.
(D) An rpt4a as2-101 seedling.
(E) A pad1 as2-101 seedling.
(F) A pbe1 as2-101 seedling. In (A) to (F), arrowheads indicate the needle-like leaves, and arrows point to the lotus leaves.
(G) Frequencies of lotus- and needle-like structures. Note that only the first six rosette leaves in each plant were scored. n, total leaves analyzed.
(H) Frequencies of plants with lotus- and needle-like cauline leaves and/or the cauline leaf having an ectopic leaf appearing on the leaf abaxial side.
DISCUSSION
The 26S Proteasome Action Implicates Posttranslational Regulation for Leaf Patterning
Leaf asymmetric growth initiates by partitioning primordia into distinct domains along their adaxial/abaxial axis, and this process has been demonstrated to be controlled by a regulatory network consisting of a number of components (Engstrom et al., 2004). These include the increasing numbers of putative transcription factors (Byrne et al., 2000; Kerstetter et al., 2001; McConnell et al., 2001; Iwakawa et al., 2002), miRNA165/166 (Emery et al., 2003; Kidner and Martienssen, 2003; Bao et al., 2004; Zhong and Ye, 2004), tasiR-ARF (Peragine et al., 2004; Allen et al., 2005; Yoshikawa et al., 2005), and proteins that are involved in RNA silencing (Li et al., 2005; Garcia et al., 2006; Xu et al., 2006; Yang et al., 2006). Previous findings, based on the nature of the identified polarity components, demonstrate the regulation of leaf patterning at both transcriptional and posttranscriptional levels. In this study, we provide genetic evidence that the protein degradation pathway mediated by the 26S proteasome also participates in the leaf adaxial/abaxial polarity formation, suggesting that posttranslational regulation is required for the leaf patterning.
Results of this study also show that the 26S proteasome protein degradation pathway acts in parallel with other pathways, including the AS1-AS2 transcriptional factor pathway and the posttranscriptional gene silencing pathway. Plants with loss of function in the AE3 gene resulted in aberrant leaf adaxial identity, but double mutants ae3 as2(1), ae3 rdr6, and ae3 rev exhibited more severe defects in adaxial leaf identity than those in each of the single mutants. These results suggest that during leaf development, different genetic pathways act synergistically in normal leaf adaxial/abaxial polarity establishment.
The Biological Significance of 26S Protein Degradation in Leaf Patterning
It was previously proposed that abaxial cell fate might be a default in the absence of activity by the adaxial-promoting genes (Sussex, 1954; Eshed et al., 2001; Bowman et al., 2002). On the other hand, some key adaxial-promoting genes for adaxial leaf identity, such as PHB, PHV, and REV, may regulate adaxial fate only at very critical stages during leaf development. For example, these genes are expressed in the adaxial domain only at about P2 to P5 leaf development stages (McConnell et al., 2001; Li et al., 2005). Before or after these stages, they are expressed either throughout the entire leaf primordium or are restricted to the vascular tissues. Because determination of adaxial fate is susceptible to the timing of development, leaves may have developed a very efficient system for the simultaneous regulation of some key components at different levels during certain developmental stages. The system must include initiation or suspension of the genes that are critical in promoting leaf adaxial fates (regulation at the transcriptional level), elimination of these genes' transcripts by RNA silencing (regulation at the posttranscriptional level), and destruction of these genes' products by protein degradation (regulation at the posttranslational level).
Functions of the 26S Proteasome Subunits in Leaf Polarity Formation
In addition to the critical role of the 26S holoenzyme in the degradation of ubiquitylated proteins, the 19S RP is also known to play roles independent of protein degradation by regulating gene expression in yeast (Ferdous et al., 2001; Gillette et al., 2004; Lee et al., 2005). In these studies, we found that mutated genes that encode putative subunits in 19S lid, 19S base, and 20S core subcomplexes all resulted in the comparable leaf phenotypic changes. These results strongly indicate that the proteolytic function of the 26S holoenzyme, rather than the 19S function alone, is involved in leaf polarity formation. We noticed that the phenotypes of different double mutant combinations of as2 with different 26S subunit mutations were not the same in severity. It is possible that the allelic effects may exist among the subunit mutants used in our double mutant constructions. In addition, most Arabidopsis 26S subunits are known to have two isoforms (Fu et al., 1998, 1999; Yang et al., 2004), and they may share partially redundant functions. Therefore, the more the functions are redundant between the two related isoforms, the less severe phenotypes the double mutants have.
