Abstract
Legumes develop different types of lateral organs from their primary root, lateral roots and nodules, the latter depending on a symbiotic interaction with Sinorhizobium meliloti. Phytohormones have been shown to function in the control of these organogeneses. However, related signaling pathways have not been identified in legumes. We cloned and characterized the expression of Medicago truncatula genes encoding members of cytokinin signaling pathways. RNA interference of the cytokinin receptor homolog Cytokinin Response1 (Mt CRE1) led to cytokinin-insensitive roots, which showed an increased number of lateral roots and a strong reduction in nodulation. Both the progression of S. meliloti infection and nodule primordia formation were affected. We also identified two cytokinin signaling response regulator genes, Mt RR1 and Mt RR4, which are induced early during the symbiotic interaction. Induction of these genes by S. meliloti infection is altered in mutants affected in the Nod factor signaling pathway; conversely, cytokinin regulation of the early nodulin Nodule Inception1 (Mt NIN) depends on Mt CRE1. Hence, cytokinin signaling mediated by a single receptor, Mt CRE1, leads to an opposite control of symbiotic nodule and lateral root organogenesis. Mt NIN, Mt RR1, and Mt RR4 define a common pathway activated during early S. meliloti interaction, allowing crosstalk between plant cytokinins and bacterial Nod factors signals.
INTRODUCTION
Two types of root lateral organs can develop on legume roots: lateral roots and nitrogen-fixing nodules. In the case of nodulation, an interaction with specific rhizobia microsymbionts is required (Geurts et al., 2005). After a specific molecular exchange of signaling molecules, including bacterial Nod factors, rhizobia provoke several early responses on plant roots, including root hair curling, progression of infection threads in root hairs toward inner cortical cells, reactivation of cell division in the outer cortex, and amyloplast deposition. Finally, in indeterminate nodules, a meristem is developed that will lead to nodule differentiation. Non-nodulating mutants (nod−) impaired in the Nod factor signaling pathway have been recently characterized, such as dmi (for doesn't make infections), nsp (for nodulation signaling pathway), or nin (for nodule inception) (Schauser et al., 1999; Catoira et al., 2000; Geurts et al., 2005; Kalo et al., 2005). In addition, plants affected in both lateral root and nodule development have been identified, for example, the har1 (for hypernodulation aberrant root formation) mutation in Lotus japonicus (Wopereis et al., 2000) and the nip/latd (for numerous infections and polyphenolics/root lateral organs deficient) mutation in Medicago truncatula (Veereshlingam et al., 2004; Bright et al., 2005). Furthermore, several early nodulin genes are also expressed in lateral roots (Hirsch and LaRue, 1997). These data indicate that common regulatory pathways may be shared in both types of root-derived organogenesis.
The plant hormone cytokinin is implicated in the regulation of many physiological processes during plant development, growth, and adaptation to environmental conditions, such as nutrient starvation (Martin et al., 2000; Mok and Mok, 2001). Physiological studies based on exogenous cytokinin application or alteration of cytokinin metabolism, by overexpression of isopentenyl transferase or cytokinin oxidase (CKX), revealed a role for this hormone in the control of root architecture and development (Lorteau et al., 2001; Werner et al., 2001, 2003; Lohar et al., 2004). Exogenous application of cytokinins on legume roots induced responses similar to rhizobial Nod factors, including cortical cell divisions, amyloplast deposition, and induction of early nodulin gene expression (Cooper and Long, 1994; Bauer et al., 1996; Fang and Hirsch, 1998; Lorteau et al., 2001). Similarly, overexpression of a heterologous CKX gene in L. japonicus leads to changes in both lateral roots and nodule numbers (Lohar et al., 2004).
In Arabidopsis thaliana, a model has been proposed for cytokinin perception and signaling involving a His-Asp multistep phosphorelay. This transduction pathway comprises a His kinase (HK) receptor and the downstream elements His phosphotransfer proteins (HPs) and response regulators (RRs; Sheen, 2002; Heyl and Schmulling, 2003). AHK4/CRE1/WOL (for Arabidopsis HISTIDINE KINASE4/CYTOKININ RESPONSE1/WOODEN LEG) was initially identified as a cytokinin receptor based upon its structure and its biochemical ability to bind cytokinins and to mediate activation of cytokinin responses (Mahonen et al., 2000; Inoue et al., 2001; Suzuki et al., 2001). Expression of AHK4/CRE1 is induced by application of exogenous cytokinins (Mahonen et al., 2000; Ueguchi et al., 2001b; Che et al., 2002; Rashotte et al., 2003), and two other closely related genes, AHK2 and AHK3, have also been characterized as cytokinin receptors (Ueguchi et al., 2001a; Yamada et al., 2001). Five Arabidopsis genes (AHP1–AHP5) encoding His phosphotransfer proteins have been identified that could activate downstream elements, such as the response regulators (ARRs). In Arabidopsis, this latter family comprises 22 genes (ARR1–ARR22) grouped into two main subfamilies: A-type ARRs contain only a receiver domain, whereas B-type ARRs, which also contain a MYB-type DNA binding domain, act as transcription factors (Heyl and Schmulling, 2003). In contrast with B-type ARRs, A-type ARRs are rapidly and strongly induced by cytokinins, indicating that these genes are primary cytokinin response targets (Brandstatter and Kieber, 1998; Taniguchi et al., 1998; Kiba et al., 1999). Mutations in Arabidopsis cytokinin signaling genes, either in AHK or ARR families, lead to alterations in root architecture. The analysis of multiple mutants of AHKs or ARRs revealed overlapping but distinct roles for these cytokinin signaling genes in root growth and architecture (Higuchi et al., 2004; Nishimura et al., 2004; To et al., 2004; Mason et al., 2005; Riefler et al., 2006). However, no legume cytokinin signaling pathway has been characterized so far. Increasing knowledge on cytokinin perception and transduction pathways is therefore crucial to understanding how the development of common and legume-specific root lateral organs is integrated. Moreover, as Nod factors and cytokinin lead to common cellular and molecular responses (Bauer et al., 1996; Hirsch et al., 1997), potential crosstalk between cytokinin signaling and other pathways, such as bacterial Nod factor signaling, could be identified.
In this work, we linked three specific cytokinin signaling genes, an HK (Mt CRE1) and two response regulators Mt RR4 (A type) and Mt RR1 (B type), to the symbiotic interaction. Mt CRE1 RNA interference (RNAi) roots showed strong reduction in their cytokinin sensitivity and in their primary response to this hormone. These roots also showed a significant increase in lateral root development and a reduction in nodulation, associated with perturbations in both infection and nodule primordia formation. Two cytokinin signaling genes, Mt RR1 and Mt RR4, were activated during nodule formation in a Nod factor–dependent manner. On the other hand, the early nodulin Mt NIN was rapidly induced by cytokinins, similar to the A-type Mt RR cytokinin primary response genes. Regulation of Mt RR1 and Mt RR4 expression by Sinorhizobium meliloti is affected in nod− mutants, whereas cytokinin induction of Mt NIN depends on Mt CRE1. Our results therefore demonstrate that the Mt CRE1 cytokinin receptor regulates nodule and lateral root organogenesis in an opposite manner and indicate that Mt NIN, Mt RR1, and Mt RR4 are involved in crosstalk between Nod factors and cytokinin signaling pathways.
RESULTS
Identification of Genes Related to Cytokinin Signaling Pathways in M. truncatula
To identify M. truncatula genes corresponding to Arabidopsis cytokinin signaling genes (AHKs, AHPs, and ARRs), an in silico search was performed on The Institute for Genomic Research (TIGR) EST database and on genomic sequences available from the National Center for Biotechnology Information (NCBI) database (see Supplemental Table 1 online). Unrooted relationship trees were produced for each family (see Supplemental Figure 1 online). Thirteen TIGR TCs (for tentative consensus) were found corresponding to five TCs for AHKs (Mt HK1–Mt HK5), three TCs for AHPs (Mt HP1–Mt HP3), and five TCs for ARRs (Mt RR1–Mt RR5). The genomic sequence found for one Mt HK allowed us to group three of the proposed TIGR Mt HK TCs (Mt HK1, Mt HK4, and Mt HK5) into a single genomic locus closely related to AHK4/CRE1/WOL (see Supplemental Figure 1A online). This gene was therefore referred to as Mt CRE1. For the Mt RR gene family, A- and B-type RR subfamilies could be identified (two and three M. truncatula genes identified, respectively; see Supplemental Figure 1C online).
