Abstract
Key requirements for microbes to initiate and establish mutualistic symbiotic interactions with plants are evasion of potential host defense responses and strict control of microbial growth. Reactive oxygen species (ROS) produced by a specific NADPH oxidase isoform, NoxA, regulate hyphal growth in the mutualistic interaction between the fungal endophyte Epichloë festucae and its grass host Lolium perenne. Unlike mammalian systems, little is known about the fungal NADPH oxidase complex and its response to differentiation signals. We identify an E. festucae p67phox-like regulator, NoxR, dispensable in culture but essential in planta for the symbiotic interaction. Plants infected with a noxR deletion mutant show severe stunting and premature senescence, whereas hyphae in the meristematic tissues show increased branching leading to increased fungal colonization of pseudostem and leaf blade tissue. Inhibition of ROS production or overexpression of noxR recapitulates the hyperbranching phenotype in culture. NoxR interacts in vitro with the small GTP binding protein RacA and requires a functional RacA binding site to complement the noxR mutant and restore the wild-type plant interaction phenotype. These results show that NoxR is a key regulator of NoxA in symbiosis, where it acts together with RacA to spatially regulate ROS production and control hyphal branching and patterning.
INTRODUCTION
Generation of a burst of reactive oxygen species (ROS) catalyzed by NADPH oxidase (Nox) is an important defense mechanism for both mammals and plants against invading microbes (Doke, 1983; Reeves et al., 2002; Lambeth, 2004; Torres and Dangl, 2005). In phagocytes, activation of the membrane-associated NADPH oxidase (gp91phox/NOX2) to produce ROS in response to microbial or inflammation signals involves cytosolic recruitment of a heterotrimeric phagocyte oxidase complex (phox) together with the small GTP binding protein Rac2 (Diebold and Bokoch, 2001; Lambeth, 2004). The discovery of new functional members of the Nox family has led to the recognition that ROS production is a ubiquitous eukaryotic signaling system to control various differentiation processes, including cell proliferation, apoptosis, and hormone responses in animals (Suh et al., 1999; Lambeth et al., 2000; Lardy et al., 2005), programmed cell death and root hair growth in plants (Foreman et al., 2003; Carol et al., 2005), and sexual development and spore germination in filamentous fungi (Lara-Ortiz et al., 2003; Malagnac et al., 2004; Aguirre et al., 2005).
We recently demonstrated that fungal production of ROS is critical for maintaining a mutualistic symbiotic interaction between the clavicipitaceous fungal endophyte Epichloë festucae and its grass host Lolium perenne (Tanaka et al., 2006). In wild-type associations, E. festucae grows systemically in intercellular spaces of vegetative and reproductive tissues as infrequently branched hyphae (Tan et al., 2001; Christensen et al., 2002). Inactivation of a specific Nox isoform, NoxA, resulted in unregulated growth of the fungal endophyte in vegetative tissue and premature senescence of the host plant (Tanaka et al., 2006). Cerium reactive deposits indicative of H2O2 production were detected in the endophyte extracellular matrix (ECM) and at the interface between the ECM and host cell walls of meristematic tissue in wild-type associations but were depleted in noxA mutant associations. These results demonstrated that fungal production of ROS is critical in maintaining a mutualistic interaction between E. festucae and perennial ryegrass.
Unlike mammalian systems, very little is known about the composition of the fungal Nox complex and how it responds to differentiation signals. Production of superoxide by human gp91phox in a cell-free system proceeds in two discrete steps (Diebold and Bokoch, 2001). Electrons are initially transferred from NADPH to cytochrome b–associated flavin adenine dinucleotide (step 1), followed by transfer to the heme groups of cytochrome b (step 2), which can directly reduce oxygen. Human Rac2 is involved in both steps but requires interaction with p67phox in step 2. Therefore, human Rac2 and p67phox are both essential for catalytic activation of gp91phox. While homologs of Rac2 have been identified in several filamentous fungi (Weinzierl et al., 2002; Boyce et al., 2003; Chen and Dickman, 2004), there are no reports to date of fungal homologs of p67phox or any of the other gp91phox regulatory component genes, including p22phox, p40phox, and p47phox (Aguirre et al., 2005). The objective of this study was to identify other components of the Nox complex in E. festucae and to test whether they play a role in regulating NoxA-catalyzed production of ROS in planta and in maintenance of the symbiotic interaction between this fungus and its host grass perennial ryegrass.