RPN8a encoded by AE3 also has its isoform RPN8b, and the amino acid sequence identity between the two proteins is rather high (94%). In addition, these two genes have a similar expression pattern (see Supplemental Figure 2 online). To determine whether and how AE3/RPN8a and RPN8b are functionally redundant, we performed double mutant construction by crossing rpn8b to ae3-1. However, the double mutant ae3-1 rpn8b was not viable. These results indicate that the AE3/RPN8a and RPN8b pair is functionally redundant in fundamental activity. It was reported that in most subunit gene pairs of the Arabidopsis 26S proteasome, one gene was usually more highly expressed than the other (Yang et al., 2004). RPN8a may have a higher expression level than RPN8b; therefore, only rpn8a mutations have a significant impact on leaf adaxial/abaxial polarity formation. However, we cannot rule out another possibility (i.e., that the effectiveness of RPN8a and b subunits may not be the same in recognition of some specific targets). We noted that RPN8b transcript levels were elevated in different tissues in the rpn8a mutant plants (see Supplemental Figure 2 online), whereas the rpn8a phenotypes could not be rescued.
To elucidate the molecular mechanism by which the 26S proteasome acts in establishment of proper leaf polarity, it is important to identify target proteins that play roles in the process. From different pathways that modulate the leaf adaxial/abaxial polarity, we found that the miR165/166 and ta-siRNA pathways may not be involved in the severe ae3 as2 phenotypes. Our miRNA filter hybridization analyses revealed that the miR165/166 levels were not markedly altered in ae3 and ae3 as2 compared with those in the wild type and the as2 single mutants, respectively (see Supplemental Figure 3 online). In addition, the vegetative phase change phenotypes, which depend on the elevated ARF3/ETT and ARF4 expression levels (Fahlgren et al., 2006), were not observed in the ae3 single and ae3 as2 double mutants (see Supplemental Figure 4 online). These results narrowed our target searching to the pathways that are required for normal leaf adaxial/abaxial polarity establishment.
METHODS
Plant Materials and Growth Conditions
The as1-101, as2-101, and rdr6-3 mutants were generated and described previously (Sun et al., 2000, 2002; Xu et al., 2002; Li et al., 2005). Mutagenesis screens for as2 enhancer mutants were described in our previous work (Li et al., 2005). To test whether the phenotypes of the new as2 enhancer were due to a second locus mutation, we pollinated the new mutant flowers with wild-type Ler pollen. The F1 progeny showed wild-type phenotypes, and F2 plants segregated with a distribution of 373 wild type, 133 as2-101, 120 plants that showed novel phenotypes with long and narrow rosette leaves, and 41 new as2 enhancer mutants, close to a 9:3:3:1 ratio. Therefore, the mutant with novel phenotypes was designated ae3-1. The ae3-1 mutant was backcrossed to wild-type Ler three times before detailed phenotypic analyses. Seeds of ae3-2 (SALK_143140), ae3-3 (SALK_151595), rpn8b (SALK_128568), rpn1a (SALK_129604), rpn9a (SALK_147710), rpt4a (SALK_052372), rpt5a (SALK_046321), pad1 (SALK_047984), and pbe1 (SALK_053781) were obtained from the ABRC. Identification of the insertional homozygous single mutants are shown in Supplemental Figure 5 online. Seeds of rev-9 and hlr-2 were kindly provided by J. Bowman (University of California, Davis) and K. Okada (Ueda et al., 2004), respectively. Plant growth was according to our previous conditions (Chen et al., 2000).
Map-Based Cloning
Mapping of the AE3 locus was performed by analysis of an F2 population from a cross between ae3-1 and the polymorphic Col ecotype. The AE3 locus was mapped onto the upper arm of chromosome 5, near the simple sequence length polymorphism marker nga225. A set of simple sequence length polymorphism markers was used to detect polymorphisms between the Col and Ler ecotypes, and the AE3 locus was mapped to a 43-kb area on BAC MJJ3. Sequencing of the candidate genes within this region revealed that ae3 carried a mutation at the first nucleotide of the second intron of the gene At5g05780, which is predicted to encode RPN8a, a 26S proteasome subunit.
