Abstract
We isolated two T-DNA insertion mutants of Arabidopsis thaliana GLUTATHIONE PEROXIDASE3 (ATGPX3) that exhibited a higher rate of water loss under drought stress, higher sensitivity to H2O2 treatment during seed germination and seedling development, and enhanced production of H2O2 in guard cells. By contrast, lines engineered to overexpress ATGPX3 were less sensitive to drought stress than the wild type and displayed less transpirational water loss, which resulted in higher leaf surface temperature. The atgpx3 mutation also disrupted abscisic acid (ABA) activation of calcium channels and the expression of ABA- and stress-responsive genes. ATGPX3 physically interacted with the 2C-type protein phosphatase ABA INSENSITIVE2 (ABI2) and, to a lesser extent, with ABI1. In addition, the redox states of both ATGPX3 and ABI2 were found to be regulated by H2O2. The phosphatase activity of ABI2, measured in vitro, was reduced approximately fivefold by the addition of oxidized ATGPX3. The reduced form of ABI2 was converted to the oxidized form by the addition of oxidized ATGPX3 in vitro, which might mediate ABA and oxidative signaling. These results suggest that ATGPX3 might play dual and distinctive roles in H2O2 homeostasis, acting as a general scavenger and specifically relaying the H2O2 signal as an oxidative signal transducer in ABA and drought stress signaling.
INTRODUCTION
H2O2 is generated in cells by the direct transfer of two electrons to the superoxide anion (O2.−). This is mediated by enzymes such as glycolate and glucose oxidases, or by the dismutation of O2.−, which frequently occurs in chloroplasts and peroxisomes under conditions of high light stress. In the dark, or in nonphotosynthetic tissues, H2O2 is produced mainly by leakage of the electron transport chain in mitochondria or microsomes (Vanlerberghe and McIntosh, 1997) and during fatty acid oxidation (Mittler, 2002). In the defense reaction to pathogen attack, the rate of cellular H2O2 production is enhanced dramatically as a consequence of superoxide dismutase activation, catalyzing the conversion of O2.− to H2O2 (Doke, 1985; Mittler et al., 1999), and of activation of the NADPH-dependent oxidase system (Apostol et al., 1989; Baker and Orlandi, 1995). To cope with increased levels of H2O2, plants have evolved different enzymatic and nonenzymatic mechanisms (Apel and Hirt, 2004; Mittler et al., 2004). These include free radical scavengers, such as superoxide dismutase, catalase (CAT), and peroxidases, and the enzymes involved in the ascorbate–glutathione cycle (Noctor and Foyer, 1998).
Although early studies focused on the cytotoxic properties of H2O2, which reacts with and thereby damages DNA, proteins, and lipids (Bolwell et al., 1995), recent evidence suggests that H2O2 also functions as a crucial component of plant development, growth, and stress responses, particularly those induced by environmental stimuli (Neill et al., 2002; Foyer and Noctor, 2005). Production of H2O2 has been found to be enhanced by abiotic and biotic stresses such as high light, osmotic stresses, and stresses attributable to pathogen attack (Mittler et al., 2004). Treatment with the plant hormone abscisic acid (ABA) has also been found to increase production of H2O2 within Arabidopsis thaliana and Vicia faba guard cells (Miao et al., 2000; Pei et al., 2000; Zhang et al., 2001b; Kwak et al., 2003). ABA-stimulated H2O2 accumulation subsequently induces stomatal closure via the activation of plasma membrane calcium channels (Pei et al., 2000).
Various Arabidopsis mutants have been used to dissect ABA and reactive oxygen species (ROS) signaling in guard cells. The mutants ABA insensitive1 (abi1) and abi2, defective in genes encoding protein phosphatase 2C (PP2C)-like enzymes, showed an insensitive phenotype during germination and in guard cell responses to ABA (Koornneef et al., 1984; Gosti et al., 1999). Treatment with ABA did not induce the production of H2O2 in abi1 mutants, whereas this response was not impaired in abi2-1. These data suggest that ABI1 may function upstream of ABA and ROS signaling, whereas ABI2 may function downstream. Interestingly, the recessive mutant growth controlled by abscisic acid2 (gca2) displayed insensitivity of the stomatal closure response to treatment with ABA (Pei et al., 2000). ABA increased H2O2 production in these mutants, but H2O2-induced calcium channel activation and stomatal closure were absent (Pei et al., 2000). ABA treatment is known to activate the Ser/Thr protein kinase OST1, an Arabidopsis homolog of V. faba AAPK, which plays a positive role in ABA responses in guard cells (Mustilli et al., 2002; Assmann, 2003). ABA-induced ROS production is absent from ost1 plants, although ost1 stomata still close in response to H2O2. These results suggest that H2O2 acts as a signal molecule involved in the regulation of ABA-induced stomatal closure. Signaling mediated by ROS is also known to involve a heterotrimeric G-protein (Joo et al., 2005) and protein phosphorylation modulated by a specific mitogen-activated protein kinase and protein Tyr phosphatase (Kovtun et al., 2000; Gupta and Luan, 2003; Rentel et al., 2004). An unresolved question regarding ABA and H2O2 signaling is how this oxidative signal transduces to or interacts with ABA signaling.
It is well established that glutathione peroxidases (GPXs) are key enzymes involved in scavenging oxyradicals in animals (Arthur, 2000). The GPX family of proteins can be divided into five classes according to their amino acid sequence, substrate specificities, and subcellular localization. GPXs catalyze the reduction of H2O2, organic hydroperoxides, and lipid peroxides using GSH and/or other reducing equivalents (Ursini et al., 1995). However, there is increasing evidence that GPXs also interact specifically with protein partners to confer peroxide-induced oxidation. For example, Delaunay et al. (2002) demonstrated that a thiol peroxidase, GPX3, forms a complex with the basic domain/leucine zipper transcription factor Yeast Activation Protein1 (Yap1), which is required for the H2O2-induced oxidation of this protein in Saccharomyces cerevisiae. Thus, GPX3 functions in both hydroperoxide sensing and scavenging. Another recent report implicates ethylene in delaying stomatal closure by inhibiting the ABA signaling pathway in plants (Tanaka et al., 2005). The Arabidopsis ethylene receptor ETR1 mediated H2O2 signaling in stomata guard cells and might serve as a sensor of H2O2 in plant cells (Desikan et al., 2005). By contrast, the repression of ABA-induced RAB18 expression by 1-aminocyclopropane-1-carboxylic acid treatment suggests that ethylene signaling might impair the Ca2+ influx or the S-type anion channel activation downstream of ABA signaling. Thus, ethylene does not appear to interfere with the early steps of the ABA signaling pathway (Tanaka et al., 2005).
Until recently, few reports on the existence of plant enzymatic activity similar to animal GPX have appeared (Holland et al., 1993; Beeor-Tzahar et al., 1995; Depege et al., 1998; Faltin et al., 1998; Herbette et al., 2002; Jung et al., 2002). Several cDNAs have been isolated, from diverse plant sources, exhibiting sequence similarity to animal GPX (Holland et al., 1993; Depege et al., 1998; Roeckle-Drevet et al., 1998). Most plant GPX proteins have a primary structure similar to that of the animal phospholipid hydroperoxide glutathione peroxidase (PHGPX) enzymes (Churin et al., 1999). These PHGPXs are considered the main enzymatic defense against the oxidative destruction of membranes (Ursini and Bindoli, 1987; Brigelius-Flohe et al., 1994). In addition, it has been found that GPX mRNA steady levels increase under different environmental stresses, such as pathogen infection (Criqui et al., 1992), treatment with high salt and metal concentrations (Sugimoto and Sakamoto, 1997), mechanical stimulation (Depege et al., 1998), and under conditions of aluminum toxicity (Rodriguez Milla et al., 2002). However, plant GPXs have lower activities than those of animals because they contain Cys at the putative catalytic site rather than the selenocysteine typical of animal GPXs (Holland et al., 1993; Faltin et al., 1998; Herbette et al., 2002; Jung et al., 2002). This low activity has made it difficult to clarify the physiological role of GPX in higher plants.
Previous reports showed that Arabidopsis GPX (ATGPX) might be a potential detoxifier of H2O2, using GSH directly as a reducing reagent (Eshdat et al., 1997). The ATGPX family comprises seven members, which are specifically targeted to the cytosol, chloroplasts, mitochondria, peroxisomes, and apoplast (Rodriguez Milla et al., 2003). These genes are expressed ubiquitously and are regulated by abiotic stresses through diverse signaling pathways (Rodriguez Milla et al., 2003). However, at present, the function of these enzymes in plants is not completely understood.
We report here that ATGPX3 plays dual roles, the first in the general control of H2O2 homeostasis, and the second in specifically linking ABA and H2O2 signaling in stomatal closure, thereby regulating plant water transpiration. This suggests that a conserved mechanism may exist in plants similar to the yeast H2O2 sensor/transducer (Delaunay et al., 2002), whereby ABA and H2O2 directly regulate gene transcription through GPX-mediated perception of H2O2.
RESULTS
Isolation and Characterization of T-DNA Insertion Mutants and of Transgenic Plants Overexpressing ATGPX3
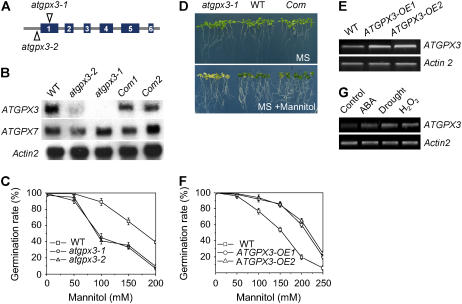
We identified two Arabidopsis lines from the SALK collection (Alonso et al., 2003), donor stock numbers SALK_071176 and SALK_001116, containing T-DNA insertions in the ATGPX3 gene (At2g43350); these lines are named atgpx3-1 and atgpx3-2. The sites of T-DNA insertion are located in the first exon of ATGPX3 in atgpx3-1 and near its initiating codon in atgpx3-2 (Figure 1A). The insertions would be expected to eliminate or strongly reduce the transcription of ATGPX3, and this was confirmed by RNA gel blot analyses (Figure 1B). The slight differences in phenotype between atgpx3-1 and atgpx3-2 may be attributed to incomplete elimination of the transcript in the latter (see below).
Figure 1.
Characterization of atgpx3 T-DNA Insertion Mutants and Overexpression Transgenic Plants.
(A) The insertion positions of T-DNA in the ATGPX3 gene.
(B) RNA gel blots show ATGPX3 expression in the wild type, atgpx3 mutants, and complemented ATGPX3 (Com) lines. Total RNA was extracted from the wild type, atgpx3-1, atgpx3-2, and complementation lines. Fifteen micrograms of total RNA was loaded in each lane. An Actin gene and ATGPX7 were used as a loading control and a positive control, respectively.
(C) Germination sensitivity to osmotic stress in atgpx3 mutants. Seeds from the wild type, atgpx3-1, and atgpx3-2 were germinated on MS agar medium or MS medium supplemented with different concentrations of mannitol for 7 d. Values are means ± sd of three independent experiments (>120 seeds per point).
(D) Complementation of the phenotype conferred by atgpx3-1 by expression of wild-type ATGPX3. After germinating for 5 d in MS medium, the seedlings were transferred to MS agar medium or MS medium containing 200 mM mannitol and grown for 15 d.
(E) Expression of ATGPX3 in two ATGPX3 overexpression transgenic lines.
(F) Germination insensitivity to osmotic stress in ATGPX3 overexpression transgenic plants. Seeds from the wild type and both overexpression lines were germinated on MS agar medium or MS medium supplemented with different concentrations of mannitol for 7 d. Values are means ± sd of three independent experiments (>120 seeds per point).