RESULTS
Isolation of a p67Phox-Like Regulator from E. festucae
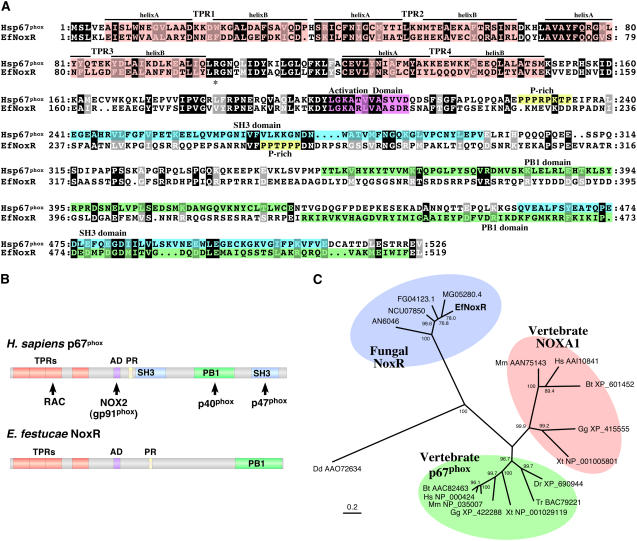
Analysis of Neurospora crassa, Aspergillus nidulans, Magnaporthe grisea, and Fusarium graminearum fungal genome databases for potential regulators of NoxA that shared homology to the human gp91phox regulatory components, including p22phox, p40phox, p47phox, and p67phox (Lambeth, 2004), failed to identify any obvious homologs of these regulators. However, in a BLASTP search of fungal genomes using p67phox as the query sequence, we did identify a gene encoding a protein with some of the motifs found in human p67phox (Figure 1A; see Supplemental Figure 1 online). A short DNA fragment (481 bp) to this p67phox-like regulatory gene was PCR amplified from E. festucae and used as a probe to isolate the full-length gene, which we have designated noxR (Nox regulator), from an E. festucae cosmid library (Tanaka et al., 2005). The N-terminal portion of NoxR is similar to human p67phox, containing four tetratricopeptide repeats (TPRs), a conserved 20–amino acid sequence between the third and fourth repeats, and a putative Nox activation domain (Figures 1A and 1B) (Han et al., 1998). By contrast, the C terminus of NoxR lacks the protein–protein interaction domains present in human p67phox, including the Scr homolog 3 and Phox and Bem1 (PB1) domains, which are required for interaction of p67phox with p47phox and p40phox, respectively (Lambeth, 2004). The absence of these domains in NoxR and the corresponding homologs identified in other fungi is consistent with the absence of p47phox and p40phox homologs in fungal genome databases. However, NoxR does possess a different type of PB1 domain in the C terminus of the protein (Figures 1A and 1B) (Noda et al., 2003). Differences in protein–protein interaction domain composition between human p67phox and NoxR suggest that fungal NoxR may have distinct regulatory partners for activation of the Nox from those found in animals. While vertebrates generally have two p67phox family members (p67phox and NOXA1), each with different Nox enzyme specificity (Lambeth, 2004), Dictyostelium discoideum has a single copy (Lardy et al., 2005), as do N. crassa, A. nidulans, M. grisea, F. graminearum, and E. festucae (Figure 1C).
Figure 1.
Comparison of the Domain Structure and Phylogenetic Relationship of Human p67phox with Fungal NoxR.
(A) Comparison of the deduced amino acid sequence of E. festucae NoxR (EfNoxR) with human p67phox (Hsp67phox). The TPR, Nox activation domain, proline-rich region (P-rich), Src homology 3 (SH3) domains, and PB1 domains are indicated.
(B) Structure of human p67phox showing domains that interact with other members of the Nox complex and domain structure of E. festucae NoxR.