For the complementation experiment, a 5.0-kb DNA fragment was PCR amplified from wild-type Col genomic DNA with KOD-PLUS polymerase (Toyobo), sequencing was verified, and the fragment was inserted into the pCAMBIA 1301 transformation vector. This resulted construct was introduced into ae3-1 plants, and 71 independent transgenic T1 plants were obtained that all showed complete rescue of the ae3-1 phenotypes.
RT-PCR
RNA extraction was performed as described previously (Xu et al., 2003) with leaves from 25-d-old seedlings, and reverse transcription was performed with 1 μg total RNA using a kit (Fermentas). PCR was performed with the following gene-specific primers: 5′-CCGGCTACAACAACGCTTACC-3′ and 5′-CTGCAAATGGCTCTTCACA-3′ for KAN1 (exon1/exon2), 5′-AGAAGCACGTTTGCATGG-3′ and 5′-CGAAACTAGTCAGTAGCC-3′ for KAN2 (exon5/3′UTR), 5′-CTTACTTCAATCCCCAGG-3′ and 5′-CTTTTGGACATGATAAACCC-3′ for FIL (exon2/exon7), 5′-CACCACCAGCCAATAGAC-3′ and 5′-ATTGGGAAAGTCTTCGTAG-3′ for YAB3 (exon3/3′UTR), 5′-GCAAAGTGGTTTTTCCTGGA-3′ and 5′-ACATCCAAATCATTCTCTCG- 3′ for AE3 (5′UTR/exon3), and 5′-TGGCATCA(T/C)ACTTTCTACAA-3′ and 5′-CCACCACT(G/A/T)AGCACAATGTT-3′ for ACTIN. Real-time PCR was performed according to our previous methods (Li et al., 2005).
In Situ Hybridization and Detection of GUS Activity
In situ hybridizations were performed as previously described (Long and Barton, 1998) using 14-d-old seedlings. FIL and REV probe preparations and histochemical detection of GUS activity were according to our previous methods (Li et al., 2005).
Microscopy
Fresh tissue from wild-type and mutant plants was examined using a SZH10 dissecting microscope (Olympus). Preparation of thin-section specimens and scanning electron microscopy were according to our previous methods (Chen et al., 2000)
Accession Numbers
Sequence data for the RPN8a protein sequence can be found in the GenBank/EMBL data libraries under accession numbers AAP86668 (Arabidopsis thaliana), BAB78487 (Oryza sativa), AAF47199 (Drosophila melanogaster), NP_002802 (Homo sapiens), and AAS56365 (Saccharomyces cerevisiae), which are used in this article.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. A Seedling of the rpt5a as2-101 Double Mutant.
Supplemental Figure 2. Expression Patterns of AE3/RPN8a and RPN8b.
Supplemental Figure 3. miR165/166 Contents in Leaves of Ler, as2-101, ae3-1, and ae3-1 as2-101.
Supplemental Figure 4. Analyses of Vegetative Phase Length of the Wild Type and Mutants.
Supplemental Figure 5. Identification of the Insertional Mutants.