Functional Characterization of Cytokinin Signaling Pathways in M. truncatula Roots
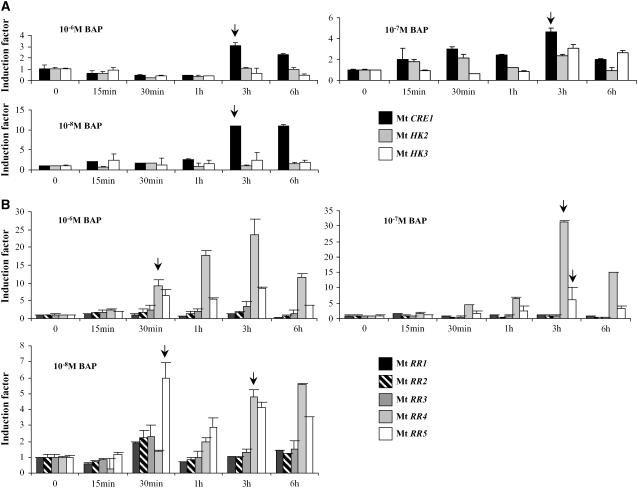
As a first characterization, we checked the transcriptional regulation of the putative cytokinin signaling genes identified in response to a short cytokinin treatment using real-time RT-PCR (Figure 1). Among those genes, only one Mt HK gene (Mt CRE1; Figure 1A, arrows) and two A-type Mt RR genes (Mt RR4 and Mt RR5; Figure 1B, arrows) were rapidly and strongly induced by this hormone at different concentrations. The identification of cytokinin-responsive genes in M. truncatula roots indicates a global conservation of the transcriptional regulation of these gene families in Arabidopsis and M. truncatula.
Figure 1.
Cytokinin Signaling Genes Are Transcriptionally Induced in Response to Short-Term Cytokinin Treatments.
Real-time RT-PCR analysis of Mt HK (A) and A- and B-type Mt RR (B) induction in response to the cytokinin BAP at different concentrations (10−8, 10−7, or 10−6 M) and various incubation times (15 min, 30 min, 1 h, 3 h, or 6 h). Histograms represent the quantification of specific PCR amplification products for each gene normalized with the constitutive control Mt ACTIN11. The value of untreated roots (t = 0) is set to 1 as reference. A representative example out of three biological experiments is shown, and error bars indicate standard deviation for three technical replicates. Arrows indicate significant induction of specific genes mentioned in the text.
A second step was to link the identified genes to a functional cytokinin response. We therefore initiated a functional approach based on silencing of specific putative cytokinin receptors in M. truncatula composite plants with Agrobacterium rhizogenes–transformed roots selected on kanamycin (as described in Boisson Dernier et al., 2001). For this, we first compared cytokinin sensitivity and expression levels of cytokinin signaling genes in A. rhizogenes–transformed control roots (i.e., roots expressing an RNAi construct targeting the gene coding for β-glucuronidase [GUS]) versus wild-type roots (i.e., nontransformed seedling roots). Dose–response curves to monitor effects of different benzyl-amino-purine (BAP) concentrations ranging from 10−8 to 10−6 M on main root growth showed a similar response for both root types and an arrest of root growth after 10−7 or 10−6 M BAP treatments (data not shown). Moreover, real-time RT-PCR analysis of Mt HK and Mt RR genes revealed similar expression levels of these genes in both root types (see Supplemental Figure 2 online). We concluded that A. rhizogenes–transformed roots display similar cytokinin responses as wild-type roots.
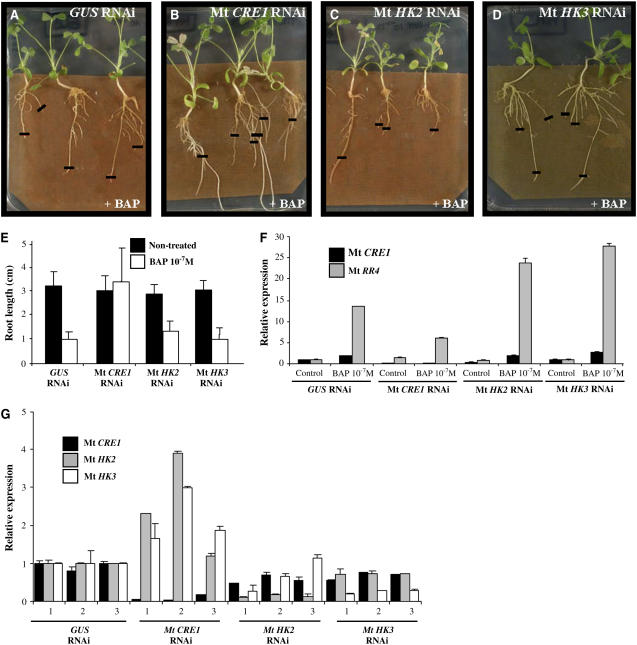
Sensitivity of Mt HK RNAi roots to exogenous cytokinins was then examined and compared with Mt GUS RNAi roots. Two weeks after A. rhizogenes infection, composite plants were transferred to a medium containing 10−7 M BAP and incubated for six more days. Measurements of >150 independent transgenic roots per construct (from seven biological experiments) revealed that Mt CRE1 RNAi roots were able to grow similarly to nontreated Mt CRE1 RNAi roots (Figures 2B and 2E; analysis of variance [ANOVA] P < 0.001). By contrast, Mt HK2 RNAi, Mt HK3 RNAi, and GUS RNAi roots showed significant growth arrest (Figures 2A to 2E). Reduction in cytokinin sensitivity of Mt CRE1 RNAi roots was further confirmed by monitoring the expression of a primary cytokinin response gene, Mt RR4. A minimum of 15 independent transgenic roots per construct were pooled to perform real-time RT-PCR analyses. In Mt CRE1 RNAi roots, the A-type response regulator Mt RR4 was weakly induced by cytokinins, whereas a strong induction was observed in GUS RNAi, Mt HK2 RNAi, and Mt HK3 RNAi roots (Figure 2F). These results indicate that Mt CRE1 RNAi roots show a downregulated cytokinin response, correlating with the observed cytokinin insensitivity.
Figure 2.
Mt CRE1 RNAi Roots Are Cytokinin Insensitive.
(A) to (D) Representative examples of root growth observed in A. rhizogenes–transformed M. truncatula roots expressing GUS RNAi, Mt CRE1 RNAi, Mt HK2 RNAi, or Mt HK3 RNAi constructs 6 d after transfer to 10−7 M BAP. Black lines indicate the position of root tips at the moment of transfer. Cytokinin insensitivity is visualized by the root's ability to grow on this inhibitory concentration of cytokinin. The transgenic roots were selected on kanamycin as described by Boisson-Dernier et al. (2001).
(E) Mean length of independent transgenic roots transformed with these different constructs and grown 6 d on media with or without 10−7 M BAP. A representative example out of seven biological experiments is shown (n > 30 per construct and condition), and error bars represent standard deviation (ANOVA, P < 0.001)
(F) Expression analysis of Mt CRE1 and a cytokinin-inducible A-type response regulator, Mt RR4. Real-time RT-PCR analysis was performed on A. rhizogenes–transformed M. truncatula roots carrying either a control (GUS RNAi) or Mt HKs RNAi constructs (n > 15) after a short-term cytokinin treatment (10−7 M BAP for 1 h). Histogram represents the quantification of specific PCR amplification products normalized to the constitutive control Mt ACTIN11. The value of control transgenic roots (i.e., non-BAP-treated GUS RNAi root) is set at 1. A representative example out of three biological experiments is shown, and error bars represent standard deviation for three technical replicates.
(G) Expression analysis of Mt CRE1, Mt HK2, and Mt HK3. Real-time RT-PCR analysis was performed on independent A. rhizogenes–transformed M. truncatula roots carrying either a control (GUS RNAi) or Mt HK RNAi constructs and grown 7 d on 10−7 M BAP. Histograms represent the quantification of each specific PCR amplification product normalized to the constitutive control Mt ACTIN11. The value of control transgenic roots (i.e., GUS RNAi) is set up at 1 as reference. A representative example out of three biological experiments is shown, and error bars represent standard deviation for three technical replicates.
The strength and specificity of the downregulation induced in Mt CRE1 RNAi roots was also correlated with the cytokinin-insensitive phenotype of several independent transgenic roots. Representative examples (Figure 2G) indicate that the Mt CRE1 RNAi construct is efficient and does not affect Mt HK2 or Mt HK3 genes. Higher Mt HK2 and Mt HK3 expression levels could even be detected in some Mt CRE1 RNAi roots, suggesting a molecular compensation effect. Altogether, a correlation has been established between reduction in Mt CRE1 (but not Mt HK2 or Mt HK3) expression levels, cytokinin insensitivity of root growth, and downregulation of a cytokinin primary response.