(C) Phylogenetic relationships of vertebrate p67phox and Nox activator 1 (NOXA1) and fungal NoxR proteins. Ef, E. festucae; FG, F. graminearum; MG, M. grisea; AN, A. nidulans; NCU, N. crassa; Dd, D. discoideum; Dr, Danio rerio; Tr, Takifugu rubripes; Gg, Gallus gallus; Hs, Homo sapiens; Mm, Mus musculus; Xt, Xenopus tropicalis. Accession numbers are shown for the animal sequences and gene identifiers for the fungal sequences. The scale bar corresponds to 0.2 estimated amino acid substitutions per site. Numbers at the nodes indicate the percentage of 1000 bootstrap replicates that supported each labeled interior branch.
E. festucae noxR Is Essential for Maintaining a Mutualistic Interaction with Perennial Ryegrass
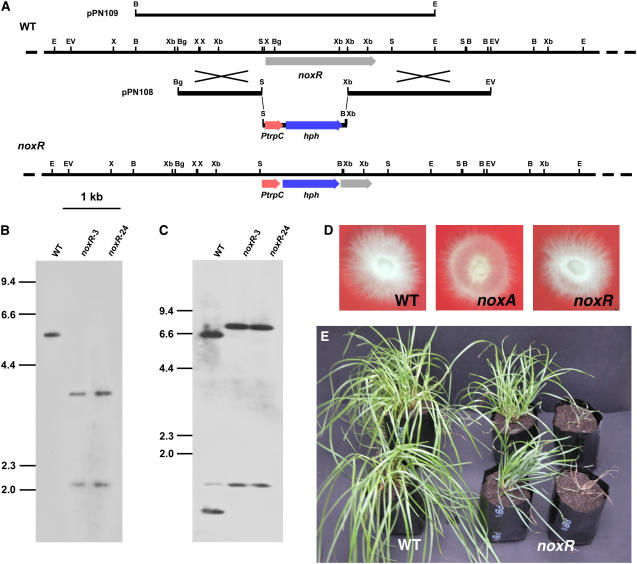
To investigate the biological role of noxR, a replacement construct, pPN108, was prepared, and a PCR-generated linear fragment of this plasmid (Figure 2A) was recombined into the genome of Fl1. PCR screening of 24 transformants identified six that had patterns consistent with targeted replacement events. DNA gel blot analysis of genomic digests of two transformants probed with the noxR replacement construct confirmed that both contained a replacement at the noxR locus (Figures 2B and 2C). In contrast with the noxA mutant that showed a reduction in formation of aerial hyphae in axenic culture (Tanaka et al., 2006), there is no detectable difference in colony morphology between the wild type and the noxR mutant (Figure 2D).
Figure 2.
Deletion of E. festucae noxR and Symbiotic Phenotype.
(A) Physical map of the E. festucae noxR wild-type genomic region, linear insert of replacement construct pPN108, E. festucae noxR mutant genomic region, and insert of complementation construct pPN109 showing restriction enzyme sites for BamHI (B), BglII (Bg), EcoRI (E), EcoRV (EV), SacI (S), XbaI (Xb), and XhoI (X).
(B) and (C) Autoradiographs of DNA gel blots of BamHI (B) or XhoI (C) genomic digests (2 μg) of E. festucae wild type and noxR deletion strains noxR-3 (PN2479) and noxR-24 (PN2497) probed with [32P]-labeled noxR replacement insert, amplified from pPN108 with primers pII99-2 and pII99-3.
(D) Colony morphology of E. festucae wild-type strain Fl1, noxA mutant strain PN2327, and noxR mutant strain PN2497 on potato dextrose (PD) agar medium after 7 d.
(E) Phenotype of perennial ryegrass plants infected with E. festucae wild type and noxR mutant strain PN2497.