Supplementary Material
Acknowledgments
We thank J. Bowman for rev-9 seeds, K. Okada for hlr-2 seeds, the ABRC for ae3-2, ae3-3, rpn8b, rpt2a, rpt4a, rpt5a, rpn1a, rpn9a, pad1, and pbe1 seeds, X. Gao, J. Mao, H. Dai, and Y. Dou for their assistance with the scanning electron microscopy, and L. Xu for useful discussion. This research was supported by grants from the Chinese National Scientific Foundation (30421001, 30370751, and 90208009) and the Shanghai Scientific Committee to H.H. and is also partially supported by a grant from the Shanghai Institute for Biological Sciences for the Plant Reproductive Development to H. Ma.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hai Huang (hhuang@sippe.ac.cn).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Allen, E., Xie, Z., Gustafson, A.M., and Carrington, J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221. [DOI] [PubMed] [Google Scholar]
- Bao, N., Lye, K.W., and Barton, M.K. (2004). MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell 7 653–662. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Eshed, Y., and Baum, S.F. (2002). Establishment of polarity in angiosperm lateral organs. Trends Genet. 18 134–141. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). ASYMMETRIC LEAVES1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971. [DOI] [PubMed] [Google Scholar]
- Chen, C., Wang, S., and Huang, H. (2000). LEUNIG has multiple functions in gynoecium development in Arabidopsis. Genesis 26 42–54. [DOI] [PubMed] [Google Scholar]
- Chen, Q., Atkinson, A., Otsuga, D., Christensen, T., Reynolds, L., and Drews, G.N. (1999). The Arabidopsis FILAMENTOUS FLOWER gene is required for flower formation. Development 126 2715–2726. [DOI] [PubMed] [Google Scholar]
- Emery, J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F., and Bowman, J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13 1768–1774. [DOI] [PubMed] [Google Scholar]
- Engstrom, E.M., Izhaki, A., and Bowman, J.L. (2004). Promoter bashing, microRNAs, and Knox genes. New insights, regulators, and targets-of-regulation in the establishment of lateral organ polarity in Arabidopsis. Plant Physiol. 135 685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., Perea, J.V., and Bowman, J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11 1251–1260. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Izhaki, A., Baum, S.F., Floyd, S.K., and Bowman, J.L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131 2997–3006. [DOI] [PubMed] [Google Scholar]
- Fahlgren, N., Montgomery, T.A., Howell, M.D., Allen, E., Dvorak, S.K., Alexander, A.L., and Carrington, J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16 939–944. [DOI] [PubMed] [Google Scholar]
- Ferdous, A., Gonzalez, F., Sun, L., Kodadek, T., and Johnston, S.A. (2001). The 19S regulatory particle of the proteasome is required for efficient transcription elongation by RNA polymerase II. Mol. Cell 7 981–991. [DOI] [PubMed] [Google Scholar]
- Fu, H., Doelling, J.H., Arendt, C.S., Hochstrasser, M., and Vierstra, R.D. (1998). Molecular organization of the 20S proteasome gene family from Arabidopsis thaliana. Genetics 149 677–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., Doelling, J.H., Rubin, D.M., and Vierstra, R.D. (1999). Structural and functional analysis of the six regulatory particle triple-A ATPase subunits from the Arabidopsis 26S proteasome. Plant J. 18 529–539. [DOI] [PubMed] [Google Scholar]
- Garcia, D., Collier, S.A., Byrne, M.E., and Martienssen, R.A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16 933–938. [DOI] [PubMed] [Google Scholar]
- Gillette, T.G., Gonzalez, F., Delahodde, A., Johnston, S.A., and Kodadek, T. (2004). Physical and functional association of RNA polymerase II and the proteasome. Proc. Natl. Acad. Sci. USA 101 5904–5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman, M.H., Rubin, D.M., Fried, V.A., and Finley, D. (1998). The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 18 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawker, N.P., and Bowman, J.L. (2004). Roles for class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol. 135 2261–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, A. (2000). Development of symmetry in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 349–370. [DOI] [PubMed] [Google Scholar]
- Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., Hasebe, M., Soma, T., Ikezaki, M., Machida, C., and Machida, Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43 467–478. [DOI] [PubMed] [Google Scholar]
- Kerstetter, R.A., Bollman, K., Taylor, R.A., Bomblies, K., and Poethig, R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411 706–709. [DOI] [PubMed] [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2003). Macro effects of microRNAs in plants. Trends Genet. 19 13–16. [DOI] [PubMed] [Google Scholar]