As ethylene perception is also mediated by a family of HKs related to cytokinin receptor genes (Bleecker and Schaller, 1996; Gamble et al., 1998), we investigated whether Mt CRE1 RNAi roots could be perturbed in ethylene perception or root responses to this phytohormone. Ethylene sensitivity of roots expressing GUS or Mt CRE1 RNAi constructs was monitored by measuring root growth inhibition induced by the biosynthetic precursor of ethylene 1-aminocyclopropane-1-carboxylic acid (ACC) at two concentrations (10−5 and 10−6 M; see Supplemental Figure 3 online). Growth of both Mt CRE1 and GUS RNAi roots was similarly inhibited in the presence of ACC compared to untreated roots. Altogether, these results suggest that Mt CRE1 plays a specific role in cytokinin perception in M. truncatula roots.
Lateral Root Density Is Enhanced in Mt CRE1 RNAi Roots
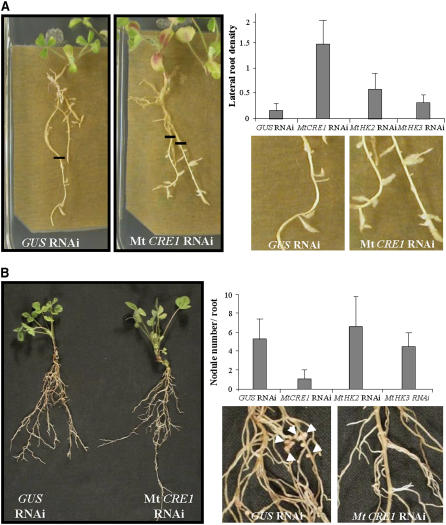
Since Mt CRE1 is crucial for root growth in response to exogenous cytokinin application, we analyzed the involvement of this signaling pathway in M. truncatula root architecture. A problem with analysis of lateral root phenotypes in A. rhizogenes–infected transgenic roots is that their architecture is heterogeneous between independent clones and experiments. For this reason, we developed a medium (lateral root inductive medium [LRIM]) to synchronously induce the formation of lateral roots in seedlings (see Supplemental Figure 4 online) and A. rhizogenes–infected roots (Figure 3A). After kanamycin selection, composite plants were therefore transferred on LRIM, and the position of the root apex was labeled to identify the newly grown root regions after 6 d (as illustrated in Figure 3A). In this way, lateral root density could be determined in these equivalent root regions independently of the initial size, developmental stage, or architecture of the A. rhizogenes–transformed roots. Analysis of independent transgenic roots (n > 70 per construct and condition) in three different biological experiments revealed a significant increase (ANOVA, P < 0.001) of lateral root density for Mt CRE1 RNAi roots compared with GUS, Mt HK2 RNAi, or Mt HK3 RNAi roots (Figure 3A, graph). This result indicates that besides its function in main root growth, Mt CRE1 is also involved in the negative control of lateral root development in M. truncatula.
Figure 3.
Mt CRE1 RNAi Roots Are Affected in Lateral Root and Nodule Development.
(A) Representative examples of root growth observed in A. rhizogenes–transformed M. truncatula roots expressing GUS RNAi (left panel and close-up) and Mt CRE1 RNAi (right panel and close-up) constructs 6 d after transfer to LRIM. Black lines indicate the position of root tips at the moment of transfer. Lateral root density is calculated in A. rhizogenes–transformed M. truncatula roots in the newly grown root region 6 d after transfer to LRIM. Histograms represent lateral root densities in one (n > 25) out of three biological experiments for GUS RNAi, Mt CRE1 RNAi, Mt HK2 RNAi, and Mt HK3 RNAi roots. Error bars indicate standard deviations (ANOVA, P < 0.001).
(B) One-month-old transgenic roots expressing GUS RNAi or Mt CRE1 RNAi constructs were S. meliloti–inoculated in a greenhouse. General view of GUS RNAi and Mt CRE1 RNAi composite plants (left panel) and close-up of S. meliloti–inoculated GUS RNAi (close-up, left panel) and Mt CRE1 RNAi (close-up, right panel) roots. Arrowheads indicate nodule locations. Quantification of nodules was done on GUS RNAi, Mt CRE1 RNAi, Mt HK2 RNAi, and Mt HK3 RNAi roots 15 DAI with S. meliloti 2011 strain. Histogram represents nodule numbers per transgenic root in one out of five biological experiments (n > 25). Error bars represent standard deviations (ANOVA, P < 0.001).
Mt CRE1 RNAi Roots Are Strongly Impaired in Nodulation
We next determined whether Mt CRE1 could also be involved in a legume-specific root lateral organogenesis (i.e., nodule formation). The ability of Mt CRE1 roots to form nodules was tested after transfer on a nitrogen-deprived medium and subsequent inoculation of A. rhizogenes–transformed roots with a bacterial suspension of S. meliloti 2011 wild-type strain. Nodule numbers in transformed roots were determined 15 d after inoculation (DAI) on at least 120 independent transgenic roots (in five biological experiments) from composite plants grown either in greenhouse conditions (representative plants illustrated in Figure 3B) or in vitro (representative experiment in Figure 3B, graph). Independent of the growth conditions used, nodulation of Mt CRE1 RNAi roots was strongly and significantly (ANOVA, P < 0.001) impaired compared either with wild-type seedlings, GUS RNAi control, Mt HK2 RNAi, or Mt HK3 RNAi roots (Figure 3B, graph). In the in vitro growth conditions, we observed that 50% of the Mt CRE1 RNAi roots did not develop any nodule, whereas 100% of GUS RNAi, Mt HK2, or Mt HK3 RNAi roots contained nodules.
These data indicate that Mt CRE1–mediated cytokinin signaling is required for efficient nodulation of M. truncatula roots. Even though Mt HK2 and Mt HK3 may also contribute to root lateral organogeneses, the Mt CRE1–dependent pathway has a major opposite effect on the development of lateral roots and root nodules.
Both S. meliloti Infection and Early Nodule Organogenesis Are Perturbed in Mt CRE1 RNAi Roots
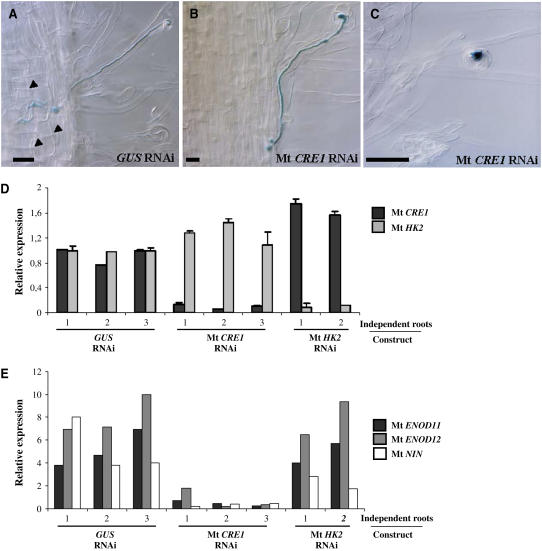
To determine how cytokinin signaling mediated by Mt CRE1 is involved in nodulation, we analyzed different steps of the symbiotic interaction. An S. meliloti 2011 strain carrying a ProHemA:LACZ reporter gene (Ardourel et al., 1994) was used to follow the early stages of infection in A. rhizogenes–transformed roots. Infection events could occur in the Mt CRE1 RNAi roots; however, progression of infection threads was rapidly blocked, either in roots hairs or in root epidermis compared with GUS RNAi roots (Figures 4A to 4C). In nodule-deficient roots, no infection threads could reach cortical cells, whereas ∼75% of the infections reached the cortex in control GUS RNAi roots (Table 1). Moreover, no amyloplast deposition or cortical cell division (Figure 4A) could be observed in these Mt CRE1 RNAi roots (Figure 4B). The few nodules contained in certain Mt CRE1 RNAi roots are similar to the wild type as shown by the use of the S. meliloti strain expressing the LACZ reporter gene and lugol staining (see Supplemental Figure 5 online). This suggests that these infection events are likely due to escapes of the Mt CRE1 RNAi silencing effect.
Figure 4.
Early Nodulation Is Perturbed in Mt CRE1 RNAi Roots.
(A) to (C) Histochemical staining of an S. meliloti strain expressing a ProHemA:LACZ reporter construct to follow early events of nodulation. Representative infection threads observed in an S. meliloti–inoculated GUS RNAi root (A) and nodule-deficient Mt CRE1 RNAi roots ([B] and [C]) are shown. Arrowheads indicate cortical cell divisions. Note the lack of cortical cell divisions in Mt CRE1 RNAi roots. Bars = 25 μm.