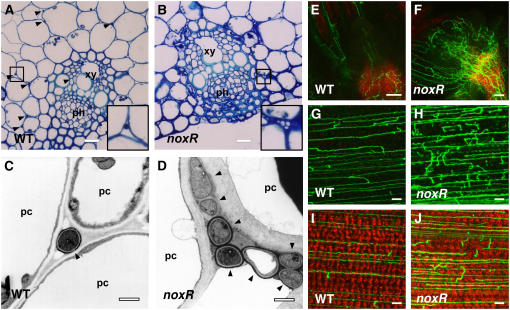
In contrast with the normal growth of host plants infected with wild-type strain Fl1, perennial ryegrass plants infected with the noxR mutant were stunted, had an increase in tiller number, and showed precocious senescence (Figure 2E). Light microscopy analysis of toluidine blue–stained cross sections of pseudostem tissue showed that large numbers of hyphae are present in older leaves of plants infected with the noxR mutant, with extensive colonization of the vascular bundles, including both xylem and phloem (Figure 3B). In leaves infected with wild-type Fl1, hyphae are much less frequent and rarely found in vascular bundles of both young and old plant tissue (Figure 3A). Wild-type hyphae were well stained compared with mutant hyphae (Figures 3A and 3B), indicating that mutant hyphae are relatively devoid of cytoplasm. Transmission electron microscopy analysis of hyphae in the outer leaf sheath confirmed that mutant hyphae were more abundant in the intercellular space than the wild type, were cytoplasmically less dense, and occasionally had large vacuoles (Figures 3C and 3D). Blebbing of the plant plasma membrane adjacent to the multiple hyphae found in mutant associations was frequently observed (Figure 3D).
Figure 3.
In Planta Phenotype of E. festucae noxR Mutant.
(A) and (B) Light micrographs of transverse sections of the outer leaves of perennial ryegrass infected with the wild type (A) or noxR mutant strain PN2497 (B) stained with toluidine blue. Endophyte hyphae are indicated by arrowheads in (A). Insets show higher magnification images of the endophyte hyphae indicated by squares in the main panels. xy, xylem; ph, phloem. Bars = 20 μm.
(C) and (D) Transmission electron micrographs of cross sections of E. festucae Fl1 wild-type (C) and noxR mutant strain PN2497 (D) hyphae (arrowheads) in the intercellular space of perennial ryegrass. pc, plant cell. Bars = 1 μm.
(E) to (J) Confocal depth series images of hyphal morphology of GFP expressing E. festucae wild-type strain PN2467 ([E], [G], and [I]) and noxR mutant strain PN2489 ([F], [H], and [J]) in perennial ryegrass meristematic tissue ([E] and [F]), pseudostem ([G] and [H]), and leaf blade ([I] and [J]). The red background in (E), (F), (I), and (J) is chlorophyll autofluorescence. Bars = 50 μm in (E) and (F) and 10 μm in (G) to (J).
To examine the differences in host colonization by wild-type strain Fl1 and the noxR mutant in living plant tissue, constitutively expressing green fluorescent protein (GFP) was introduced into cells of the noxR mutant by transforming protoplasts with pPN83. Perennial ryegrass symbiotic associations were established with a GFP-expressing noxR transformant and a previously isolated GFP-containing wild-type strain. Plants infected with the GFP-containing noxR mutant showed the typical stunted phenotype previously observed for the noxR mutant (Figure 2E), whereas associations with the GFP-containing wild-type strain had a phenotype similar to Fl1. Hyphal growth within these associations was examined by confocal microscopy. The most dramatic difference in phenotype between the noxR mutant and wild-type strain in planta is the increase in hyphal branching and biomass in the meristematic tissue at the base of the plant (Figures 3E and 3F). More extensive hyphal colonization of both pseudostem and leaf blade tissue was observed for noxR associations (Figures 3H and 3J) compared with wild-type associations (Figures 3G and 3I), reflecting the fact that hyphae in the meristematic tissue are the source of inoculum for the leaves that emerge from the axillary buds of the meristem.
E. festucae noxR Is Preferentially Expressed in Planta
Semiquantitative RT-PCR expression analysis showed that noxR is weakly expressed in axenic culture but strongly expressed in planta (Figure 4). As previously shown (Tanaka et al., 2006), noxA is also preferentially expressed in planta, whereas the steady state levels of noxB and racA (see later) transcripts found in planta are similar to the levels found in mycelial cultures, using cDNA dilutions of the latter that were normalized using β-tubulin (tubB) cDNA to take into account the low biomass of endophyte found in planta (Tanaka et al., 2005, 2006). We previously showed that no tubB product was obtained using cDNA from uninfected perennial ryegrass (Tanaka et al., 2005).
Figure 4.