- Lee, D., Ezhkova, E., Li, B., Pattenden, S.G., Tansey, W.P., and Workman, J.L. (2005). The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell 123 423–436. [DOI] [PubMed] [Google Scholar]
- Li, H., Xu, L., Wang, H., Yuan, Z., Cao, X., Yang, Z., Zhang, D., Xu, Y., and Huang, H. (2005). The putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and microRNA165/166 in Arabidopsis leaf development. Plant Cell 17 2157–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W.C., Shuai, B., and Springer, P.S. (2003). The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A., and Barton, M.K. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125 3027–3035. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411 709–713. [DOI] [PubMed] [Google Scholar]
- Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H.L., and Poethig, R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Y., Sun, Y., Xu, L., Xu, Y., and Huang, H. (2004). ERECTA is required for protection against heat-stress in the AS1/ AS2 pathway to regulate adaxial-abaxial leaf polarity in Arabidopsis. Planta 219 270–276. [DOI] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. (2002). Prediction of plant microRNA targets. Cell 110 513–520. [DOI] [PubMed] [Google Scholar]
- Rinaldi, T., Pick, E., Gambadoro, A., Zilli, S., Maytal-Kivity, V., Frontali, L., and Glickman, M.H. (2004). Participation of the proteasomal lid subunit Rpn11 in mitochondrial morphology and function is mapped to a distinct C-terminal domain. Biochem. J. 381 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, S., Watanabe, K., Goto, K., Liu, Y.G., Shibata, D., Kanaya, E., Morita, E.H., and Okada, K. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126 4117–4128. [DOI] [PubMed] [Google Scholar]
- Smalle, J., Kurepa, J., Yang, P., Babiychuk, E., Kushnir, S., Durski, A., and Vierstra, R.D. (2002). Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J., Kurepa, J., Yang, P., Emborg, T.J., Babiychuk, E., Kushnir, S., and Vierstra, R.D. (2003). The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15 965–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J., and Vierstra, R.D. (2004). The ubiquitin 26s proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55 555–590. [DOI] [PubMed] [Google Scholar]
- Sun, Y., Zhang, W., Li, F., Guo, Y., Liu, T., and Huang, H. (2000). Identification and genetic mapping of four novel genes that regulate leaf development in Arabidopsis. Cell Res. 10 325–335. [DOI] [PubMed] [Google Scholar]
- Sun, Y., Zhou, Q., Zhang, W., Fu, Y., and Huang, H. (2002). ASYMMETRIC LEAVES1, an Arabidopsis gene that is involved in the control of cell differentiation in leaves. Planta 214 694–702. [DOI] [PubMed] [Google Scholar]
- Sussex, I.M. (1954). Experiments on the cause of dorsaiventrality in leaves. Nature 174 351–352. [Google Scholar]
- Ueda, M., Matsui, K., Ishiguro, S., Sano, R., Wada, T., Paponov, I., Palme, K., and Okada, K. (2004). The HALTED ROOT gene encoding the 26S proteasome subunit RPT2a is essential for the maintenance of Arabidopsis meristems. Development 131 2101–2111. [DOI] [PubMed] [Google Scholar]
- Voges, D., Zwickl, P., and Baumeister, W. (1999). The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68 1015–1068. [DOI] [PubMed] [Google Scholar]
- Xu, L., Xu, Y., Dong, A., Sun, Y., Pi, L., and Huang, H. (2003). Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130 4097–4107. [DOI] [PubMed] [Google Scholar]
- Xu, L., Yang, L., Pi, L., Liu, Q., Ling, Q., Wang, H., Poethig, R.S., and Huang, H. (2006). Genetic interaction between the AS1-AS2 and RDR6-SGS3-AGO7 pathways for leaf morphogenesis. Plant Cell Physiol. 47 853–863. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Sun, Y., Liang, W., and Huang, H. (2002). The Arabidopsis AS2 gene encoding a predicted leucine-zipper protein is required for the leaf polarity formation. Acta Bot. Sin. 44 1194–1202. [Google Scholar]
- Yang, L., Huang, W., Wang, H., Cai, R., Xu, Y., and Huang, H. (2006). Characterizations of a hypomorphic argonaute1 mutant reveal novel AGO1 functions in Arabidopsis lateral organ development. Plant Mol. Biol. 61 63–78. [DOI] [PubMed] [Google Scholar]
- Yang, P., Fu, H., Walker, J., Papa, C.M., Smalle, J., Ju, Y.M., and Vierstra, R.D. (2004). Purification of the Arabidopsis 26 S proteasome: Biochemical and molecular analyses revealed the presence of multiple isoforms. J. Biol. Chem. 279 6401–6413. [DOI] [PubMed] [Google Scholar]
- Yoshikawa, M., Peragine, A., Park, M.Y., and Poethig, R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.H. (2004). Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol. 45 369–385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.