(G) RT-PCR analysis of ATGPX3 gene transcripts in response to stress conditions. Arabidopsis seedlings were grown on MS medium plates for 15 d. Wild-type plants were treated with 60 μM ABA, drought stress, or 5 mM H2O2 for 4 h. Total RNA was isolated from treated and untreated wild-type plants. Actin2 primer was used as an internal control.
Compared with the wild type, seed germination of atgpx3-1 and atgpx3-2 is more sensitive to mannitol, and this effect is dose-dependent (Figure 1C). The leaves of mutant seedlings, maintained for prolonged periods on Murashige and Skoog (1962) (MS) medium containing 200 mM mannitol, became chlorotic (Figure 1D). To exclude the possibility that a second site mutation closely linked to the ATGPX3 T-DNA insertion might be responsible for the phenotype, the coding sequence of wild-type ATGPX3 was placed under the control of the constitutive 35S promoter of Cauliflower mosaic virus and was expressed in homozygous atgpx3 plants. Transgenic lines resistant to both hygromycin B (transgene T-DNA) and kanamycin (knockout T-DNA) were phenotypically indistinguishable from wild-type lines. A transgenic ATGPX3-1 plant complemented with the complete wild-type ATGPX3 gene was obtained, and its self progeny (T2) was analyzed. As shown in Figures 1B and 1D, the complementation lines displayed a wild-type phenotype regarding seed germination and leaf color under osmotic stress; other phenotypes, such as stomatal closure and activity of calcium channels in response to ABA (see below), caused by the defects in ATGPX3 were also recovered. Therefore, we conclude that the disruption of the ATGPX3 gene is responsible for the phenotypes observed in the two mutant lines.
To further investigate the function of ATGPX3 in vivo, we also overexpressed the gene by placing its complete coding sequence under the transcriptional regulation of the constitutive super promoter (Narasimhulu et al., 1996). Fifteen T3 homozygous transgenic lines of Arabidopsis were recovered, and representative lines with high ATGPX3 expression (Figure 1E) were used for detailed analyses. As done for the atgpx3 mutants described in Figure 1C, we first tested whether osmotic stress tolerance was affected by overexpression of ATGPX3. Germination of wild-type seeds was arrested in the MS medium containing 200 mM mannitol. By contrast, the seeds of plants overexpressing ATGPX3 germinated normally under these conditions (see Supplemental Figure 1 online). The effect of mannitol on the inhibition of wild-type seed germination was significant between concentrations of 100 and 250 mM mannitol (Figure 1F; significant difference according to the Steel-Dwass test [P < 0.05]).
Next, we tested whether ABA, H2O2, and drought stresses induced ATGPX3 expression by RT-PCR analysis using total RNA isolated from Arabidopsis seedlings (Figure 1G). In these experiments, seedlings were exposed to stress conditions for only 4 h to avoid possible secondary effects. The data indicate that ABA, H2O2, and drought stresses induced the accumulation of ATGPX3 mRNAs.
atgpx3 Mutants Are Sensitive to and Produce More H2O2
Previous analyses had suggested that GPX is one of the key enzymes involved in scavenging oxyradicals in animals (Arthur, 2000), and ATGPX3 was considered to be a putative peroxidase in Arabidopsis (Rodriguez Milla et al., 2003). To examine whether a deficiency of ATGPX3 might affect the activity of scavenging enzymes, we investigated the sensitivity of atgpx3-1 and atgpx3-2 to H2O2. The cotyledons of atgpx3-1 and atgpx3-2 plants were bleached by 3 mM H2O2 in the medium (Figure 2A), and the growth of true leaves was delayed when the seeds were permitted to grow for 15 d after germination. Compared with the wild type, true leaf development of atgpx3 mutant plants was inhibited by ∼40% (Figure 2B).
Figure 2.
The atgpx3 Mutants Are More Sensitive to H2O2 in Terms of Seedling Growth and Produce More H2O2 in Guard Cells.
(A) Sensitivity to H2O2 during seedling development. Seeds from the wild type, atgpx3-1, and atgpx3-2 were germinated and allowed to grow on vertical agar medium containing MS nutrients (top panel) or MS nutrients supplemented with 3 mM H2O2 (bottom panel). The photographs were taken at 10 d after seed imbibition.
(B) Seedlings with true leaves over total number of seeds planted on MS medium supplemented with H2O2 at the indicated concentrations. Data represent means ± sd of four independent experiments (>120 seeds per point).
(C) Exogenous ABA-induced production of ROS in guard cells. A pair of guard cells from the wild type and mutants loaded with H2DCF-DA before and 5 min after the addition of 10 μM ABA. The micrographs show representative fluorescence images from guard cells of the wild type and atgpx3 mutants in three independent experiments. The pseudocolor key is shown in the bottom right panel and was applied to pixel intensity (0 to 255) for all fluorescence images. Bar = 10 μm for all images.
(D) Effects of ABA on the DCF fluorescence in guard cells of wild-type and atgpx3 plants. The time points represent means ± se from measurements of pixel intensity in whole cells determined before and 5 min after ABA (10 μM) treatment in three independent experiments (for detailed steps for measuring pixel intensity, see Methods). For the wild type, n = 150 cells before ABA treatment, n = 110 cells after ABA treatment; for atgpx3-1, n = 110 cells before ABA treatment, n = 150 cells after ABA treatment; and for atgpx3-2, n = 96 cells before ABA treatment, n = 105 cells after ABA treatment.
(E) Histochemical localization of ABA-induced ROS production in leaves of wild-type and atgpx3 plants visualized using DAB. The detached leaves of plants were treated with 10 μM ABA for 10 min and then stained with DAB. Shown are representative leaves from three independent experiments.
(F) Assays of ATGPX3 peroxidase activity. Line A, complete reaction without thioredoxin; line B, complete reaction without ATGPX3; line C, complete assay in the presence of GSH, without thioredoxin and thioredoxin reductase; line D, complete assay in the presence of ATGPX3, thioredoxin, thioredoxin reductase, NADPH, and H2O2.
(G) In vitro reduction of ATGPX3. Lane 1, oxidized ATGPX3; lane 2, reduced ATGPX3; lane 3, oxidized ATGPX3, thioredoxin, thioredoxin reductase, and NADPH; lane 4, reduced ATGPX3, H2O2, thioredoxin, thioredoxin reductase, and NADPH; lane 5, oxidized ATGPX3, GSH, glutathione reductase, and NADPH. Shown is a representative gel from three independent experiments.
The abilities of atgpx3-1, atgpx3-2, and wild-type plants to produce and scavenge H2O2 after ABA treatment were determined more directly by monitoring H2O2 production using 2′,7′-dichlorofluorescein (H2DCF). The nonpolar diacetate ester of H2DCF (H2DCF-DA) freely permeates the cell (Allan and Fluhr, 1997; Zhang et al., 2001b), where it is hydrolyzed into the more polar, nonfluorescent compound H2DCF, which therefore is trapped. Subsequent oxidation of H2DCF by H2O2, catalyzed by peroxidases, yields DCF, which is highly fluorescent (Cathcart et al., 1983). H2DCF-DA loads readily into guard cells, and its optical properties are amenable to analysis using laser scanning confocal microscopy. Exogenous application of ABA enhanced the relative fluorescence intensity of DCF in guard cells from atgpx3-1, atgpx3-2, and wild-type cells, which is represented by pixel intensity averaged over whole cells (Figure 2C). As expected, the relative fluorescence emission increased more quickly in the guard cells of both atgpx3 mutant plants with 10 μM ABA treatment compared with the wild-type plants (Figure 2D; a significant difference according to the Stell-Dwass test [P < 0.03]). After a 5-min treatment with 10 μM ABA, the pixel intensity of fluorescence emission in guard cells from atgpx3-1 and atgpx3-2 plants was 94 and 76% higher than that of wild-type plants, respectively.
The 3,5-diaminobenzidine (DAB) uptake method was also used to examine the production of ROS in leaves of wild-type and atgpx3 mutants after ABA treatment. ABA-induced ROS increases were observed in the leaves of wild-type, atgpx3-1, and atgpx3-2 plants (Figure 2E). ROS were clearly distinguishable as dark brown deposits localized in veins and some areas near the veins of leaves. Importantly, the leaves of atgpx3-1 and atgpx3-2 stained more intensely than did wild-type leaves. Thus, both cellular and histochemical analyses revealed that ATGPX3 plays an important role as a scavenger in the control of H2O2 homeostasis.
We also tested the peroxidase activity of ATGPX3 in vitro using different substrates. With thioredoxin as a substrate, significant peroxidase activity was observed in the presence of thioredoxin reductase. By contrast, with GSH as a substrate, ATGPX3 did not show any detectable peroxidase activity either in the presence or absence of glutathione reductase (Figure 2F, curve D versus curves A, B, and C; significant difference according to the Stell-Dwass test [P < 0.03]). Moreover, in a native PAGE assay system, reduced ATGPX3-GST (for glutathione S-transferase) migrates more slowly than the oxidized form because free SH groups were modified by iodoacetamide (Kobayashi et al., 1997) (Figure 2G). The oxidized form of peroxidase becomes the reduced form with thioredoxin and the thioredoxin reductase system, but not with the GSH system (Figure 2G, lanes 3 and 5). Even after the addition of H2O2 in the thioredoxin system, the reduced form of GPX3 was not able to turn into the oxidized form (Figure 2G, lane 4). These data suggest that thioredoxin, not GSH, may be the physiological electron donor system for ATGPX3. These findings are consistent with previous reports in which many plant GPX-like proteins were found to function as antioxidant enzymes, having PHGPX and thioredoxin activities (Herbette et al., 2002; Comtois et al., 2003).
Deficiency of ATGPX3 Impairs the ABA- and H2O2-Regulated Stomatal Responses
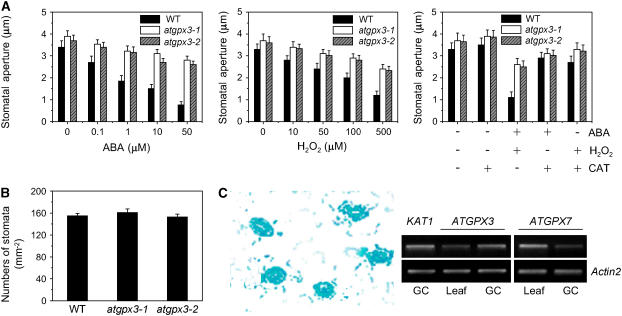
Several lines of evidence indicate that H2O2 may serve as a second messenger for ABA action in guard cells (Guan et al., 2000; Pei et al., 2000; Zhang et al., 2001b; Foyer and Noctor, 2005). Having determined that atgpx3-1 and atgpx3-2 produced more H2O2 than did wild-type plants, we expected that mutation of ATGPX3 might also disturb stomatal closing in response to ABA. Indeed, measurements of stomatal aperture showed that the degree of opening of the mutant stomata without ABA treatment was slightly larger than that seen in the wild type (Figure 3A). Stomatal closing in atgpx3 plants was less sensitive to ABA than that in wild-type plants at 1, 10, and 50 μM ABA (Figure 3A, left panel [P < 0.03]). The stomatal aperture of atgpx3-1 was 3.21 ± 0.25 μm, or 82% of the control value at 2 h after treatment with 1 μM ABA; that of the wild type was 1.85 ± 0.25 μm, or 54% of the control value at the same time point. Likewise, the atgpx3 mutations resulted in an impaired response of guard cells to H2O2 (Figure 3A, middle panel). However, both ABA- and H2O2-induced stomatal closure was partially recovered by the addition of CAT in the solution (Figure 3A, right panel [P < 0.02]). By contrast, atgpx3-1 and agpx3-2 did not exhibit a significant difference in stomatal density compared with the wild type (Figure 3B). Together, these data suggest that the ATGPX3 protein is involved in ABA-mediated H2O2 production, which controls stomatal behavior and transpirational water loss.