(D) Real-time RT-PCR analysis of Mt CRE1 and Mt HK2 expression in representative independent transgenic roots expressing GUS RNAi, Mt CRE1 RNAi, or Mt HK2 RNAi constructs 15 DAI by S. meliloti. Histograms represent the quantification of each specific PCR amplification product normalized to the constitutive control Mt ACTIN11. The value of control transgenic roots (i.e., GUS RNAi) is set to 1 as reference. Error bars represent standard deviation for three technical replicates.
(E) Expression analysis of the early nodulation markers Mt ENOD11, Mt ENOD12, and Mt NIN by semiquantitative RT-PCR in independent transgenic roots expressing GUS RNAi, Mt CRE1 RNAi, or Mt HK2 RNAi constructs 15 DAI by S. meliloti. Histogram represents the quantification of each specific PCR amplification product normalized to the constitutive control Mt ACTIN11.
Table 1.
Cellular Localization of Infection Thread Ends in GUS RNAi and Mt CRE1 RNAi Roots to Monitor Infection Progression
| GUS RNAi | Mt CRE1 RNAi | |
|---|---|---|
| Root hairs | 2 | 13 |
| Root epidermis | 5 | 7 |
| Outer cortex | 6 | 0 |
| Inner cortex | 7 | 0 |
| Nodule primordia formed | 10 | 0 |
| Total number of infections scored | 30 | 20 |
We then analyzed the expression of Mt HKs and nodulation marker genes associated with early stages of nodule formation (early nodulins Mt ENOD11, Mt ENOD12, and Mt NIN; Geurts et al., 2005) on several representative independent A. rhizogenes–transformed roots per construct using RT-PCR analysis (Figures 4D and 4E). First, we could correlate the absence of nodules in Mt CRE1 RNAi roots to a specific and efficient reduction in the expression of this gene (Figure 4D). In uninoculated roots, these three early nodulin genes were not expressed at significant levels (data not shown). High levels of expression of these early nodulin genes were detected in GUS RNAi and Mt HK2 RNAi inoculated roots in contrast with Mt CRE1 RNAi roots (Figure 4E), suggesting that no or limited induction of early nodulin genes occurred in these roots.
Altogether, Mt CRE1 is required at an early stage of the epidermal infection process and/or in cortical events associated with nodule organogenesis. Cytokinin signaling mediated by Mt CRE1 is thus a novel and crucial plant regulatory pathway for the early stages of the symbiotic interaction.
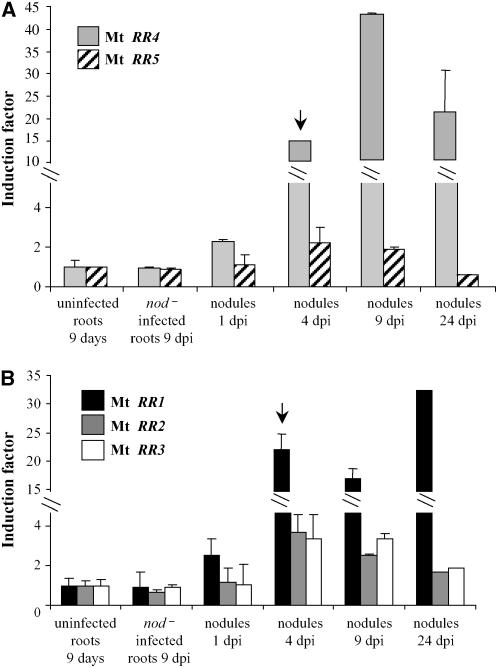
Mt RR1, Mt RR4, and Mt NIN Define Crosstalk between Nod Factors and Cytokinin Signaling Pathways
Our data open the question of an interaction between cytokinin and Nod factor signaling pathways. To further test the hypothesis of such crosstalk, we analyzed the regulation of cytokinin signaling genes during nodule development in wild-type plants (Figure 5). Two specific Mt RRs, an A type (Mt RR4) and a B type (Mt RR1), are differentially expressed from the early stages of nodule development (nodule primordia; Figures 5A and 5B, arrows). No induction was observed in roots inoculated with a nod− derivative S. meliloti strain, suggesting that regulation of Mt RR1 and Mt RR4 expression was dependent on the presence of Nod factors. However, we cannot rule out that other Mt RR genes also may be regulated at different time points, based on the observed variations in the cytokinin responses among these gene families (Figure 1).
Figure 5.
Differential Expression of Cytokinin-Related Response Regulator Genes during Nodule Formation.
Real-time RT-PCR analysis of roots or nodules at different days after S. meliloti infection (dpi). Expression analysis of A-type (A) and B-type (B) response regulator genes. Histograms represent the quantification of specific PCR amplification products normalized to the constitutive control Mt EF1α. The value for uninfected control roots is set to 1 as reference. A representative example out of three biological experiments is shown, and error bars represent standard deviation for three technical replicates. Arrows indicate significant induction of specific genes mentioned in the text.
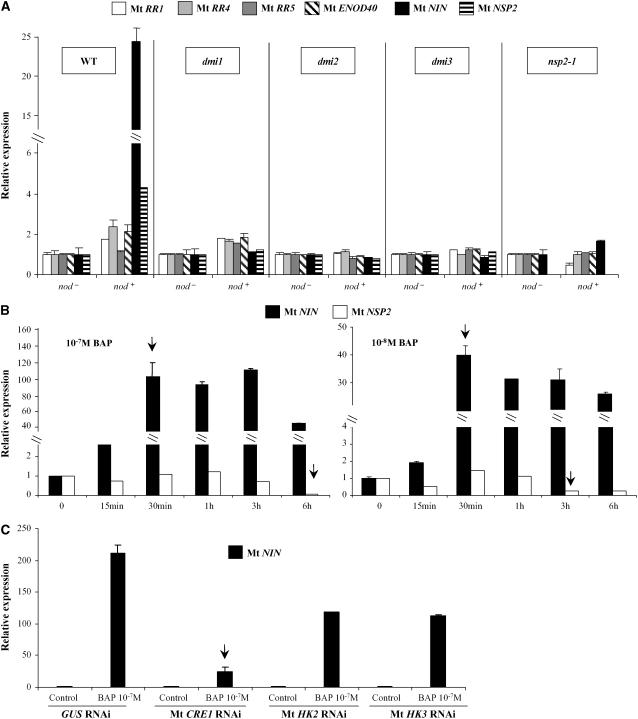
We therefore studied the expression of downstream elements of the Nod factor signaling pathway, such as Mt NSP2, Mt NIN, and Mt ENOD40 (Schauser et al., 1999; Geurts et al., 2005; Kalo et al., 2005), and the Mt RR genes regulated by cytokinins only (Mt RR5), by nodulation signals (Mt RR1), or by both (Mt RR4). These expression studies (Figure 6) were performed in various Nod factor signaling mutants: dmi1, dmi2, dmi3, (Catoira et al., 2000) and nsp2 (Kalo et al., 2005). After S. meliloti inoculation (1 DAI with a nod− or a nod+ strain), induction of Mt RR1 and Mt RR4 (but not Mt RR5, as previously mentioned in Figure 5) as well as Mt NSP2, Mt NIN, and Mt ENOD40 were at least strongly reduced in these nod− mutants (Figure 6A). Short-term cytokinin (10−7 M BAP, 3 or 6 h) inductions of both A-type Mt RRs and Mt ENOD40 were still detected in these mutants, although variations in the induction levels at different time points were observed (see Supplemental Figure 6 online). These data indicate that rapid induction of MtRR1 and MtRR4 by S. meliloti depends on the Nod factor signaling pathway.
Figure 6.
Crosstalk between Cytokinin and Nod Factor Signaling Pathways.
(A) Real-time RT-PCR analysis of several nodulation and cytokinin early response genes in wild-type and Nod factor signaling pathway mutant (dmi1-Y6, dmi2-TR25, dmi3-TRV25, and nsp2-1) roots 1 DAI with S. meliloti nod+ or nod− strain.
(B) Real-time RT-PCR analysis of Mt NIN and Mt NSP2 in response to short-term cytokinin treatments: BAP at different concentrations (10−8 or 10−7 M) and various incubations times (15 min, 30 min, 1 h, 3 h, or 6 h).
(C) Real-time RT-PCR analysis of Mt NIN expression in response to short-term cytokinin treatments (BAP 10−7 M for 1 h) in GUS RNAi, Mt CRE1 RNAi, Mt HK2 RNAi, or Mt HK3 RNAi A. rhizogenes–transformed M. truncatula roots (n > 15).