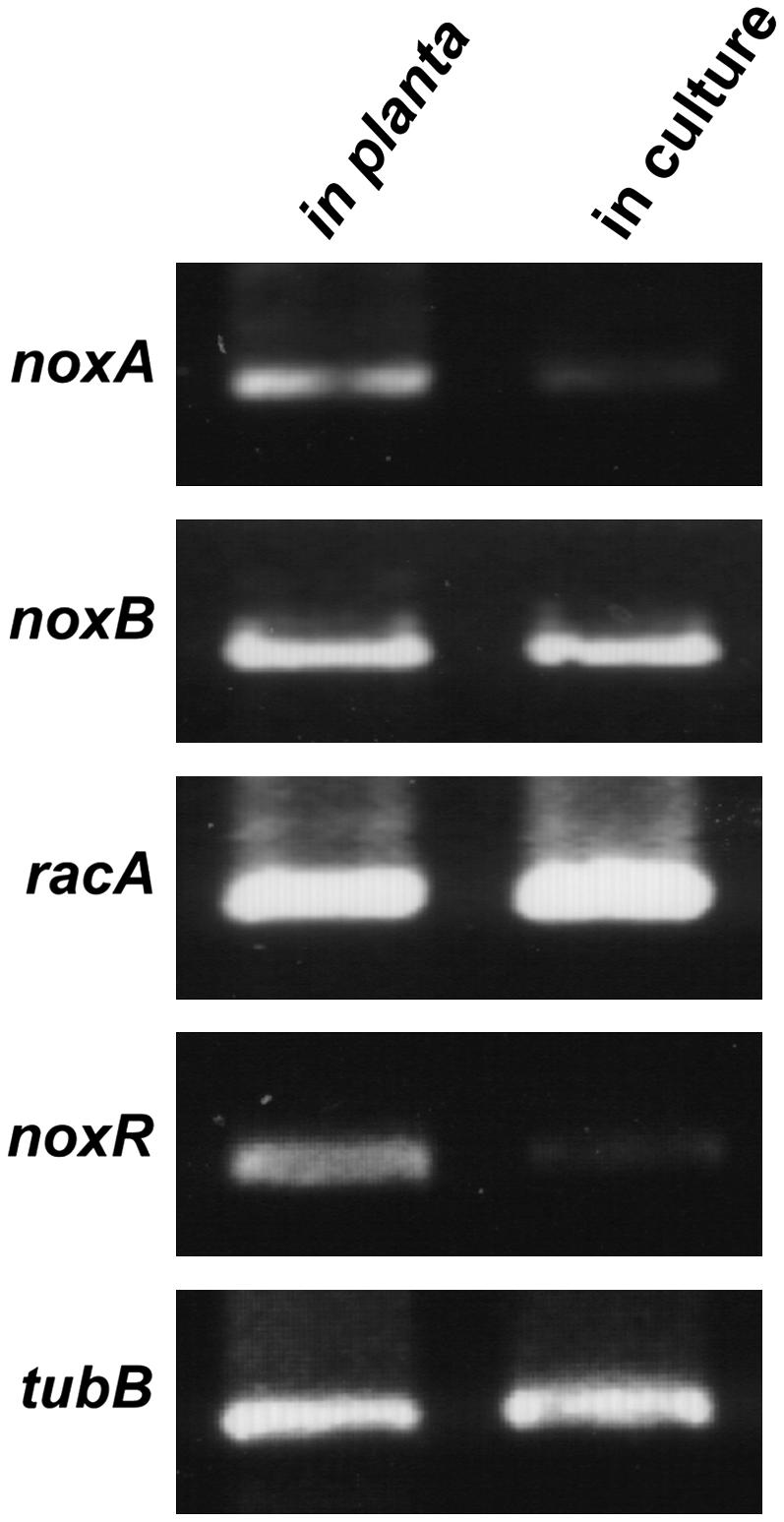
Expression of noxR Is Preferentially Induced in Planta.
Total RNA was isolated from perennial ryegrass pseudostems infected with E. festucae Fl1 (in planta) or from mycelia of Fl1 grown in axenic culture (in culture) and used for cDNA synthesis. RT-PCR was performed with primers specific for E. festucae noxA, noxB, racA, noxR, and tubB. To compensate for differences in endophyte biomass in planta versus in culture, a 1:80 dilution of Fl1 cDNA was used compared with undiluted pseudostem template.