Figure 3.
atgpx3 Mutants Impair ABA- and H2O2-Induced Stomatal Closure but Not Stomatal Density.
(A) Effects of ABA and H2O2 on stomatal closure in the wild type, atgpx3-1, and atgpx3-2. Stomatal apertures were measured on epidermal peels of the wild type and atgpx3-1. Values are means ± sd of 50 measurements from three independent experiments. Left panel, ABA-induced stomatal closing; middle panel, H2O2-induced stomatal closing; right panel, effects of CAT on ABA- (10 μM) and H2O2- (100 μM) induced stomatal closing.
(B) Stomatal density in the epidermis of the abaxial surface of rosette leaves from the wild type, atgpx3-1, and atgpx3-2. Stomatal density is presented as stomatal numbers per square millimeter ± se, determined from leaves of five individual wild-type and mutant plants. Five independent counts were performed on each leaf.
(C) Expression of ATGPX3-GUS in guard cells and RT-PCR analysis. Left panel, GUS activity in guard cells of wild-type plants expressing the GUS reporter gene under the control of the ATGPX3 promoter; right panel, ATGPX3 transcript detected by RT-PCR analysis of 10 μg of guard cell–enriched total RNA (GC). Ten micrograms of total RNA from whole leaves was run in parallel as a control (Leaf). KAT1 and ATGPX7 were used as positive controls in guard cells.
Consistent with a role in stomatal signal transduction, high levels of glucuronidase (GUS) expression were detected in the guard cells of ATGPX3 promoter–GUS transgenic plants (Figure 3C, left panel). RT-PCR analysis of total RNA extracted from guard cell–enriched leaf epidermal strips confirmed the expression of ATGPX3 in guard cells (Figure 3C, right panel). By contrast, the expression of ATGPX7 was not restricted to guard cells. The strong expression of ATGPX3 in guard cells is consistent with the suggestion that ATGPX3 functions in stomatal regulation to control water loss.
Deficiency and Overexpression of ATGPX3 Reduced and Enhanced Drought Stress Tolerance, Respectively
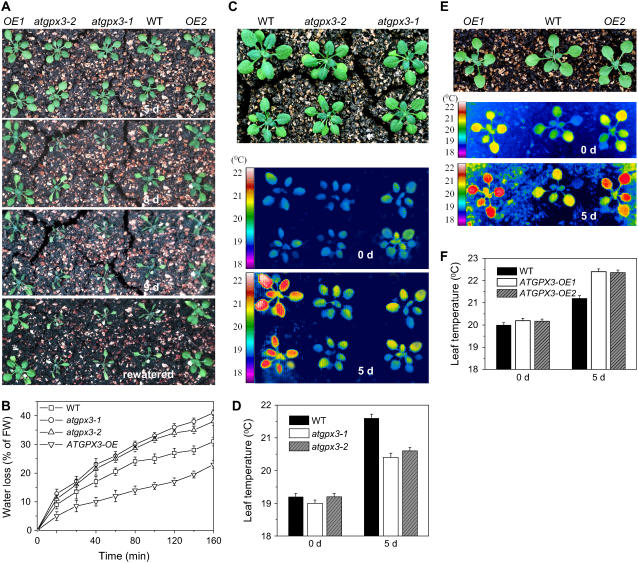
In response to drought, plants typically synthesize ABA, which triggers the closing of stomata, thus reducing water loss and enhancing drought stress resistance (Schroeder et al., 2001; Luan, 2002). To test whether ATGPX3 might be involved in plant drought stress responses, atgpx3 mutants, overexpression lines, and wild-type plants were grown in soil in continuous light for 10 d. After withholding water for 5 d, the visible phenotypes of atgpx3 mutants and overexpression lines were no different from those of the wild type (Figure 4A, top panel). However, by day 7 after treatment, the mutant plants began to display symptoms of dehydration, and permanent wilting was observed by day 8. Although wild-type plants also displayed dehydration symptoms, wilting was only temporary (Figure 4A, second panel). By contrast, overexpression lines of ATGPX3 were found to grow normally for the first 8 d of drought treatment and did not wilt until day 9. These results imply that the drought tolerance in ATGPX3 transgenic plants was increased to that of wild-type plants and was much greater than that of the mutants. Rewatering did not allow the recovery of atgpx3-1 and atgpx3-2 plants drought-treated for 9 d, whereas the overexpression lines, treated similarly, appeared normal (Figure 4A, bottom panel). Furthermore, wilting of atgpx3-2 occurred 1 d later than in atgpx3-1 (see Supplemental Figures 2A and 2C online), probably as a result of the fact that atgpx3-2 is only a partial knockdown allele. We subsequently compared the rates of water loss in rosette leaves and found that atgpx3-1 and atgpx3-2 mutant leaves lost water slightly faster than did wild-type leaves (Figure 4B; significant difference according to the Stell-Dwass test [P < 0.04]), whereas water loss was significantly lower in ATGPX3 overexpression lines compared with the wild type (Figure 4B).
Figure 4.
Responses of atgpx3 Mutants and Overexpression Lines to Drought Stress.
(A) Analysis of the drought stress sensitivity of Arabidopsis seedlings. Seven-day-old Arabidopsis seedlings were transferred to soil and grown for 10 d in a growth chamber, after which watering was stopped for the drought stress treatment. The photographs were taken (from top to bottom) 5, 8, and 9 d later and again 1 d after being rewatered. OE, overexpression.
(B) Transpirational water loss in wild-type, atgpx3, and overexpression lines at the indicated time points after detachment. Water loss is expressed as the percentage of initial fresh weight (FW). Values are means ± sd of four samples (each sample had six leaves).
(C) False-color infrared images of drought-stressed plantlets. The thermal images show leaf temperature profiles of 14-d-old wild-type, atgpx3-1, and atgpx3-2 plantlets at the start of drought treatment (middle panel) and 5 d later (bottom panel). The top panel shows the 5-d drought-stressed plants.
(D) Temperature of the leaf surface in ATGPX3 mutants quantified by infrared thermal imaging. Data are means ± sd (n = 20 plants for each condition; data are from ∼4000 measurements of square pixels from multiple leaves of each plant).
(E) False-color infrared images of drought-stressed wild-type and ATGPX3 transgenic plants. The same experimental procedures were used as described for (C). The middle and bottom panels show leaf temperature profiles at the start of drought treatment (middle panel) and 5 d later (bottom panel). The top panel shows the 5-d drought-stressed plants.
(F) Temperature of the leaf surface in ATGPX3 transgenic plants quantified by infrared thermal imaging upon drought for 5 d. Data are means ± sd (n = 20 plants for each condition; values are from ∼4000 measurements of square pixels from multiple leaves of each plant in three independent experiments).
Correspondingly, upon drought treatment, the leaf temperature of atgpx3-1 and atgpx3-2 plantlets was found to be ∼1°C lower than that of the wild type, as recorded by infrared thermography (Figures 4C and 4D; see Supplemental Figure 2B online). Upon drought stress for 5 d, the leaf temperature of ATGPX3 overexpression plants was ∼1.2°C higher than that of the wild type (Figures 4E and 4F; significant difference according to the Stell-Dwass test [P < 0.03]). These temperature differences reflect the role of ATGPX3 in the regulation of stomatal aperture to control transpirational water loss. These results are consistent with the reduced sensitivity of stomatal closure to ABA and H2O2 in atgpx3 mutant plants compared with wild-type plants. Together, these data imply that ATGPX3 is involved in the regulation of drought signaling in guard cells.
ATGPX3 Interacts with ABI1 and ABI2
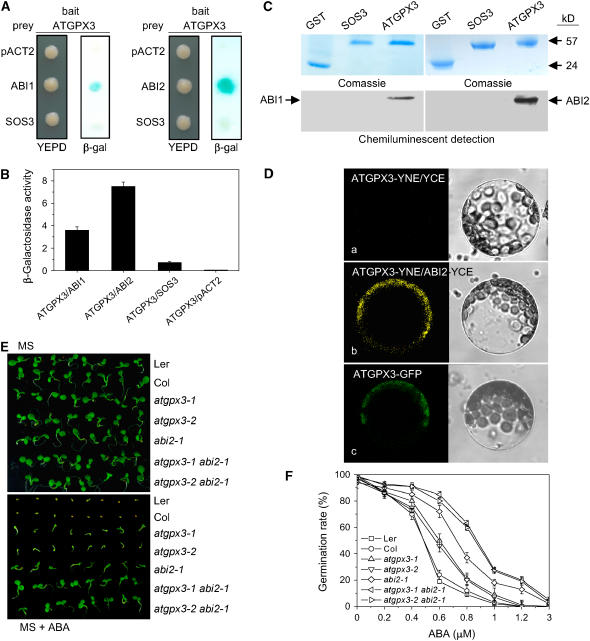
ABI1 and ABI2 are two protein Ser/Thr phosphatases of type 2C that act as key regulators in the responses of Arabidopsis to ABA (Merlot et al., 2001) and H2O2 (Meinhard and Grill, 2001; Murata et al., 2001; Meinhard et al., 2002). Having established that ATGPX3 is involved in ABA signaling, we hypothesized that ATGPX3 might interfere with the ABA response through modulation of the activities of ABI1 or ABI2. As a first test of this hypothesis, we examined whether ATGPX3 and ABI1 or ABI2 might interact physically using the yeast two-hybrid system. Figures 5A and 5B show that ATGPX3 interacts weakly with ABI1 and strongly with ABI2, the colored β-galactosidase product appearing at 6 and 0.5 h, respectively, when the ATGPX3 bait was cotransformed with the ABI2 or ABI1 prey. As a negative control, the Salt Overly Sensitive3 (SOS3) protein (Liu and Zhu, 1998) was almost completely ineffective at activating β-galactosidase reporter expression (Figures 5A and 5B).
Figure 5.
ATGPX3 Specifically Interacts with ABI2 and ABI1.
(A) ATGPX3 strongly interacted with ABI2 or ABI1 in the yeast two-hybrid system. Yeast strains containing pAS-ATGPX3 as bait and pACT-ABI2/1 as prey were grown on YEPD medium lacking Trp and Leu for 48 h (left panel) and were assayed for LacZ expression by a filter-lift assay (right panel). pACT-SOS3 and the empty prey vector were used as negative controls. Blue color indicates interaction. β-gal, β-galactosidase activity.
(B) Quantitative analysis of β-galactosidase activity of the yeast strains in liquid culture showing the interaction between ATGPX3 and ABI2 or ABI1 and with the control partner SOS3. Values are means of data from three independent experiments. Error bars indicate sd.
(C) Biotinylated Lys-labeled ABI1/2 protein was pulled down by GST-ATGPX3 but not by GST-SOS3. GST was used as a negative control.
(D) In vivo interaction between ATGPX3 and ABI2 as determined using bimolecular fluorescence complementation. a, control (SYNE-ATGPX3 and SYCE); b, the YFP signal in the cytoplasm indicates a positive interaction between ATGPX3 and ABI2; c, the ATGPX3-GFP protein is localized in the cytoplasm. Left panel, fluorescence images under confocal microscopy; right panel, bright-field images of the cell.
(E) The phenotypes of atgpx3 and abi2 single mutants and double mutants (atgpx3-1 abi2-1 and atgpx3-2 abi2-1). F2 seeds from the crosses between the respective mutants were planted on MS agar medium (top plate) or MS agar medium supplemented 0.5 μM ABA (bottom plate) and allowed to grow for 2 weeks before the photographs were taken. Col, Columbia; Ler, Landsberg erecta.
(F) Comparisons of germination rates of the wild type, atgpx3-1, atgpx3-2, abi2-1, and the double mutants atgpx3-1 abi2-1 and atgpx3-2 abi2-1 after exposure to different concentrations of ABA for 10 d. Values are means ± sd of three independent experiments (>120 seeds per point).