In all cases, the histogram represents the quantification of specific PCR amplification products for each gene normalized with the constitutive control Mt ACTIN11. The value of wild-type, mutant, or transgenic nontreated roots (S. meliloti nod− strain or no BAP [e.g., t = 0]) is set to 1 as reference. A representative example out of three biological experiments is shown, and error bars represent standard deviation for three technical replicates. Arrows indicate significant induction of specific genes mentioned in the text.
We then analyzed the induction by a short-term cytokinin treatment of Mt NSP2 and Mt NIN transcription factors (Figure 6B). Mt NIN was rapidly (after 30 min) and strongly induced after 10−8 or 10−7 M BAP treatments (Figure 6B, arrows). By contrast, Mt NSP2 expression was not modified at early time points but was repressed later (after 3 and 6 h of cytokinin treatment; Figure 6B, arrows). The Mt NIN expression pattern was therefore similar to those of the A-type response regulator genes (Figure 1), indicating that this nodulin gene may be a primary cytokinin response gene. Short-term cytokinin induction of Mt NIN expression was strongly reduced in Mt CRE1 RNAi roots, revealing the major role of the Mt CRE1–mediated pathway in the regulation of Mt NIN expression (Figure 6C, arrow). Nevertheless, Mt HK2 and Mt HK3 may contribute to Mt NIN regulation.
These results suggest a crosstalk between bacteria-derived Nod factors and cytokinin signaling pathways through the cross-regulation of Mt NIN, Mt RR1, and Mt RR4 expression.
DISCUSSION
In this work, we have shown that a unique homolog of an Arabidopsis cytokinin receptor, Mt CRE1, is required for the regulation of several aspects of root architecture in the model legume species M. truncatula. This gene controls secondary root organogenesis, with a positive effect on symbiotic nodule development and a negative impact on lateral root formation. Crosstalk between the cytokinin and Nod factor signaling pathways involving Mt RR1, Mt RR4, and Mt NIN could be identified.
In Arabidopsis, several HKs display distinct specificities regarding their involvement in root development. The closest homolog of Mt CRE1, AHK4/CRE1/WOL, was initially associated with root development, as the wol allele only showed a root phenotype (Scheres et al., 1995; Mahonen et al., 2000). By contrast, mutations in the other AHKs, ahk2 and ahk3, showed no root phenotype, whereas an ahk2 ahk3 double mutant was semidwarfed and its roots, despite a normal response to cytokinins, showed increased branching (Nishimura et al., 2004; Riefler et al., 2006). This led to the hypothesis that in Arabidopsis, different cytokinin receptors contribute to the growth of the root system. In M. truncatula, silencing of the single Mt CRE1 cytokinin receptor affects various aspects of root architecture and growth, including exogenous cytokinin perception, lateral root formation and nodulation. However, due to the heterogeneity of RNAi approaches in A. rhizogenes–transformed roots, together with the specific setup used to monitor lateral root induction, we may have overlooked contributions of the other Mt HKs to the regulation of the root system. Our results therefore suggest that Mt CRE1 is a major player for regulation of root architecture and cytokinin perception in M. truncatula roots, even though additive effects of other Mt HKs cannot be ruled out.
Several lines of evidence exist for an implication of cytokinins in nodulation, mainly derived from exogenous application experiments (Cooper and Long, 1994; Lorteau et al., 2001). Cytokinin induction of the expression of several early nodulin genes was reported earlier (Hirsch and Fang, 1994; Fang and Hirsch, 1998; Jimenez-Zurdo et al., 2000; Mathesius et al., 2000), and our work adds Mt NIN to this list. Moreover, cytokinin application mimics early nodulation responses, such as amyloplast deposition and cortical cell division (Bauer et al., 1996). More recently, reduction of cytokinin content in transgenic roots overexpressing a heterologous CKX gene revealed an effect on the number of nodules formed (Lohar et al., 2004). Altogether, these results suggest that cytokinins are involved in symbiotic nodule organogenesis. However, the signaling pathway involved, its relation with Nod factor signaling, and the precise nodulation step(s) involving cytokinins were not known. The cytokinin-insensitive roots obtained by decreasing Mt CRE1 expression showed a strong reduction in their ability to form symbiotic nodules. This phenotype affects early stages of the symbiotic interaction, the infection process, with infection threads blocked in root hairs and root epidermal cells, as well as the induction of cortical cell division, amyloplast deposition, and expression of early nodulins. It is however difficult to determine whether infection or primordia organogenesis is primarily affected and what the causal relationships between these two processes might be. As cortical cell division precedes infection thread formation (Timmers et al., 1999), it may be required for infection thread growth. In the case of Mt CRE1 RNAi symbiotic interaction, infection threads are formed but their growth is rapidly blocked in the epidermis. It is therefore likely that the inability of cortical cells to divide and the lack of primordia formation in these roots induce premature termination of infection thread growth. Cytokinin perception and signaling through Mt CRE1 may then have a primary function in controlling cortical cell division, and infection thread progression in the epidermis would therefore be blocked as a consequence. This scenario is in agreement with recent results obtained with a ProMtCRE1:GUS fusion that revealed weak expression in the cortex but strongly increased in patches of cortical cells after S. meliloti inoculation (Lohar et al., 2006).
Our expression analysis showed that one A-type response regulator, Mt RR4, and one B-type response regulator, Mt RR1, were early nodulation markers. Mt RR4 early induction was consistent with the results obtained by Lohar et al. (2004) using the heterologous Arabidopsis ProARR5:GUS reporter (closely related to the Mt RR4 gene; see Supplemental Figure 1 online) and a transcriptomic approach (Lohar et al., 2006). Our analysis suggests that one A-type RR is upregulated during nodule development. We cannot, however, exclude that Mt RR5 may also be involved in the nodulation process. Surprisingly, the only other cytokinin signaling gene found to be strongly upregulated at early stages of nodulation is Mt RR1, a B-type RR. Interestingly, in Arabidopsis, some B-type ARRs are able to bind specific boxes in promoters of A-type ARRs, leading to their rapid and direct transcriptional activation upon cytokinin application (Heyl and Schmulling, 2003). Our data suggest that S. meliloti induction of Mt RR1 transcription factor expression could mediate a downstream activation of Mt RR4. Again, we cannot exclude that other B-type Mt RRs may be involved in nodule development. Indeed, A. rhizogenes–tranformed M. truncatula roots expressing an Mt RR1 RNAi construct did not lead to any major root cytokinin sensitivity or nodulation phenotypes (see Supplemental Figure 7 online). This suggests either that functional redundancy occurs between B-type RRs or that RR1 is not a limiting factor in the regulation of cytokinin responses. Alternatively, Mt RR1 may not be involved in these processes, despite its differential expression.
To identify new putative cytokinin response genes and to test the hypothesis of crosstalk between cytokinin and Nod factor signaling pathways, we analyzed early cytokinin induction of crucial genes of the Nod factor signaling pathway (Geurts et al., 2005). Both the Mt NSP2 and Mt NIN genes are regulated by cytokinins. The late downregulation of Mt NSP2 (Figure 6B) occurs at the same time as the activation of Mt CRE1 expression (Figure 1), suggesting a role of this transcription factor in secondary effects of cytokinin action. By contrast, Mt NIN was rapidly induced by short-term cytokinin treatments, suggesting that this gene may act downstream of this putative crosstalk. Mt NIN is indeed induced as early as Mt RR4 and Mt RR5 (after 30 min of BAP treatment) and, consequently, may be a cytokinin primary response gene. Recently, a subset of Arabidopsis AP2 transcription factors was identified as rapidly induced by cytokinins through an AHK/AHP-dependent pathway and referred as CYTOKININ RESPONSE FACTOR (CRF) genes (Rashotte et al., 2006). Similarly, in our experiments, cytokinin regulation of Mt NIN expression is dependent on Mt HKs. It would be of interest to test a possible relocalization of Mt NIN to the nucleus upon cytokinin application, as recently observed for the CRFs. On the other hand, expression of cytokinin signaling genes (like Mt RR1 and Mt RR4) after S. meliloti short-term inoculation was dependent on Nod factor signaling, further supporting the existence of crosstalk between Nod factors and Mt CRE1–mediated cytokinin signaling pathways. Since certain nod− mutants are also affected in the mycorrhizal symbiosis (dmis; Geurts et al., 2005), these cytokinin signaling genes also may be involved in other types of plant–microbe interactions.
The activation of a specific cytokinin signaling pathway may act as an endogenous relay of early organogenesis-related nodulation responses, such as cortical cell division and formation of a carbon sink. Mt RR1 may be involved in the activation of at least some A-type Mt RR, such as Mt RR4, as well as early nodulins. Interestingly, in a minimal promoter region necessary for Mt ENOD11 expression during S. meliloti infection, ARR1 binding boxes were found, and this organization was conserved in another early nodulin promoter, Mt ENOD12, showing similar transcriptional regulation (Boisson-Dernier et al., 2005).