NoxR Is Required for ROS Production in Planta but Is Dispensable in Culture
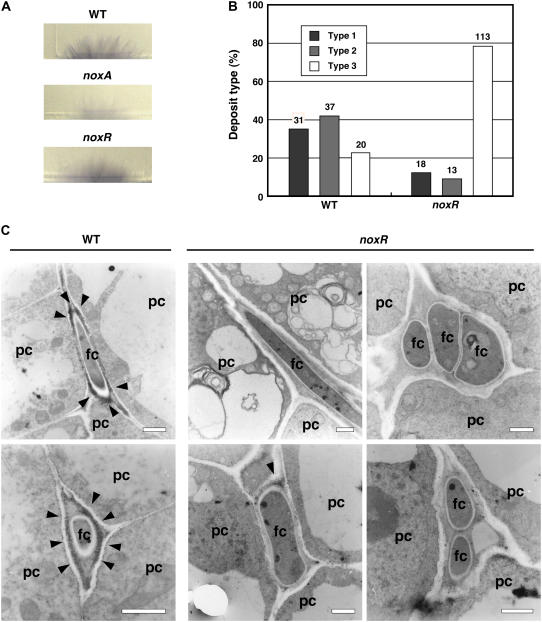
To test whether NoxR is required for activation of Nox protein(s), ROS production in the noxR mutant was examined both in axenic culture and in the host plant. Production of ROS (superoxide) in culture was assessed by reduction of nitroblue tetrazolium (NBT). NBT staining showed that ROS production of the noxR mutant was comparable to the wild type but was reduced in the noxA mutant (Figure 5A) (Tanaka et al., 2006), indicating that NoxR is dispensable for ROS production in culture. ROS production in the host plant was determined cytochemically using transmission electron microscopy to locate deposits of electron-dense cerium perhydroxides formed by the reaction of cerium ions with H2O2 (Shinogi et al., 2003). In host meristematic tissue infected with the wild type, cerium-reactive deposits were observed predominantly in the ECM surrounding the hyphal cell walls and at the interface between the ECM and the host cell walls (Figure 5C). By contrast, cerium-reactive deposits were rarely observed in meristematic tissue containing the noxR mutant (Figure 5C). Analysis of the distribution patterns of cerium-reactive deposits observed in meristematic tissues infected with either the wild type or the noxR mutant showed that 78% of the cells examined in tissue containing the noxR mutant were devoid of deposits, whereas 77% of the images examined for the Fl1 wild-type interaction showed some type of cerium-reactive deposit in the ECM and at the interface between the ECM and plant cell walls (Figure 5B). These results indicate that NoxR has been specifically recruited to activate NoxA in the symbiosis.
Figure 5.
NoxR Is an in Planta–Specific Regulator of Fungal ROS Production.
(A) Superoxide production by mycelia of E. festucae wild type and noxA and noxR mutant strains PN2327 and PN2497 grown on PD agar medium for 5 d and stained with 0.05% (w/v) NBT solution for 7 h.
(B) Distribution of different types of cerium chloride–reactive deposits in meristematic tissue of perennial ryegrass infected with E. festucae or noxR mutant. Type 1, cerium chloride–reactive deposits in the ECM of the endophyte; Type 2, cerium chloride–reactive deposits in the ECM of endophyte and around the plant host cell wall; Type 3, no cerium chloride–reactive deposits detected. The number of fungal cells of each particular type is given above each column.
(C) Transmission electron micrographs of H2O2 localization in the intercellular space of meristematic tissue of perennial ryegrass infected with E. festucae wild type or noxR mutant. Cerium chloride–reactive deposits indicative of H2O2 production are indicated by arrowheads. pc, plant cell; fc, fungal cell. Bars = 1 μm.