To further confirm the interaction between ATGPX3 and ABI2, we performed a GST pull-down assay. The biotinylated Lys-labeled ABI2 and ABI1 proteins were incubated together with GST-ATGPX3 or GST-SOS3 or GST as controls. ABI2 and ABI1 were capable of binding to ATGPX3, but SOS3 and GST were not (Figure 5C). The binding of ABI1 to ATGPX3 was weaker than that of ABI2. These results demonstrate that ATGPX3 is able to strongly interact with ABI2.
To test whether the ATGPX3 and ABI2 proteins can also interact in vivo, their interaction in plant cells was visualized using the bimolecular fluorescence complementation assay (Walter et al., 2004; Kapoor et al., 2005). For this, ATGPX3 was translationally fused to the N-terminal 86–amino acid portion of yellow fluorescent protein (YFP; pSPYNE-GPX3) and ABI2 was translationally fused to the C-terminal 155–amino acid portion of YFP (pSPYCE-ABI2). As shown in Figure 5D, yellow fluorescence was seen in cytoplasm when protoplasts of Arabidopsis leaves were cotransformed into pSPYCE-GPX3 and pSPYNE-ABI2 (Figure 5D, b). No fluorescence signal was measured when the pSPYCE vector was cotransformed with pSPYNE-GPX3 (Figure 5D, a) or when pSPYNE was cotransformed with the pSPYCE-ABI2 vector (see Supplemental Figure 3 online). Consistent with the localization of the interaction signal, the confocal image of protoplasts transformed with pGFP-ATGPX3 showed that the ATGPX3 fusion protein was localized in the cytoplasm (Figure 5D, c). These results suggest that ATGPX3 and ABI2 interact in vivo in plant cells.
To provide further genetic evidence that ATGPX3 is involved in the regulation of ABA signaling through ABI1 or ABI2, double mutants for atgpx3 and abi2-1 were constructed by crossing abi2-1 with atgpx3-1 and atgpx3-2 and screening the resulting F2 population for homozygosity at both loci. On MS medium lacking ABA, the double or single atgpx3 and abi2-1 lines germinated and grew similarly to the wild type (Figure 5E, top panel). However, when the seeds were planted on MS medium containing 0.5 μM ABA, both atgpx3 and abi2-1 single mutants were found to be insensitive to ABA, and the double mutants were even more insensitive to ABA than either abi2-1 or atgpx3 mutants (Figure 5E, bottom panel). The stronger insensitivity to ABA in double mutants was indicated by the higher chlorophyll content and longer root growth in the presence of 0.5 μM ABA. The degree of seed germination insensitivity to ABA was dose-dependent (Figure 5F). Thus, these results strongly support the suggestion that ATGPX3 plays an important role in regulating the activities of ABI2 under oxidative stress in vivo.
H2O2 Regulates the Redox States of Both ATGPX3 and ABI2, and ATGPX3 Inhibits the PP2C Activity of ABI2
ABI2 is believed to exert negative regulation on ABA action (Leung et al., 1997). Transient inactivation of this protein phosphatase by H2O2 represents a likely target for redox regulation of a hormonal signaling pathway (Meinhard et al., 2002). Our data indicated that ATGPX3 strongly interacts with ABI2, implying that the redox states of both ATGPX3 and ABI2 might be coupled. To test this possibility, we performed a series of experiments to examine the ATGPX3 activity and redox states of GST-ATGPX3 and ABI2 in vitro.
First, the redox states of GST-ATGPX3 with and without H2O2 were examined by native PAGE, in which the reduced and oxidized forms can be separated as a result of modification of the reduced Cys residues (Kishigami et al., 1995; Inaba and Ito, 2002). Without prior treatment of cells with H2O2, the extracted GST-ATGPX3 migrated as a single band. By contrast, for cells treated with H2O2, a faster mobility band was observed (Figure 6A, top panel). Consistent with these results, FLAG-tagged ATGPX3 (ATGPX3-FLAG) from protoplasts of Arabidopsis leaves transfected with plasmid encoding ATGPX3 displayed both oxidized and redox forms on native PAGE blots using the anti-FLAG antibody (Figure 6B), indicating that the ATGPX3 protein exists in both states in planta. We subsequently investigated the redox regulation of GST-ABI2 by H2O2 with the same procedures used for the analysis of ATGPX3. The protein phosphatase activity of ABI2 was highly sensitive to H2O2. ABI2 protein untreated with H2O2 migrated as a single band of reduced form. However, upon 5 mM H2O2 challenge for 5 min, ABI2 migrated as two bands, indicating that ABI2 was partially oxidized (Figure 6A, middle panel). Treatment of GST protein with H2O2 did not alter the GST state (Figure 6A, bottom panel).
Figure 6.
Redox Regulation of ATGPX3 and ABI2, Inactivation of PP2C Activities by ATGPX3, and Alterations of Gene Transcription in atgpx3 Mutants.
(A) In vitro analysis of the ATGPX3 and ABI2 redox states. GST-ATGPX3 and GST-ABI2 extracts from E. coli were exposed to H2O2 (5 mM) for 5 min and analyzed under nonreducing conditions as indicated.
(B) Redox states of ATGPX3 in planta. ATGPX3-FLAG was transiently expressed in Arabidopsis protoplasts. The protein was extracted from 106 protoplasts, and protein gel blot analysis was performed using anti-FLAG antibody. Lane 1, control (empty vector); lane 2, ATGPX-FLAG. The gel shown is representative of five independent experiments.
(C) ATGPX3 is required for the oxidation of ABI2 in vitro. To make reduced proteins, extracts from E. coli BL21 strains carrying GST-ATGPX3/ABI2 were incubated with anti-GST-bound Sepharose beads for 4 h under reducing conditions (2 mM DTT) at 4°C. The eluate containing expressed GST-ATGPX3 and GST-ABI2 was applied to analyze redox state. To make oxidative ATGPX3 and ABI2, H2O2 (5 mM) was added to purified reduced GST-ATGPX3 and GST-ABI2 under anaerobiosis. The reaction was stopped by N-ethylmaleimide (10 mM) after 10 min, and the ATGPX3 redox state was monitored by immunoblotting. The gel shown is a representative image from three independent experiments.
(D) ATGPX3 inactivated ABI2/1 activity. In vitro PP2C activity was assayed by measuring the remaining 32P in the substrate casein. The data are presented as relative PP2C activity from three independent experiments. Error bars indicate sd.
(E) Expression of ABA- and stress-responsive genes in the wild type and atgpx3 mutants. Total RNA was extracted from wild-type and atgpx3-1 and atgpx3-2 seedlings. Real-time PCR was performed in three independent experiments. Error bars indicate sd. An Actin2 primer was used in the PCR as an internal control.
To determine whether the changes of ATGPX3 and ABI2 redox states were coupled, we incubated a purified and oxidized preparation of ABI2 with bacterial extracts expressing the GST-ATGPX3. Examination by native PAGE showed that the reduced form of ATGPX3 was converted to the oxidized form (Figure 6C, left panel; see Supplemental Figure 4, left panel, online). Likewise, we found that the reduced form of ABI2 was converted to the oxidized form by the addition of oxidized ATGPX3 (Figure 6C, middle panel; see Supplemental Figure 4, middle panel, online). It should be noted that no oxidizing reagent had been added to either the medium or the buffers used. It is intriguing that the role of ABI2 interacting with ATGPX3 is similar to that of H2O2.
To investigate the physiological relevance of the interaction between ATGPX3 and ABI2 or ABI1, GST-ABI2 and -ABI1 recombinant proteins were produced in Escherichia coli, and their PP2C activities were assayed in vitro using 32P-labeled casein as substrate with or without oxidized ATGPX3. As shown in Figure 6D, ATGPX3 reduced ∼40 and 70% of the phosphatase activity of ABI1 or ABI2. Thus, the inactivation of PP2C activities resulting from ATGPX3 might function as a feedback loop to control the redox states of the protein and ABA signaling. These data are consistent with the greater ABA insensitivity shown in atgpx3 abi2-1 double mutants.
To determine whether the expression of ABA- and stress-responsive genes is affected by the ATGPX3 mutation, we used real-time PCR analysis to compare several stress-induced genes in the wild type and the ATGPX3 mutants. Previous studies showed that the expression of RD29A, RbohD, and PR1 was induced by ABA and drought, oxidative, and pathogen stress, respectively (Yamaguchi-Shinozaki and Shinozaki, 1994; Sticher et al., 1997; Torres et al., 2002; Kwak et al., 2003). We found that the steady state levels of both RbohD and PR1 gene transcripts were lower in the mutant than in the wild type, whereas RD29A transcription appeared higher in mutants (Figure 6E). These findings suggest that ATGPX3 may have a general role in ABA- and stress-mediated gene expression in plants.
atgpx3-1 Mutants Disrupt the ABA- and H2O2-Activated Calcium Channels in the Plasma Membrane
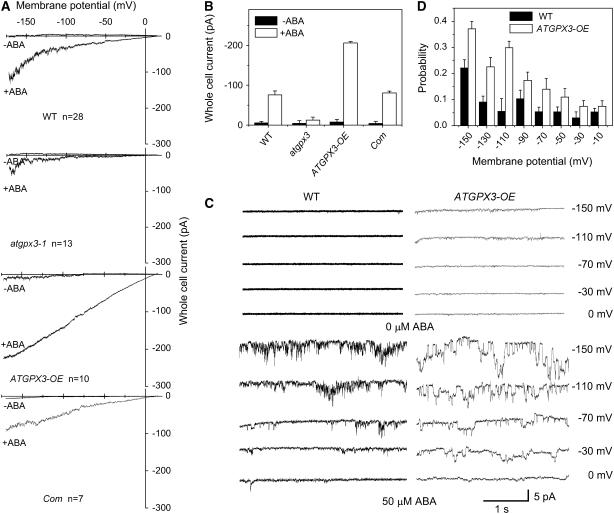
H2O2 is known to induce an increase in the cytosolic calcium concentration, mediating stomatal closure in Arabidopsis plants (McAinsh et al., 1996; Pei et al., 2000). Previous studies have shown that abi1-1 and abi2-1 differentially disrupt ABA activation of Ca2+ channels in guard cells requiring NAD(P)H-dependent ROS production (Murata et al., 2001). We have shown an ABA impaired stomatal closure in atgpx3-1 mutant plants. We hypothesize that ATGPX3 may be involved in ABA-mediated H2O2 production, which causes a change of calcium current in guard cells. To test this hypothesis, patch-clamp experiments were performed with guard cell protoplasts using Ca2+ salt solutions to determine the whole cell current of calcium. In the whole cell patch-clamp recording using conditions described previously (Allen et al., 1999; Pei et al., 2000), +10 mV was close to the equilibrium potential for Ca2+ and Ba2+ compared with that of chloride ions (∼−58.5 mV, obtained from the calculation according to the Nernst formula in these experimental conditions) and hyperpolarization did not activate a Ca2+ current in either wild-type or atgpx3-1 guard cells (n = 35). However, in the presence of ABA in both the bathing solution and cytosolic solutions, large whole cell currents were activated by hyperpolarization in the wild-type guard cells (n = 28) (Figures 7A and 7B). Activation of calcium channels in guard cells was found for ABA concentrations ranging from 5 to 50 μM. By contrast, in response to ABA treatment, only a very small background current was found in guard cells from the atgpx3-1 mutant (n = 13). To confirm the fact that the mutation of ATGPX3 is responsible for disruption of the ABA-activated calcium channel, protoplasts from the complementation lines were used to measured calcium channel activity. We found that the current of calcium channels in response to ABA in the complementation line was almost equal to that in the wild type (n = 10), suggesting that the ABA-activated calcium current was recovered in transgenic plants containing the wild-type ATGPX3 (Figure 7A, top and bottom panels). Not surprisingly, when the guard cells from the ATGPX3 overexpression transgenic plants were exposed to 50 μM ABA, the calcium current was greatly increased compared with that in the wild-type (n = 10) (Figure 7A). On average, at −150 mV, currents in wild-type, atgpx3-1, complementation, and overexpression transgenic plants were 70.3 ± 3, 28.6 ± 2, 69.2 ± 2, and 197 ± 7 pA, respectively (P < 0.05) (Figure 7B). Similar results were obtained in excised patches with the voltage under direct experimental control. The analysis of open probabilities of single-channel events indicated that the open probability values increased by approximately fivefold in the overexpression lines at −110 mV (Figures 7C and 7D). These data suggest that ATGPX3 is involved in the regulation of ABA-activated calcium signaling in guard cells.