Interactions between lateral root and nodulation pathways are supported by physiological experiments (Nutman, 1948) as well as the existence of mutants affected in both processes, such as har1 or latd/nip (Wopereis et al., 2000; Veereshlingam et al., 2004; Bright et al., 2005). More recently, Nod factor effects on lateral root formation have been described in certain symbiotic mutants (Olah et al., 2005). It also has been documented in many plants that several hormones can interact together to regulate lateral root organogenesis (Casimiro et al., 2003). A nonexhaustive list includes a role for auxin/cytokinin balance, ethylene, and abscisic acid in the control of root architecture. In most cases, interactions have been reported at the level of hormone synthesis (Cary et al., 1995; Swarup et al., 2002), but interactions that provoke changes in the signaling response may also occur. Our results indicate that Mt CRE1–silenced roots are affected in cytokinin sensitivity but show normal responses to ethylene. However, affecting cytokinin signaling could induce indirect effects on phytohormone concentration (including cytokinins themselves, as recently shown in Arabidopsis; Riefler et al., 2006), signaling, or transport. The phenotypes of Mt CRE1 RNAi roots are consistent with a general role for cytokinins in the control of the initiation of lateral root organs and more generally on the control of root architecture in response to environmental conditions and root apical dominance (Franco-Zorrilla et al., 2002; Aloni et al., 2006). It is well known that a primary target of cytokinin action is cell cycle control (Redig et al., 1996; Zhang et al., 1996). Our results from Mt CRE1–silenced roots could be integrated by proposing that reduced nodulation may rely on a block of cortical cell divisions, whereas increased lateral root formation may be related to an activation of pericycle divisions. Tissue-specific activation of cytokinin signaling pathways therefore may be critical for the regulation of root lateral organogenesis in legumes.
METHODS
Plant Material
Medicago truncatula cv Jemalong A17 seeds were sterilized for 20 min in bleach (12% [v/v] sodium hypochlorite). After washing with sterilized water, seeds were sown on 1% agar plates and stored for 2 d at 4°C before incubating overnight at 24°C in the dark to ensure uniform germination. Germinated seedlings were transferred to square plates containing appropriate medium (see below) and grown vertically in chambers at 24°C under long-day conditions (16 h light/8 h dark).
Nod factor signaling pathway (nod−) mutant alleles used in these studies were dmi1-Y6, dmi2-TR25, dmi3-TRV25 (dmi1, 2, and 3; Catoira et al., 2000), and nsp2-1 (Oldroyd and Long, 2003; Kalo et al., 2005).
Cytokinin Treatments
Fifteen germinated seedlings were placed in flasks with 30 mL of low-nitrogen liquid medium (Blondon, 1964) and grown in a shaking incubator (125 rpm) at 24°C under long-day conditions (16 h light/8 h dark). After 5 d, seedlings were treated with various concentrations (10−6, 10−7, and 10−8 M) of BAP for various incubation times (0, 15 min, 30 min, 1 h, 3 h, and 6 h) and maintained under the same growth conditions. Roots were collected at the indicated time points and immediately frozen in liquid nitrogen for RNA extraction. Three independent biological experiments were performed.
Nodulation Experiments
Germinated seeds were grown in vitro on Fahraeus medium without nitrogen (Truchet et al., 1985). Roots were inoculated with 10 mL of Sinorhizobium meliloti suspension (OD600 = 0.05) per plate for 1 h. Different bacterial strains were used: a wild-type Sm2011 strain and a nodA− Sm2011 derivative (GMI5382; Debelle et al., 1986). Nodules were collected 4 DAI (primordium stage), 9 DAI (non-nitrogen-fixing nodules), and 24 DAI (nitrogen-fixing nodules). Noninoculated roots grown in the same conditions were also used as control. Roots (excluding the root tip region) or nodules were immediately frozen in liquid nitrogen for subsequent RNA extraction. For experiments using the nod− mutant, S. meliloti–infected roots were collected at 1 or 9 DAI. A minimum of 15 plants was used for each time point, and three biological experiments were performed.
Bioinformatic Analyses
To identify candidate genes related to cytokinin signaling in M. truncatula, BLASTN and BLASTX searches were performed on the TIGR database (http://www.tigr.org/tdb/tgi/mtgi/; release 8.0, January, 2005) for ESTs and on the NCBI database (http://www.ncbi.nlm.nih.gov/BLAST/) for available genomic sequences (October, 2005; see Supplemental Table 1 online). Arabidopsis thaliana sequences used were described by Hwang et al. (2002). Protein sequence comparison and unrooted tree visualization were done using ClustalW (http://www.ebi.ac.uk/clustalw/) and TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) software, respectively.
Gene Expression Analysis
Total RNAs were extracted from frozen roots using the RNeasy plant mini kit (Qiagen). First-strand cDNA was synthesized from 1.5 μg of total RNA using the Superscript II first-strand synthesis system (Invitrogen).
Primer design was performed using Primer3 software (available at http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Primers sequences used are listed in Supplemental Table 2 online. To check specificity of the designed primers, all amplicons were sequenced and BLAST searches were done against the TIGR database.
Real-time RT-PCR reactions were performed using the LightCycler FastStart DNA Master SYBR Green I kit (Roche) on a Roche LightCycler apparatus according to manufacturer's instructions. Cycling conditions were as follows: 95°C for 10 min, 50 cycles at 95°C for 5 s, 58°C for 5 s, and 72°C for 15 s. PCR amplification specificity was verified by a dissociation curve (55 to 95°C), and primer combinations showing a minimum amplification efficiency of 92% were used in real-time RT-PCR experiments (see Supplemental Table 2 online). A negative control without cDNA template was always included for each primer combination. Technical replicates on three independent syntheses of cDNA (derived from the same RNA sample) and three independent biological experiments were performed in all cases. Ratios were done with constitutive controls for gene expression to normalize the data between different biological conditions. Mt ACTIN11 gene was used for experiments involving hormonal treatments on roots or S. meliloti–infected roots and Mt EF1α gene for experiments including nodule samples (primers shown in Supplemental Table 2 online). The ratio value of the experimental control condition was set up to 1 as a reference to determine relative expression or induction factors.
For the few genes for which efficient primers for real-time RT-PCR could not be obtained, a semiquantitative RT-PCR was performed on several dilutions (1:3, 1:9, 1:27, and 1:81) of cDNAs using the primer pairs listed in Supplemental Table 2 online. Intensity of amplicon bands on ethidium bromide–stained agarose gels was quantified for the different cDNA dilutions using ImageQuant software (Molecular Dynamics), and ratio to Mt ACTIN11 (used to normalize the amount of template cDNA) was calculated.
Agrobacterium rhizogenes Root Transformation
Specific sequences of Mt CRE1, Mt HK2, Mt HK3, and GUS were amplified by PCR and cloned into the pFRN destination vector (derived from pFGC5941; NCBI accession number AY310901) using Gateway technology (Invitrogen). Primers used were as follows: Mt CRE1-FOR, 5′-GGTGATCATGAAGCCACTGA-3′, and Mt CRE1-REV, 5′-TCCATGAACGAAGCATCAAA-3′; Mt HK2-FOR, 5′-ACAAAACCACTCAGGGCAAG-3′, and Mt HK2-REV, 5′-GAAACAGGCGTCGAACTGAT-3′; Mt HK3-FOR, 5′-TGCTCCTGTCATCTTTGCAC-3′ and Mt HK3-REV, 5′-CCCACCAAGATACCCATCAG-3′; GUS-FOR, 5′-GGCCAGCGTATCGTGCTGCG-3′, and GUS-REV, 5′-GGTCGTGCACCATCAGCACG-3′.
The resulting constructs were introduced into Agrobacterium rhizogenes ARqua1 (Smr-derivative strain of A4T; Quandt et al., 1993) and used for M. truncatula root transformation. The transgenic roots were obtained after kanamycin selection (25 mg/L) as described by Boisson-Dernier et al. (2001). For all A. rhizogenes–transformed roots experiments, at least three biological experiments were performed. Specificity and efficiency of silencing was checked for each construct using real-time RT-PCR (see above) on several representative individual clones.
Treatments of A. rhizogenes–Transformed Roots
For root sensitivity assays to exogenous cytokinin or ethylene, A. rhizogenes–infected transgenic roots were selected 2 weeks on kanamycin, and composite plants were then transferred onto growth papers (Mega International) placed on Fahraeus medium and supplemented or not with 10−7 M BAP of the ethylene precursor ACC at 10−5 or 10−6 M. Position of root tips was marked at the time of transfer, and root growth from this point was measured after 6 d using Scion software (available at http://www.scioncorp.com/). Seven biological experiments were performed, and at least 150 independent transgenic roots per construct and per condition were analyzed.
For the induction of lateral roots, A. rhizogenes–infected transgenic roots were selected 2 weeks on kanamycin, and composite plants were then transferred onto growth papers (Mega International) on LRIM. LRIM is a rich medium (Soluplant 18.6.26 [Duclos International] and 0.9% Kalys HP696 Agar) supplemented with 3% sucrose to induce lateral root development (S. Gonzalez-Rizzo, unpublished results). The position of root tips was marked at the time of transfer. Primary root length and lateral root number were measured from this mark after 6 d using Scion software, and lateral root density was then calculated (lateral root number/centimeter of main root length). In these conditions, lateral root density is independent of the initial status (length and branching) of the transgenic roots. Three biological experiments were performed, and a minimum of 70 independent transgenic roots per construct and per condition was measured.
For nodulation assays, A. rhizogenes–infected transgenic roots were selected 2 weeks on kanamycin, and composite plants were then transferred onto growth papers (Mega International) on Fahraeus medium without nitrogen during 4 d. Thereafter, the transgenic roots were inoculated as described before with an S. meliloti 2011 strain. Nodulation was evaluated at 15 DAI. In a second set of experiments, composite plants obtained in vitro were transferred to the greenhouse (16 h light/8 h dark, 22°C, 60 to 70% hygrometry) on a perlite:sand 4:1 mixed substrate imbibed with Fahraeus without nitrogen liquid medium, and nodule number was determined at 30 DAI. Five biological experiments were performed, and a minimum of 120 independent transgenic roots per construct and per condition were analyzed.
Histochemical Assay
An S. meliloti 2011 derivative strain (GMI6526; Ardourel et al., 1994) carrying the pXLGD4 plasmid containing a ProHemA:LACZ transcriptional fusion was used to inoculate transgenic roots and follow the early stages of this infection process. Roots were stained for β-galactosidase activity as described by Ardourel et al. (1994). Observations were performed using a Reichert Polyvar microscope equipped with a Nikon digital DXM1200 camera.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL and TIGR data libraries under accession numbers provided in Supplemental Table 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Mt HK, Mt HP, and Mt RR Accession Numbers.
Supplemental Table 2. Primers Used in Real-Time and Semiquantitative RT-PCR Experiments.
Supplemental Figure 1. Unrooted Relationship Trees of Arabidopsis and M. truncatula Cytokinin Signaling Genes.
Supplemental Figure 2. Expression Analysis of Cytokinin Signaling Genes in Wild-Type versus A. rhizogenes–Transformed GUS RNAi Roots.
Supplemental Figure 3. Effect of Ethylene on Mt CRE1 RNAi Root Growth.
Supplemental Figure 4. Induction of Lateral Roots in M. truncatula Seedlings.
Supplemental Figure 5. Structure of Nodules Formed on A. rhizogenes–Transformed GUS RNAi or Mt CRE1 RNAi Roots.
Supplemental Figure 6. Real-Time RT-PCR Analysis of Nodulation and/or Cytokinin Early Response Genes in Response to Short-Term Cytokinin Treatment.
Supplemental Figure 7. Nodule Number in Mt RR1 RNAi and Control Roots.
Supplementary Material
Acknowledgments
We thank D. Barker, F. de Carvalho-Niebel, and A. Andriankaja for their advice concerning the A. rhizogenes root transformation protocol for M. truncatula cv Jemalong and for providing the Sm2011 nod− GMI5382 strain; A. Niebel for the GMI6526 S. meliloti strain; C. Gough for the dmi1-Y6, dmi2-TR25, and dmi3-TRV25 M. truncatula seeds; G. Oldroyd for nsp2-1 seeds; M. Denekamp for the pFRN plasmid; C. Capdevielle, K. Aftis, J. Plet, and C. Laffont for help on certain experiments; L. Troussard for sequence analysis; the Imaging and Cell Biology platform of IFR87 (FR-W2251) “La plante et son environnement” (supported by “Action de Soutien à la Technologie et la Recherche en Essonne, Conseil de l'Essonne, France”); and A. Maizel and D. Couch for careful reading of the manuscript. S.G.-R. was the recipient of a grant from Consejo Nacional de Ciencia y Tecnología, Mexico.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Martin Crespi (crespi@isv.cnrs-gif.fr).
Online version contains Web-only data.
References
- Aloni, R., Aloni, E., Langhans, M., and Ullrich, C.I. (2006). Role of cytokinin and auxin in shaping root architecture: Regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. (Lond.) 97 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardourel, M., Demont, N., Debelle, F., Maillet, F., de Billy, F., Prome, J.C., Denarie, J., and Truchet, G. (1994). Rhizobium meliloti lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6 1357–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, P., Ratet, P., Crespi, M., Schultze, M., and Kondorosi, A. (1996). Nod factors and cytokinins induce similar cortical cell division, amyloplast deposition and Msenod12A expression patterns in alfalfa roots. Plant J. 10 91–105. [Google Scholar]
- Bleecker, A.B., and Schaller, G.E. (1996). The mechanism of ethylene perception. Plant Physiol. 11 653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondon, F. (1964). Contribution à l'étude du développement de graminées fourragères: Ray-grass et dactyle. Rev. Gen. Bot. 71 293–381. [Google Scholar]
- Boisson-Dernier, A., Andriankaja, A., Chabaud, M., Niebel, A., Journet, E.P., Barker, D.G., and de Carvalho-Niebel, F. (2005). MtENOD11 gene activation during rhizobial infection and mycorrhizal arbuscule development requires a common AT-rich-containing regulatory sequence. Mol. Plant Microbe Interact. 18 1269–1276. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier, A., Chabaud, M., Garcia, F., Becard, G., Rosenberg, C., and Barker, D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14 695–700. [DOI] [PubMed] [Google Scholar]
- Brandstatter, I., and Kieber, J.J. (1998). Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright, L.J., Liang, Y., Mitchell, D.M., and Harris, J.M. (2005). The LATD gene of Medicago truncatula is required for both nodule and root development. Mol. Plant Microbe Interact. 18 521–532. [DOI] [PubMed] [Google Scholar]
- Cary, A.J., Liu, W., and Howell, S.H. (1995). Cytokinin action is coupled to ethylene in its effects on the inhibition of root and hypocotyl elongation in Arabidopsis thaliana seedlings. Plant Physiol. 107 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro, I., Beeckman, T., Graham, N., Bhalerao, R., Zhang, H., Casero, P., Sandberg, G., and Bennett, M.J. (2003). Dissecting Arabidopsis lateral root development. Trends Plant Sci. 8 165–171. [DOI] [PubMed] [Google Scholar]
- Catoira, R., Galera, C., de Billy, F., Penmetsa, R.V., Journet, E.P., Maillet, F., Rosenberg, C., Cook, D., Gough, C., and Dénarié, J. (2000). Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12 1647–1666.11006338 [Google Scholar]
- Che, P., Gingerich, D.J., Lall, S., and Howell, S.H. (2002). Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell 14 2771–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, J.B., and Long, S. (1994). Morphogenetic rescue of Rhizobium meliloti nodulation mutants by trans-zeatin secretion. Plant Cell 6 215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debelle, F., Rosenberg, C., Vasse, J., Maillet, F., Martinez, E., Denarie, J., and Truchet, G. (1986). Assignment of symbiotic developmental phenotypes to common and specific nodulation (nod) genetic loci of Rhizobium meliloti. J. Bacteriol. 168 1075–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Y., and Hirsch, A.M. (1998). Studying early nodulin gene ENOD40 expression and induction by nodulation factor and cytokinin in transgenic alfalfa. Plant Physiol. 116 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla, J.M., Martin, A.C., Solano, R., Rubio, V., Leyva, A., and Paz-Ares, J. (2002). Mutations at CRE1 impair cytokinin-induced repression of phosphate starvation responses in Arabidopsis. Plant J. 32 353–360. [DOI] [PubMed] [Google Scholar]
- Gamble, R.L., Coonfield, M.L., and Schaller, G.E. (1998). Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc. Natl. Acad. Sci. USA 95 7825–7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts, R., Fedorova, E., and Bisseling, T. (2005). Nod factor signaling genes and their function in the early stages of Rhizobium infection. Curr. Opin. Plant Biol. 8 346–352. [DOI] [PubMed] [Google Scholar]
- Heyl, A., and Schmulling, T.R. (2003). Cytokinin signal perception and transduction. Curr. Opin. Plant Biol. 6 480–488. [DOI] [PubMed] [Google Scholar]