Figure 7.
Changes of Calcium Channel Activity in Guard Cells of ATGPX3-Deficient Mutant and Overexpression Plants.
(A) ABA (50 μM) failed to activate Ca2+ channel currents in the atgpx3-1 mutant but greatly increased Ca2+ channel currents of guard cells in ATGPX3 overexpression transgenic lines.
(B) Effects of ATGPX3 mutation and overexpression on calcium channel activity at –150 mV (n = 35 [wild type], 28 [wild type + ABA], 13 [atgpx3-1 + ABA], 7 [ATGPX3 overexpression + ABA], and 10 [complementation line + ABA]). Error bars indicate sd.
(C) Single-channel current of calcium from guard cells of an ATGPX3 overexpression plant.
(D) Dependence of channel open probability on pressure in the pipette. The open probability gradually increased with negative pressure. Values are means ± sd (n = 3 or 4).
DISCUSSION
In this study, our data provide genetic evidence that ATGPX3 plays a dual role within plants, first in the control of H2O2 homeostasis, and second in the transduction of an H2O2 signal in guard cells that mediates stomatal regulation in response to ABA and drought stress. The mechanism of oxidative signal-sensing and -relaying described here appears to be coupled to ABI1 and ABI2 PP2C, which are known to be important players in ABA signaling (Meinhard et al., 2002).
Upon exposure to H2O2, the ATGPX3 mutants showed increased sensitivity during vegetative growth (Figures 2A and 2B). With thioredoxin as a substrate, a significant level of peroxidase activity was observed in the presence of thioredoxin reductase (Figure 2F). Consistent with this, the H2O2 production induced by ABA was increased significantly in guard cells of atgpx3 mutants (Figures 2C to 2E), suggesting that loss-of-function mutation in ATGPX3 impairs H2O2 homeostasis in guard cells. Earlier studies had demonstrated the generation of H2O2 and its effects on stomatal closure (McAinsh et al., 1996; Allan and Fluhr, 1997; Lee et al., 1999; Miao et al., 2000; Zhang et al., 2001b). Moreover, the balance of superoxide dismutase, ascorbate peroxidase, and CAT activities will be crucial for suppressing toxic ROS levels in a cell. GPXs are commonly considered to be important ROS scavengers because of their broader substrate specificities and stronger affinity for H2O2 than CATs (Brigelius-Flohe and Flohe, 2003). In addition, very strong expression of ATGPX3 promoter–GUS in transgenic plants was observed in guard cells of the leaf epidermis (Figure 3C), implying that ATGPX3 also detoxifies H2O2 to H2O in guard cells. Hence, it is possible that ATGPX3 acts as a key regulator that specifically modulates H2O2 homeostasis in guard cells.
ATGPX3 is not only critical for scavenging H2O2 but is an essential element of the ABA signaling pathway that mediates stomatal regulation in response to drought stress. Compared with wild-type plants, atgpx3 plants displayed impaired ABA- and H2O2-induced stomatal closure (Figure 3A), faster water loss, and lower temperatures of leaves in response to water deficit stress (Figures 4A to 4D). By contrast, the water loss of detached leaves was significantly lower, and leaf surface temperature was higher in ATGPX3 overexpression lines than in wild-type plants (Figures 4B, 4E, and 4F). Consistently, defects of ATGPX3 reduced drought stress tolerance, whereas ATGPX3 overexpression in transgenic plants enhanced resistance to drought stress (Figure 4A). Previous data have demonstrated ABA-induced H2O2 generation in guard cells of Vicia and Arabidopsis (Miao et al., 2000; Pei et al., 2000; Zhang et al., 2001b), mediating ABA signaling, thereby activating plasma membrane Ca2+ and K+ channels to permit Ca2+ and K+ influx in stomatal closure (An et al., 2000; Pei et al., 2000; Zhang et al., 2001a). Thus, ATGPX3 might play an important role in ABA-mediated stomatal closure under drought stress.
The finding that ATGPX3 is involved in the ABA signaling of guard cells raises the questions of how ABA and H2O2 signals are transduced and the identities of the components of the cascade involved in this process. The abi1-1, abi2-1, and gca2 mutations impair ABA-induced stomatal closure in Arabidopsis. The three mutations were shown to affect a signaling cascade that involves ABA-induced ROS production (Pei et al., 2000; Murata et al., 2001; Mustilli et al., 2002). Transient inactivation of ABI2 phosphatase by H2O2 would allow or enhance the ABA-dependent signaling process (Meinhard et al., 2002). Our data indicated that ATGPX3 interacted strongly with ABI2 but only relatively weakly with ABI1 (Figures 5A to 5C). Interestingly, both ATGPX3 and ABI2 redox states were altered after exposure to H2O2, and the changes in the redox state of ATGPX3 and ABI2 were coupled, suggesting that ABI2 could reduce oxidized ATGPX3, at least in vitro (Figures 6A and 6C). Furthermore, in enzymatic assays using recombinant proteins, the PP2C activity of the ABI2 and ABI1 proteins in the presence of ATGPX3 was ∼70 and 40% lower than that of the ABI2 or ABI1 protein alone (Figure 6D). Compared with the single mutants of atgpx3 and abi2-1, their double mutant exhibited greater insensitivity in response to ABA (Figures 5E and 5F). These results suggest that, in the ABA signaling pathway, ABI2 represents a likely target for redox regulation by the oxidized form of ATGPX3. Previous evidence has implicated H2O2 as an intercellular messenger that modulates the extent of protein phosphorylation on Ser/Thr or Tyr residues in animal cells (Sundaresan et al., 1995; Bae et al., 1997; Liu et al., 2000). The human protein Tyr phosphatase PTP1B was modified by H2O2 at the active site Cys (van Montfort et al., 2003). The link between ABA-induced H2O2 production and protein phosphorylation is further strengthened by our observations. However, as shown for the Yap1 system in yeast (Delaunay et al., 2002), the biochemical and structural basis of ATGPX-like enzymes as a hydroperoxide sensor or redox transducer remains to be established in plant cells.
Calcium-permeable channels activated by ROS have been shown to function in the ABA signaling network in Arabidopsis guard cells (Pei et al., 2000; Murata et al., 2001). Increases in cytosolic calcium are triggered by ROS production during root hair formation and ABA signaling (Foreman et al., 2003; Kwak et al., 2003) and also occur in the early events during pathogen responses (Dangl and Jones, 2001). Like abi2-1, the atgpx3 mutations impair ABA-induced stomatal closure and the activation of Ca2+-permeable channels in the plasma membrane (Figure 7A). By contrast, calcium current was greatly increased compared with that in the wild type in the overexpression lines of ATGPX3 (Figures 7A to 7D). One possibility is that the atgpx3 mutation affects an early or the initial event of H2O2 and ABA signaling, thereby blocking the activation of Ca2+ channels within guard cells and disturbing stomatal closure. The intimate connection between ATGPX3 and Ca2+ channel activity implies that intracellular H2O2 homeostasis may be monitored and sensed in the plant cells by GPX(s).
H2O2 regulates the expression of various genes, including those encoding antioxidant enzymes and modulating H2O2 generation in cells (Mittler et al., 2004; Wang et al., 2006). A global transcriptome analysis using microarrays revealed that 1 to 2% of genes are changed in H2O2-treated Arabidopsis (Desikan et al., 2001). The fact that alterations of RD29A, RbohD, and PR-1 transcript levels are observed in atgpx3-1 and atgpx3-2 suggests that the atgpx3-1 mutation affects ABA and H2O2 signaling (Figure 6E). Thus, ATGPX3 must directly or indirectly regulate the upstream signaling events that control gene expression. The bacterial H2O2 sensor, OxyR, as a transcription factor directly regulates gene expression (Zheng et al., 1998; Kim et al., 2002). The model of the GPX-based sensor postulated that the redox state of GPX3 and Yap1 are coupled, the latter modulating gene expression. Unlike bacterial and yeast cells, however, our database survey in the Arabidopsis genome indicated that plant cells lack orthologs of OxyR or Yap1-like transcription factors. Alternatively, a Yap1-interacting protein was found, designated CEO1, which is thought to be a transcription regulator in Arabidopsis (Belles-Boix et al., 2000). The CEO1 protein could interact with several transcription factors, such as DREB2A (Ahlfors et al., 2004), which suggests that it might regulate gene expression via the function or activity of these transcription factors. Our studies have also shown that ATGPX3 interacts with CEO1 in the yeast two-hybrid system (Y. Miao and C.-P. Song, unpublished data). It is possible that the CEO1 protein and its partner(s) might be the plant counterparts of yeast Yap1 (Belles-Boix et al., 2000; Overmyer et al., 2000; Ahlfors et al., 2004).
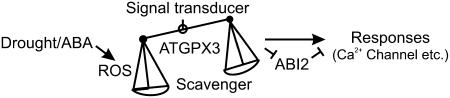
On the basis of the results presented here as well as those reported previously (Allen et al., 1999; Murata et al., 2001), we present a model describing the interrelationships among ATGPX3, ABI2, the Ca2+ channel, and the oxidative signal transduction pathway (Figure 8). This model, together with other recent studies of the role of PP2C-type enzymes mediating phosphorylation in H2O2 and ABA signaling (Merlot et al., 2001; Kuhn et al., 2006; Mishra et al., 2006), allows us to fully describe the oxidative and ABA signal transduction pathway from sensing to the response in plants.
Figure 8.
Model Showing the Putative Signal Transduction Pathway Mediated by ATGPX3.
For details, see the text. Arrows indicate positive regulation, and open blocks indicate negative regulation.
In summary, we have demonstrated that the ATGPX3 function in H2O2 signal transduction is closely related to the activity of ABI2, which directly influences Ca2+ channel activity (Murata et al., 2001). Thus, ATGPX3 might sense and transduce the H2O2 signal to downstream components via ABI2, thereby controlling the Ca2+ channel. Further research is needed to ascertain how the thiol group of these proteins might be involved in the regulation of the redox state and also the critical concentration of H2O2 in the cell for the protein to respond as a sensor rather than simply act in its general scavenging function.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia was used in this study. For seed germination, all seeds were sterilized and kept for 4 d at 4°C in the dark to break dormancy. The seeds then were sown on 0.6% agar-containing MS medium containing different levels of mannitol, H2O2, or ABA as indicated. The plates were incubated at 22 ± 2°C with a 16-h-light photoperiod. For seedling growth, 5-d-old seedlings from the germination medium were transferred to 1.2% agar supplemented with various mannitol concentrations (placed vertically). For morphological examination of aerial parts, seedlings were transferred to soil at 7 to 10 d after germination and placed in a growth chamber at 22°C under a 16-h-light/8-h-dark photoperiod and 70% RH.
Identification and Isolation of atgpx3 Mutants
T-DNA insertion mutants (SALK_071176 and SALK_001116) of the ATGPX3 gene generated by the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/) were obtained from the ABRC (Ohio State University). The seeds were planted on agar plates containing MS medium and kanamycin, and the kanamycin-resistant plants were transferred to soil. Seeds were harvested separately from individual plants. Subsequently, to confirm the mutant line as homozygous, PCR was performed with the genomic DNA of atgpx3-1 and atgpx3-2 mutants using gene-specific oligonucleotides (LP, 5′-TTGACTATAAGAAGCACTTTCCCG-3′; RP, 5′-GCATGGTTTGACGATTTTGTGA-3′; LP, 5′-TGGATGATTGTTCCACCGTCG-3′; RP, 5′-GGCACTCCTCTTGAAGACCAGAA-3′) and left-border-specific primers (LBal, 5′-TGGTTCACGTAGTGGGCCATCG-3′; LBb1, 5′-GCGTGGACCGCTTGCTGCAACT-3′).
Plasmid Constructs
Gene-specific cDNA fragments of ATGPX3 were amplified by PCR using the following primer pairs: forward primer 5′-TACCCGGGATGCCTAGATCAAGCAG-3′ and reverse primer 5′-CTGGATCCTCAAGCAGATGCCAATA-3′. The forward primer contains a SacI restriction site, and the reverse primer contains a XbaI restriction site, which are underlined. The PCR fragments of ATGPX3 were first cloned into pBIB between the SacI and XbaI sites. For yeast two-hybrid experiments, the ATGPX3 coding region was amplified by PCR with primers containing restriction sites and was cloned in frame between the SmaI and SalI sites of pAS2, to form pAS-ATGPX3 as the bait. For the in vivo interaction assay, the PCR fragments of ATGPX3 and ABI2 were cloned into pSPYNE and pSPYCE (for split YFP N-terminal/C-terminal fragment expression), respectively. For the subcellular localization of ATGPX3, the cDNA for ATGPX3 was amplified by PCR, digested with XbaI and BamHI, and inserted into pBI-GFP. For ATGPX3 promoter–GUS constructs, a 450-bp ATGPX3 promoter fragment was obtained by PCR for plant transformation. PCR was performed using the primer pair 5′-CGCGAATTCTGGAGTCGGGACACTATAG-3′ and 5′-GCGGTCGACAACACAGTTAGTCTTCTTC-3′. The forward primer contains a EcoRI restriction site, and the reverse primer contains a SalI restriction site, which are underlined. The promoter fragments were then cloned into pCAMBIA1381 between the EcoRI and SalI sites.
Plant Transformation and Protoplast Transfection
The constructs were introduced into Agrobacterium tumefaciens strain GV3101 and transformed by floral infiltration into wild-type Arabidopsis (Columbia ecotype) (for gene overexpression and the GUS staining assays) and atgpx3-1 mutants (for gene complementation).
For in vivo interaction measurements, the protoplasts isolated from Arabidopsis leaves were transformed with plasmid combinations pSPYNE-GPX3 and pSYCE, pSYNE and pSYCE-ABI2, or pSYCE-ABI2 and pSPYNE-GPX3, according to the protocols of Merkle et al. (1996) and Negrutiu et al. (1987). For GFP-ATGPX3 constructs, the protoplast transient expression assay was performed as described by Sheen (2001). After incubation for 16 to 20 h, the fluorescence of protoplasts was measured with a laser-scanning confocal microscope (Bio-Rad). All figures show representative images from three independent experiments.
Infrared Thermography Imaging
Thermal imaging of drought-stressed plantlets was performed as described previously (Merlot et al., 2002). In brief, plantlets were first grown under well-watered conditions (22°C, 60 to 70% RH, 16-h photoperiod) for ∼14 d. Drought stress then was initiated by withholding water. Thermal images were obtained within the growth chamber (22°C, 60 to 70% RH, 16-h photoperiod) using a ThermaCAM SC3000 infrared camera (FLIR System). Images were saved on a Personal Computer Memory Card International Association card and were analyzed subsequently using version 5.31 of the public domain image-analysis program IRWin Reporter.
Epidermal Strip Bioassay and Water Loss Measurement
Stomatal bioassay experiments were performed as described (Hugouvieux et al., 2001; Zhang et al., 2001b; Song et al., 2005) with slight modifications. Epidermal strips were peeled from Arabidopsis leaves. To study the promotion of stomatal closure by ABA, stomata were opened by exposing plants for 12 h to light and high humidity and incubating the leaves for 1.5 h in stomata-opening solution containing 50 mM KCl, 10 μM CaCl2, and 10 mM MES, pH 6.15, in a growth chamber at 22 to 25°C under a photon flux density of 0.20 to 0.30 mmol·m−2·s−1. Stomatal apertures were measured 1.5 h after adding 2 μM ABA or 3 mM H2O2. The apertures of usually 50 to 60 stomata were measured in three independent experiments. Data analysis, including statistical tests, was performed using Origin software (version 6.1).
For water loss measurement, rosette leaves of wild-type, ATGPX3-deficient mutant, and overexpression lines were detached from their seedlings, placed in weighing dishes, and incubated on the laboratory bench. Losses in fresh weight was monitored at the indicated times. Water loss is expressed as the percentage of initial fresh weight.
Measurements of H2O2 Production
H2O2 production from guard cells was examined by loading epidermal peels with H2DCF-DA (Molecular Probes) (Cathcart et al., 1983; Lee et al., 1999; Zhang et al., 2001b). The epidermal strips, previously incubated for 3 h under conditions promoting stomatal opening, were placed into loading buffer with 10 or 50 mM Tris-KCl, pH 7.2, containing 50 μM H2DCF-DA. Before further experiments, peels were preincubated in the dark for 10 to 15 min, and then the epidermal tissues were collected using a nylon mesh and washed with distilled water to remove excess dye from the apoplast. Examinations of peel fluorescence were performed using a MicroRadiance laser-scanning confocal microscope (Bio-Rad). ABA and ethanol (control) were added directly to the buffer during the experiments.
Confocal fluorescence optical sections were collected from dye-loaded guard cells with the following settings: excitation, 488 nm; emission, 525 nm; power, 3%; zoom, 4; mild scanning; frame, 512 × 512; with pixel spacing of 0.183 to 0.25 μm. To extract quantitative data, pixel values were averaged over rectangular regions (4 μm2) manually located on each image. The pixel intensity from at least 110 guard cells was recorded.
H2O2 was also detected in situ by DAB staining as described previously (Thordal-Christensen et al., 1997). The terminal leaflet of the first fully expanded leaf was sampled from 20-d-old wild-type and atgpx3-1 and atgpx3-2 plants. Leaflets were collected and vacuum-infiltrated with the DAB solution (1 mg/mL, pH 3.8; Sigma-Aldrich). The sampled leaves were placed in a plastic box under high humidity until reddish-brown precipitate was observed (8 h), then fixed with a solution of 3:1:1 ethanol:lactic acid:glycerol and photographed.
Histochemical Detection of GUS Activity
Ten independent transgenic lines containing the ATGPX3 promoter–GUS constructs were tested for GUS activity by incubation of excised tissues overnight in GUS staining buffer (3 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, 0.1 M sodium phosphate buffer, pH 7.0, 0.1% Triton X-100, and 8 mM β-mercaptoethanol) at 37°C in the dark. The staining was terminated by replacement of the staining solution with 70% ethanol solution, and samples were stored at 4°C until observation with the microscope.
Analysis of Protein Redox States
To express GST-ATGPX3 in bacteria, the coding regions of ATGPX3 cDNAs were cloned in-frame into the BamHI-EcoRI sites of pGEX-2TK. The constructs were introduced into Escherichia coli BL21 cells. The transformed strains were incubated at 37°C in Luria-Bertani medium and induced for 4 h with isopropyl β-d-thiogalactopyranoside (1 mM). The recombinant GST-ATGPX3 and GST-ABI2 proteins (Guo et al., 2002) were purified using glutathione resins (BD Biosciences) according to the manufacturer's instructions. Purified recombinant proteins were used for the analysis of in vitro ATGPX3 and ABI2 redox states. Extracts were resolved by nonreducing or reducing 12% SDS-PAGE. The fused proteins were immunoblotted with the anti-GST antibody (Roche).
Immunoprecipitations
To make the ATGPX3-FLAG transient expression vector, 3×FLAG tag was fused to the 3′ end of the ATGPX3 coding region and was cloned into the plasmid pRT105-3FLAG between the XbaI and BamHI sites under the control of a double 35S promoter. The protoplast transient expression assay was performed as described by Sheen (2001). After 16 h of incubation, the protein extract was prepared using 106 protoplasts per 100 μL of extraction buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 1% Triton X-100, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 2 μM leupeptin, and 2 μM pepstatin). Two microliters of anti-FLAG (Sigma-Aldrich) was used for protein from the extracts. Protein gel blotting was performed using standard methods.
Yeast Two-Hybrid Interaction and GST Pull-Down Assays
Yeast two-hybrid interaction and protein pull-down assays were performed as described (Guo et al., 2002; Song et al., 2005). Competent cells of Saccharomyces cerevisiae strain Y190 were transformed simultaneously with pAS-ATGPX3 and pACT2-ABI1/2 (Guo et al., 2002). Yeast cells cotransformed with pAS2-1 and pACT2 without inserts were used as negative controls, whereas those cotransformed with pAS-ATGPX3 and pACT2-SOS3 (Ohta et al., 2003) were used as positive controls.
For GST pull-down assays, recombinant ATGPX3-GST and SOS3-GST (Ohta et al., 2003) fusion proteins expressed using the pGEX-2TK vector were used. Biotinylated Lys ABI1 and ABI2 proteins were produced from pET14b-ABI1 and pET14b-ABI2 (Guo et al., 2002) using an in vitro transcription and translation assay kit (TNT Quick Coupled Transcription/Translation system; Promega) with Transcend Biotinylated Lys tRNA incorporation and detection, according to the manufacturer's instructions.
Real-Time Quantitative RT-PCR
Total RNAs were isolated from 15-d-old atgpx3-1/2 mutants and also from the wild type with Trizol reagent. Reverse transcription of all RNA samples was performed with Moloney murine leukemia virus reverse transcriptase (Promega). SYBR Premix Ex Taq (Takara) was used for real-time quantitative PCR. We performed real-time PCR on a Rotor-Gene 3000 apparatus (Corbett Research) and repeated experiments three times. Cycling conditions were as follows: 5 min at 94°C; 40 cycles of 15 s at 94°C, 15 s at 53°C, and 20 s at 72°C; 300 s at 40°C; and 60 s at 55°C. This was followed by a melting-curve program (55 to 99°C, with a 5-s hold at each temperature). Specific cDNA was quantified with a standard curve based on the known amounts of amplified target gene fragments. The mean value of three replicates was normalized using Actin2 as the internal control.
Semiquantitative RT-PCR and RNA Gel Blot Analysis
RNA was isolated from 200-mg tissue samples using Trizol solution (Invitrogen). The first-strand cDNA syntheses were performed using 5 μg of total RNA and Moloney murine leukemia virus reverse transcriptase (Promega). The yield of cDNA was measured according to the PCR signal generated from the internal standard, the housekeeping gene β-Actin, amplified from 18 to 24 cycles starting with 1 μL of the cDNA solution. Cycling conditions were as follows: 5 min at 94°C; and 20 cycles of 30 s at 94°C, 30 s at 55°C, and 40 s at 72°C. The volume of each cDNA pool was adjusted to give the same exponential phase PCR signal strength for β-Actin after 20 cycles. The resulting cDNAs were subjected to PCR with primers designed to amplify ATGPX3. The expression of Actin was used as an internal control. The RT-PCR product was analyzed by electrophoresis on a 1.5% agarose gel. All PCRs were performed in triplicate.
To extract total RNA from guard cell–enriched epidermal strips, leaves were blended in a Waring blender in cold water. Epidermal strips with >95% guard cell purity were used to extract total RNA using Trizol reagent. RT-PCR was performed according to the procedures described above.