- Higuchi, M., et al. (2004). In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. USA 8 8821–8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, A.M., and Fang, Y. (1994). Plant hormones and nodulation: What's the connection? Plant Mol. Biol. 26 5–9. [DOI] [PubMed] [Google Scholar]
- Hirsch, A.M., Fang, Y., Asad, S., and Kapulnik, Y. (1997). The role of phytohormones in plant-microbe symbioses. Plant Soil 194 171–184. [Google Scholar]
- Hirsch, A.M., and LaRue, T.A. (1997). Is the legume nodule the modified roots or stem or an organ sui generis? CRC Crit. Rev. Plant Sci. 16 361–392. [Google Scholar]
- Hwang, I., Chen, H.C., and Sheen, J. (2002). Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 129 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K., and Kakimoto, T. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409 1060–1063. [DOI] [PubMed] [Google Scholar]
- Jimenez-Zurdo, J.I., Frugier, F., Crespi, M.D., and Kondorosi, A. (2000). Expression profiles of 22 novel molecular markers for organogenetic pathways acting in alfalfa nodule development. Mol. Plant Microbe Interact. 13 96–106. [DOI] [PubMed] [Google Scholar]
- Kalo, P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308 1786–1789. [DOI] [PubMed] [Google Scholar]
- Kiba, T., Taniguchi, M., Imamura, A., Ueguchi, C., Mizuno, T., and Sugiyama, T. (1999). Differential expression of genes for response regulators in response to cytokinins and nitrate in Arabidopsis thaliana. Plant Cell Physiol. 40 767–771. [DOI] [PubMed] [Google Scholar]
- Lohar, D.P., Schaff, J.E., Laskey, J.G., Kieber, J.J., Bilyeu, K.D., and Bird, D.M. (2004). Cytokinins play opposite roles in lateral root formation, and nematode and rhizobial symbioses. Plant J. 38 203–214. [DOI] [PubMed] [Google Scholar]
- Lohar, D.P., Sharopova, N., Endre, G., Penuela, S., Samac, D., Town, C., Silverstein, K.A., and VandenBosch, K.A. (2006). Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol. 140 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorteau, M.A., Ferguson, B.J., and Guinel, F.C. (2001). Effects of cytokinin on ethylene production and nodulation in pea (Pisum sativum) cv. Sparkle. Physiol. Plant. 112 421–428. [DOI] [PubMed] [Google Scholar]
- Mahonen, A.A., Bonke, M., Kauppinen, L., Riikone, M., Benfey, P.N., and Helariutta, Y. (2000). A novel two components hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 14 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, A.C., del Pozo, J.C., Iglesias, J., Rubio, V., Solano, R., de La Pena, A., Leyva, A., and Paz-Ares, J. (2000). Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 24 559–567. [DOI] [PubMed] [Google Scholar]
- Mason, M.G., Mathews, D.E., Argyros, D.A., Maxwell, B.B., Kieber, J.J., Alonso, J.M., Ecker, J.R., and Schaller, G.E. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17 3007–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius, U., Charon, C., Rolfe, B.G., Kondorosi, A., and Crespi, M. (2000). Temporal and spatial order of events during the induction of cortical cell divisions in white clover by Rhizobium leguminosarum bv. trifolii inoculation or localized cytokinin addition. Mol. Plant Microbe Interact. 13 617–628. [DOI] [PubMed] [Google Scholar]
- Mok, D.W.S., and Mok, M.C. (2001). Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 89–118. [DOI] [PubMed] [Google Scholar]
- Nishimura, C., Ohashi, Y., Sato, S., Kato, T., Tabata, S., and Ueguchi, C. (2004). Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16 1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutman, P.S. (1948). Physiological studies on nodule formation. I. The relation between nodulation and lateral root formation in red clover. Ann. Bot. (Lond.) 7 81–96. [Google Scholar]
- Olah, B., Briere, C., Becard, G., Denarie, J., and Gough, C. (2005). Nod factors and a diffusible factor from arbuscular mycorrhizal fungi stimulate lateral root formation in Medicago truncatula via the DMI1/DMI2 signaling pathway. Plant J. 44 195–207. [DOI] [PubMed] [Google Scholar]
- Oldroyd, G.E., and Long, S.R. (2003). Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod actor signaling. Plant Physiol. 131 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt, H.J., Pühler, A., and Broer, I. (1993). Transgenic root nodules of Vicia hirsuta: A fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol. Plant Microbe Interact. 6 699–706. [Google Scholar]
- Rashotte, A.M., Carson, S.D., To, J.P., and Kieber, J.J. (2003). Expression profiling of cytokinin action in Arabidopsis. Plant Physiol. 132 1998–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte, A.M., Mason, M.G., Hutchison, C.E., Ferreira, F.J., Schaller, G.E., and Kieber, J.J. (2006). A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. USA 103 11081–11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redig, P., Schmulling, T., and Van Onckelen, H. (1996). Analysis of cytokinin metabolism in ipt transgenic tobacco by liquid chromatography-tandem mass spectrometry. Plant Physiol. 112 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler, M., Novak, O., Strnad, M., and Schmulling, T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser, L., Roussis, A., Stiller, J., and Stougaard, J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195. [DOI] [PubMed] [Google Scholar]
- Scheres, B., Di Laurenzio, L., Willemsen, V., Hauser, M.T., Janmaat, K., Weisbeek, P., and Benfey, P.N. (1995). Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121 53–62. [Google Scholar]
- Sheen, J. (2002). Phosphorelay and transcription control in cytokinin signal transduction. Science 296 1650–1652. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Miwa, K., Ishikawa, K., Yamada, H., Aiba, H., and Mizuno, T. (2001). The Arabidopsis sensor His-kinase, AHK4, can respond to cytokinins. Plant Cell Physiol. 42 107–113. [DOI] [PubMed] [Google Scholar]
- Swarup, R., Parry, G., Graham, N., Allen, T., and Bennett, M. (2002). Auxin cross-talk: Integration of signaling pathways to control plant development. Plant Mol. Biol. 49 411–426. [DOI] [PubMed] [Google Scholar]
- Taniguchi, M., Kiba, T., Sakakibara, H., Ueguchi, C., Mizuno, T., and Sugiyama, T. (1998). Expression of Arabidopsis response regulator homologs is induced by cytokinins and nitrate. FEBS Lett. 16 259–262. [DOI] [PubMed] [Google Scholar]
- Timmers, A.C., Auriac, M.C., and Truchet, G. (1999). Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126 3617–3628. [DOI] [PubMed] [Google Scholar]
- To, J.P., Haberer, G., Ferreira, F.J., Deruere, J., Mason, M.G., Schaller, G.E., Alonso, J.M., Ecker, J.R., and Kieber, J.J. (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truchet, G., Debelle, F., Vasse, J., Terzaghi, B., Garnerone, A.M., Rosenberg, C., Batut, J., Maillet, F., and Denarie, J. (1985). Identification of a Rhizobium meliloti pSym2011 region controlling the host specificity of root hair curling and nodulation. J. Bacteriol. 164 1200–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi, C., Koizumi, H., Suzuki, T., and Mizuno, T. (2001. a). Novel family of sensor histidine kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 42 231–235. [DOI] [PubMed] [Google Scholar]
- Ueguchi, C., Sato, S., Kato, T., and Tabata, S. (2001. b). The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol. 42 751–755. [DOI] [PubMed] [Google Scholar]
- Veereshlingam, H., Haynes, J.G., Penmetsa, R.V., Cook, D.R., Sherrier, D.J., and Dickstein, R. (2004). nip, a symbiotic Medicago truncatula mutant that forms root nodules with aberrant infection threads and plant defense-like response. Plant Physiol. 136 3692–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, T., Motyka, V., Laucou, V., Smets, R., Van Onckelen, H., and Schmulling, T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 2532–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, T., Motyka, V., Strnad, M., and Schmulling, T. (2001). Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 28 10487–10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wopereis, J., Pajuelo, E., Dazzo, F.B., Jiang, Q., Gresshoff, P.M., De Bruijn, F.J., Stougaard, J., and Szczyglowski, K. (2000). Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant J. 23 97–114. [DOI] [PubMed] [Google Scholar]
- Yamada, H., Suzuki, T., Terada, K., Takei, K., Ishikawa, K., Miwa, K., Yamashino, T., and Mizuno, T. (2001). The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 42 1017–1023. [DOI] [PubMed] [Google Scholar]
- Zhang, X.D., Letham, D.S., Zhang, R., and Higgins, T.J. (1996). Expression of the isopentenyl transferase gene is regulated by auxin in transgenic tobacco tissues. Transgenic Res. 5 57–65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.