RNA gel blot analysis was performed as described previously (Rodriguez Milla et al., 2003). ATGPX3 and ATGPX7 full-length cDNAs were used as probes. The expression of Actin was used as a loading control.
PP2C and Peroxidase Activities
PP2C activity was determined by measuring the remaining 32P in substrates. Two microliters of casein (P-4765; Sigma-Aldrich) was phosphorylated by 2 μL of protein kinase A (P2645; Sigma-Aldrich) in a 50-μL reaction volume containing 20 mM Tris-HCl, pH 7.2, 5 mM MgCl2, 2 mM DTT, 10 μM unlabeled ATP, and 1.85 MBq [γ-32P]ATP. After incubation for 30 min at 30°C, 5-μL samples of product were used for the phosphatase assay. Reactions were started by the addition of recombinant enzymes with or without the effector ATGPX3 in a buffer containing 20 mM Tris-HCl, pH 7.2, 5 mM MgCl2, and 2 mM DTT. Reactions were terminated by spotting samples onto P81 filter papers (3698-915; Whatman) after incubation for 30 min at 30°C. After the papers were washed three times with 1% phosphoric acid and dried, a phosphorimager was used to quantify the amounts of 32P on the papers. Experiments were performed in triplicate. The relative activities of PP2C were calculated according to the formula (S − E)/S × 100%, where S stands for the amount of 32P in substrate and E stands for the remaining amount of 32P in substrate after the addition of enzymes.
ATGPX3 peroxidase activity was monitored by the spectrometric kinetic determination of NADPH consumption as described by Delaunay et al. (2002).
Patch Clamp and Data Acquisition
Arabidopsis guard cell protoplasts were isolated as described (Pandey et al., 2002). The whole cell voltage-clamp or single-channel currents of Arabidopsis guard cells were recorded with an EPC-9 amplifier (Heka Instrument) as described (Pei et al., 2000; Wang et al., 2004). Pipettes were pulled with a vertical puller (Narishige) modified for two-stage pulls. Data were analyzed using PULSEFIT 8.7, IGOR 3.0, and ORIGIN 7.0 software. TAC/TACFIT 4.0 software was used to analyze single-channel probability. The standard solution for calcium current measurements contained (in mM) 100 CaCl2, 0.1 DTT, and 10 MES-Tris, pH 5.6, in the bath and 10 BaCl2, 0.1 DTT, 4 EGTA, and 10 HEPES-Tris, pH 7.1, in the pipette. ABA and H2O2 were freshly added to bath solutions at the indicated concentrations. d-Sorbitol was used to adjust the osmolalities of pipette and bath solutions to 490 and 510 mmol/kg, respectively.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: ATGPX3, At2g43350; ATGPX7, At4g31870; ABI1, At4g26080; ABI2, At5g57050; SOS3, At5g24270; RD29A, At5g52310; RbohD, At5g47910; and PR1, At2g14610.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Response of the Overexpression of ATGPX3 to Osmotic Stress.
Supplemental Figure 2. Responses of atgpx3 Mutants to Drought Stress.
Supplemental Figure 3. No Fluorescence Signal Was Measured When pSPYNE Was Cotransformed with the pSPYCE-ABI2 Vector.
Supplemental Figure 4. Analysis of the ATGPX3 and ABI2 Redox States in Vitro.
Supplementary Material
Acknowledgments
We thank Andre Jagendorf (Cornell University), David Galbraith (University of Arizona), and Wolf Frommer (Carnegie Institution of Washington) for critical reading of the manuscript and helpful comments. We also thank Yan Guo, Zhizhong Gong, Ximing Gong, Xiao Zhang, Jie Dai, Ertao Liu, Jinggong Guo, and Kun Li for their technical support and helpful discussions. This work was supported by the National Key Basic Special Funds (Grant 2003CB114305) and the National Natural Science Foundation of China (Grants 30370765 and 30530430 to C.-P.S; Grant 30170088 to J.C.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Chun-Peng Song (songcp@henu.edu.cn).
Online version contains Web-only data.
References
- Ahlfors, R., et al. (2004). Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE protein–protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell 16 1925–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan, A.C., and Fluhr, R. (1997). Two distinct sources of elicited reactive oxygen species in tobacco epidermal cells. Plant Cell 9 1559–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, G.J., Kuchitsu, K., Chu, S.P., Murata, Y., and Schroeder, J.I. (1999). Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid–induced cytoplasmic calcium rises in guard cells. Plant Cell 11 1785–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- An, G., Song, C.P., Zhang, X., Jing, Y.C., Yang, D.M., Huang, M.J., Zhou, P.A., and Wu, C.H. (2000). Effect of peroxide generation on stomata movement and K+ channel on plasma membrane in Vicia faba guard cell. Acta Phytophysiol. Sin. 26 458–464. [Google Scholar]
- Apel, K., and Hirt, H. (2004). Active oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 373–399. [DOI] [PubMed] [Google Scholar]
- Apostol, I., Heinstein, P.F., and Low, P.S. (1989). Rapid stimulation of an oxidative burst during elicitation of cultured plant cells: Role in defense and signal transduction. Plant Physiol. 90 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur, J.R. (2000). The glutathione peroxidases. Cell. Mol. Life Sci. 57 1825–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann, S.M. (2003). OPEN STOMATA1 opens the door to ABA signaling in Arabidopsis guard cells. Trends Plant Sci. 8 151–153. [DOI] [PubMed] [Google Scholar]
- Bae, Y.S., Kang, S.W., Seo, M.S., Baines, I.C., Tekle, E., Chock, P.B., and Rhee, S.G. (1997). Epidermal growth factor (EGF)-induced generation of hydrogen peroxide. Role in EGF receptor-mediated tyrosine phosphorylation. J. Biol. Chem. 272 217–221. [PubMed] [Google Scholar]
- Baker, M.A., and Orlandi, E.W. (1995). Active oxygen in plant pathogenesis. Annu. Rev. Phytopathol. 33 299–321. [DOI] [PubMed] [Google Scholar]
- Beeor-Tzahar, T., Ben-Hayyim, G., Holland, D., Faltin, Z., and Eshdat, Y. (1995). A stress-associated citrus protein is a distinct plant phospholipid hydroperoxide glutathione peroxidase. FEBS Lett. 366 151–155. [DOI] [PubMed] [Google Scholar]
- Belles-Boix, E., Babiychuk, E., Van Montagu, M., Inzé, D., and Kushnir, S. (2000). CEO1, a new protein from Arabidopsis thaliana, protects yeast against oxidative damage. FEBS Lett. 482 19–24. [DOI] [PubMed] [Google Scholar]
- Bolwell, G.P., Butt, V.S., Davies, D.R., and Zimmerlin, A. (1995). The origin of the oxidative burst in plants. Free Radic. Res. 23 517–532. [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohe, R., et al. (1994). Phospholipid hydroperoxide glutathione peroxidase. Genomic DNA, cDNA, and deduced amino acid sequence. J. Biol. Chem. 269 7342–7348. [PubMed] [Google Scholar]
- Brigelius-Flohe, R., and Flohe, L. (2003). Is there a role of glutathione peroxidases in signaling and differentiation? Biofactors 17 93–102. [DOI] [PubMed] [Google Scholar]
- Cathcart, R., Schwiers, E., and Ames, B.N. (1983). Detection of picomole levels of hydroperoxides using a fluorescent dichlorifluorescein assay. Anal. Biochem. 134 111–116. [DOI] [PubMed] [Google Scholar]
- Churin, Y., Schilling, S., and Börner, T. (1999). A gene family encoding glutathione peroxidases in barley (Hordeum vulgare L.). FEBS Lett. 459 33–38. [DOI] [PubMed] [Google Scholar]
- Comtois, S.L., Gidley, M.D., and Kelly, D.J. (2003). Role of the thioredoxin system and the thiol-peroxidases Tpx and Bcp in mediating resistance to oxidative and nitrosative stress in Helicobacter pylori. Microbiology 149 121–129. [DOI] [PubMed] [Google Scholar]
- Criqui, M.C., Jamet, E., Parmentier, Y., Marbach, J., Durr, A., and Fleck, J. (1992). Isolation and characterization of a plant cDNA showing homology to animal glutathione peroxidases. Plant Mol. Biol. 18 623–627. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- Delaunay, A., Pflieger, D., Barrault, M., Vinh, J., and Toledano, M.B. (2002). A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111 471–481. [DOI] [PubMed] [Google Scholar]
- Depege, N., Drevet, J., and Boyer, N. (1998). Molecular cloning and characterization of tomato cDNAs encoding glutathione peroxidase-like proteins. Eur. J. Biochem. 253 445–451. [DOI] [PubMed] [Google Scholar]
- Desikan, R., Hancock, J.T., Bright, J., Harrison, J., Weir, I., Hooley, R., and Neill, S.J. (2005). A role for ETR1 in hydrogen peroxide signaling in stomatal guard cells. Plant Physiol. 137 831–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan, R., A.-H. Mackerness, S., Hancock, J.T., and Neill, S.J. (2001). Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 127 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doke, N. (1985). NADPH-dependent O2− generation in membrane fractions isolated from wounded potato tubers inoculated with Phytophthora infestans. Physiol. Plant Pathol. 27 311–322. [Google Scholar]
- Eshdat, Y., Holland, D., Faltin, Z., and Ben-Hayyim, G. (1997). Plant glutathione peroxidases. Physiol. Plant. 100 234–240. [Google Scholar]
- Faltin, Z., Camoin, L., Ben-Hayyim, G., Perl, A., Beeor-Tzahar, T., Strosberg, A.D., Holland, D., and Eshdat, Y. (1998). Cysteine is the presumed catalytic residue of Citrus sinensis phospholipids hydroperoxide glutathione peroxidase over-expressed under salt stress. Physiol. Plant. 104 741–746. [Google Scholar]
- Foreman, J., Demidchik, V., Bothwell, J.H., Mylona, P., Miedema, H., Torres, M.A., Linstead, P., Costa, S., Brownlee, C., Jones, J.D., Davies, J.M., and Dolan, L. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 27 442–446. [DOI] [PubMed] [Google Scholar]
- Foyer, C.H., and Noctor, G. (2005). Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 17 1866–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti, F., Beaudoin, N., Serizet, C., Webb, A.A.R., Vartanian, N., and Giraudat, J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11 1897–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, L., Zhao, J., and Scandalios, J.G. (2000). Cis-elements and trans-factors that regulate expression of the maize Cat1 antioxidant gene in response to ABA and osmotic stress: H2O2 is the likely intermediary signaling molecule for the response. Plant J. 22 87–95. [DOI] [PubMed] [Google Scholar]
- Guo, Y., Xiong, L.M., Song, C.P., Gong, D.M., Hafter, U., and Zhu, J.K. (2002). A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev. Cell 3 233–244. [DOI] [PubMed] [Google Scholar]
- Gupta, R., and Luan, S. (2003). Redox regulation of protein tyrosine phosphatases and MAP kinases in higher plants. Plant Physiol. 132 1149–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbette, P., Lenne, C., Leblanc, N., Julien, J.L., JoeDrevet, R., and Roeckel-Drevet, P. (2002). Two GPX-like proteins from Lycopersicon esculentum and Helianthus annuus are antioxidant enzymes with phospholipid hydroperoxide glutathione peroxidase and thioredoxin peroxidase activities. Eur. J. Biol. Chem. 269 2414–2420. [DOI] [PubMed] [Google Scholar]
- Holland, D., Ben-Hayyim, G., Faltin, Z., Camoin, L., Strosberg, A.D., and Eshdat, Y. (1993). Molecular characterization of salt-stress-associated protein in Citrus: Protein and cDNA sequence homology to mammalian glutathione peroxidase. Plant Mol. Biol. 21 923–927. [DOI] [PubMed] [Google Scholar]
- Hugouvieux, V., Kwak, J.M., and Schroeder, J.I. (2001). An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106 477–487. [DOI] [PubMed] [Google Scholar]
- Inaba, K., and Ito, K. (2002). Paradoxical redox properties of DsbB and DsbA in the protein disulfide-introducing reaction cascade. EMBO J. 21 2646–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo, J.H., Wang, S., Chen, J.G., Jones, A.M., and Fedoroff, N.V. (2005). Different signaling and cell death roles of heterotrimeric G protein α and β subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell 17 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, B.G., et al. (2002). A Chinese cabbage cDNA with high sequence identity to phospholipid hydroperoxide glutathione peroxidases encodes a novel isoform of thioredoxin-dependent peroxidase. J. Biol. Chem. 277 12572–12578. [DOI] [PubMed] [Google Scholar]
- Kapoor, A., Agarwal, M., Andreucci, A., Zheng, X., Gong, Z., Hasegawa, P.M., Bressan, R.A., and Zhu, J.-K. (2005). Mutations in a conserved replication protein suppress transcriptional gene silencing in a DNA methylation independent manner in Arabidopsis. Curr. Biol. 15 1912–1918. [DOI] [PubMed] [Google Scholar]
- Kim, S.O., Merchant, K., Nudelman, R., Beyer, W.F., Jr., Keng, T., DeAngelo, J., Hausladen, A., and Stamler, J.S. (2002). OxyR: A molecular code for redox-related signaling. Cell 109 383–396. [DOI] [PubMed] [Google Scholar]
- Kishigami, S., Akiyama, Y., and Ito, K. (1995). Redox states of DsbA in the periplasm of Escherichia coli. FEBS Lett. 364 55–58. [DOI] [PubMed] [Google Scholar]
- Kobayashi, T., Kishigami, S., Sone, M., Inokuchi, H., Mogi, T., and Ito, K. (1997). Respiratory chain is required to maintain oxidized states of the DsbA–DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc. Natl. Acad. Sci. USA 94 11857–11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Reuling, G., and Karssen, C.M. (1984). The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 61 377–383. [Google Scholar]
- Kovtun, Y., Chiu, W.L., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade. Proc. Natl. Acad. Sci. USA 97 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, J., Boisson-Dernier, A., Dizon, M.B., Maktabi, M.H., and Schroeder, J.I. (2006). The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 140 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak, J.M., Mori, I.C., Pei, Z.M., Leonhardt, N., Angel Torres, M., Dangl, J.L., Bloom, R.E., Bodde, S., Jones, J.D.G., and Schroeder, J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Choi, H., Suh, S., Doo, I.S., Oh, K.Y., Choi, E.J., Taylor, A.T.S., Low, P.S., and Lee, Y. (1999). Oligogalacturonic acid and chitosan reduce stomatal aperture by inducing the evolution of reactive oxygen species from guard cells of tomato and Commelina communis. Plant Physiol. 121 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, J., Merlot, S., and Giraudat, J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Nishitoh, H., Ichijo, H., and Kyriakis, J.M. (2000). Activation of apoptosis signal-regulating kinase 1 (ASK1) by tumor necrosis factor receptor-associated factor 2 requires prior dissociation of the ASK1 inhibitor thioredoxin. Mol. Cell. Biol. 20 2198–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., and Zhu, J.-K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280 1943–1945. [DOI] [PubMed] [Google Scholar]
- Luan, S. (2002). Signalling drought in guard cells. Plant Cell Environ. 25 229–237. [DOI] [PubMed] [Google Scholar]
- McAinsh, M.R., Clayton, H., Mansfield, T.A., and Hetherington, A.M. (1996). Changes in stomatal behaviour and guard cell cytosolic free calcium in response to oxidative stress. Plant Physiol. 111 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhard, M., and Grill, E. (2001). Hydrogen peroxide is a regulator of ABI1, a protein phosphatase 2C from Arabidopsis. FEBS Lett. 508 443–446. [DOI] [PubMed] [Google Scholar]
- Meinhard, M., Rodriguez, P.L., and Grill, E. (2002). The sensitivity of ABI2 to hydrogen peroxide links the abscisic acid-response regulator to redox signaling. Planta 214 775–782. [DOI] [PubMed] [Google Scholar]
- Merkle, T., Leclerc, D., Marshallsay, C., and Nagy, F. (1996). A plant in vitro system for the nuclear import of proteins. Plant J. 10 1177–1186. [DOI] [PubMed] [Google Scholar]
- Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., and Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25 295–303. [DOI] [PubMed] [Google Scholar]
- Merlot, S., Mustilli, A.C., Genty, B., North, H., Lefebvre, V., Sotta, B., Vavasseur, A., and Giraudat, J. (2002). Using infrared thermal imaging to isolate Arabidopsis mutants defective in stomatal regulation. Plant J. 30 601–609. [DOI] [PubMed] [Google Scholar]
- Miao, Y.C., Song, C.-P., Dong, F.C., and Wang, X.C. (2000). ABA-induced hydrogen peroxide generation in guard cells of Vicia faba. Acta Phytophysiol. Sin. 26 53–58. [Google Scholar]
- Mishra, G., Zhang, W., Deng, F., Zhao, J., and Wang, X. (2006). A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 312 264–266. [DOI] [PubMed] [Google Scholar]
- Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7 405–410. [DOI] [PubMed] [Google Scholar]
- Mittler, R., Herr, E.H., Orvar, B.L., van Camp, W., Willekens, H., Inzé, D., and Ellis, B.E. (1999). Transgenic tobacco plants with reduced capability to detoxify reactive oxygen intermediates are hyperresponsive to pathogen infection. Proc. Natl. Acad. Sci. USA 96 14165–14170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9 490–496. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Murata, Y., Pei, Z.M., Mori, I.C., and Schroeder, J.I. (2001). Abscisic acid activation of plasma membrane Ca2+ channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell 13 2513–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli, A.C., Merlot, S., Vavasseur, A., Fenzi, F., and Giraudat, J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhulu, S.B., Deng, X.-B., Sarria, R., and Gelvin, S.B. (1996). Early transcription of Agrobacterium T-DNA genes in tobacco and maize. Plant Cell 8 873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrutiu, I., Shillito, R., Potrykus, I., Biasini, G., and Sala, F. (1987). Hybrid genes in the analysis of transformation conditions. Plant Mol. Biol. 8 363–373. [DOI] [PubMed] [Google Scholar]
- Neill, N., Desikan, R., and Hancock, J. (2002). Hydrogen peroxide signaling. Curr. Opin. Plant Biol. 5 388–395. [DOI] [PubMed] [Google Scholar]
- Noctor, G., and Foyer, C.H. (1998). Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 249–279. [DOI] [PubMed] [Google Scholar]
- Ohta, M., Guo, Y., Halfter, U., and Zhu, J.-K. (2003). A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. USA 100 11771–11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer, K., Tuominen, H., Kettunen, R., Betz, C., Langebartels, C., Sandermann, H., Jr., and Kangasjärvi, J. (2000). Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 12 1849–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, S., Wang, X.-Q., Coursol, S.A., and Assmann, S.M. (2002). Preparation and application of Arabidopsis thaliana guard cell protoplasts. New Phytol. 153 517–526. [DOI] [PubMed] [Google Scholar]
- Pei, Z.M., Murata, Y., Benning, G., Thomine, S., Klusener, B., Allen, G.J., Grill, E., and Schroeder, J.I. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic signaling in guard cells. Nature 406 731–734. [DOI] [PubMed] [Google Scholar]
- Rentel, M.C., Lecourieux, D., Ouaked, F., Usher, S.L., Petersen, L., Okamoto, H., Knight, H., Peck, S.C., Grierson, C.S., Hirt, H., and Knight, M.R. (2004). OXI1 kinase is necessary for oxidative burst-mediated signaling in Arabidopsis. Nature 427 858–861. [DOI] [PubMed] [Google Scholar]
- Rodriguez Milla, M.A., Butler, E., Rodriguez Huete, A., Wilson, C.F., Anderson, O., and Gustafson, J.P. (2002). EST-based gene expression analysis under aluminum stress in rye (Secale cereale L.). Plant Physiol. 130 1706–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Milla, M.A., Maurer, A., Huete, A.R., and Gustafson, J.P. (2003). Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J. 36 602–615. [DOI] [PubMed] [Google Scholar]
- Roeckel-Drevet, P., de Gagne, G., Labrouhe, T.D., Dufaure, J.P., Nicolas, P., and Drevet, J.R. (1998). Molecular characterization of organ distribution and stress-mediated induction of two glutathione peroxidase-encoding mRNAs in sunflower (Helianthus annuus). Physiol. Plant. 103 385–394. [Google Scholar]
- Schroeder, J.I., Kwak, J.M., and Allen, G.J. (2001). Guard cell abscisic acid signalling and engineering of drought hardiness in plants. Nature 410 327–330. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127 1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Song, C.-P., Agarwal, M., Ohta, M., Guo, Y., Halfter, U., Wang, P., and Zhu, J.-K. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17 2384–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticher, L., Mauch-Mani, B., and Metraux, J.P. (1997). Systemic acquired resistance. Annu. Rev. Phytopathol. 35 235–270. [DOI] [PubMed] [Google Scholar]
- Sugimoto, M., and Sakamoto, W. (1997). Putative phospholipid hydroperoxide glutathione peroxidase gene from Arabidopsis thaliana induced by oxidative stress. Genes Genet. Syst. 72 311–316. [DOI] [PubMed] [Google Scholar]
- Sundaresan, M., Yu, Z.-X., Ferrans, V., Irani, K., and Finkel, T. (1995). Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science 270 296–299. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., Sano, T., Tamaoki, M., Nakajima, N., Kondo, N., and Hasezawa, S. (2005). Ethylene inhibits abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol. 138 2337–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen, H., Zhang, Z.G., Wei, Y.D., and Collinge, D.B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11 1187–1194. [Google Scholar]
- Torres, M.A., Dangl, J.L., and Jones, J.D.G. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursini, F., and Bindoli, A. (1987). The role of selenium peroxidases in the protection against oxidative damage of membranes. Chem. Phys. Lipids. 44 255–276. [DOI] [PubMed] [Google Scholar]
- Ursini, F., Miaorino, M., Brigelius-Flohe, R., Aumann, K.D., Roveri, A., Schomburg, D., and Flohe, L. (1995). Diversity of glutathione peroxides. Methods Enzymol. 252 38–53. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe, G.C., and McIntosh, L. (1997). Alternative oxidase: From gene to function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 703–734. [DOI] [PubMed] [Google Scholar]
- van Montfort, R.L., Congreve, M., Tisi, D., Carr, R., and Jhoti, H. (2003). Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature 423 773–777. [DOI] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Schutze, K.S., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Wang, P.C., Du, Y.Y., An, G.Y., Zhou, Y., and Song, C.-P. (2006). Analysis of global expression profiles of Arabidopsis genes under abscisic acid and H2O2 applications using Affymetrix GeneChips. J. Integr. Plant Biol. 48 62–74. [Google Scholar]
- Wang, Y.-F., Fan, L.-M., Zhang, W.-Z., Zhang, W., and Wu, W.-H. (2004). Ca2+-permeable channels in the plasma membrane of Arabidopsis pollen are regulated by actin microfilaments. Plant Physiol. 136 3892–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Miao, Y.C., An, G.Y., Zhou, Y., Shangguan, Z.P., Gao, J.F., and Song, C.P. (2001. a). K+ channels inhibited by hydrogen peroxide mediate abscisic acid signalling in Vicia guard cells. Cell Res. 11 195–202. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Zhang, L., Dong, F., Gao, J., Galbraith, D.W., and Song, C.P. (2001. b). Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 126 1438–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, M., Aslund, F., and Storz, G. (1998). Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279 1718–1721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.