Depletion of ROS in Culture Promotes Hyperbranching of E. festucae
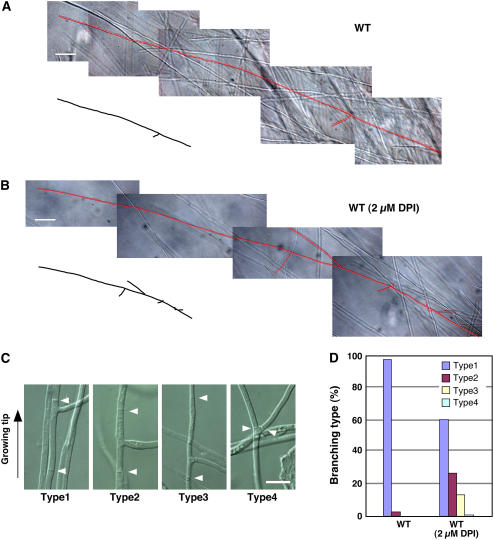
To determine whether depletion of Nox-induced ROS production in mycelia grown in axenic culture would affect hyphal growth and development, the wild-type strain was grown in the presence of diphenylene iodonium (DPI), a substrate inhibitor of Nox. This treatment reduced colony size, caused hyphae to become hyperbranched, and disrupted the highly organized pattern of branch initiation (Figures 6A and 6B). In wild-type fungal cells, lateral branching is almost always observed at the cell end proximal to the growing tip. However, the branching positions in the DPI-treated cells were more randomly distributed (Figures 6C and 6D). The smaller colony size of the wild-type strain treated with DPI indicates that ROS regulates apical tip growth and that reduced tip growth is a consequence of increased hyphal branching or vice versa. These results demonstrate that ROS depletion disrupts the spatial patterning of hyphal branching, a phenotype similar to that observed for the noxA and noxR mutants growing in the host plant.
Figure 6.
Inhibition of ROS Production in Mycelial Cultures Induces a Hyperbranched Phenotype.
(A) and (B) Light micrograph images captured with differential interference contrast (DIC) of E. festucae wild-type hyphae (highlighted in red) growing on PD agar medium without (A) and with (B) 2 μM DPI after 7 d. Bars = 50 μm.
(C) Light micrograph images captured with DIC of E. festucae wild type growing on PD agar in presence of 2 μM DPI showing four different patterns of lateral branching. Type 1, branch initiation close to apical tip proximal septum; Type 2, branch initiation approximately midway between septa; Type 3, branch initiation close to apical tip distal septum; Type 4, apical tip branching.
(D) Distribution of different branching types of E. festucae wild type grown in presence and absence of DPI.
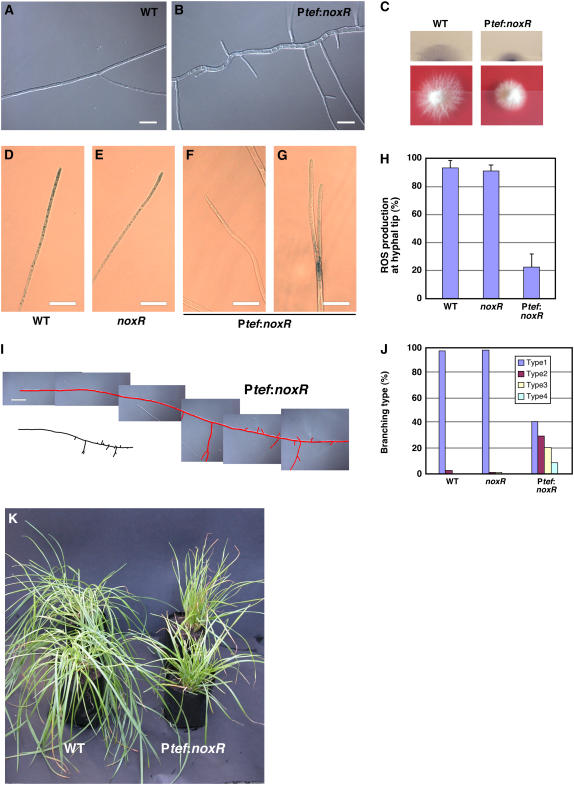
Overexpression of noxR Causes Hyphal Hyperbranching
To further elucidate the role of NoxR in fungal growth and branching, we generated an E. festucae noxR overexpressing strain, containing noxR under the control of a translation elongation factor (TEF) promoter (Vanden Wymelenberg et al., 1997). Ptef-noxR transformants of E. festucae formed smaller, more compact colonies than the wild type and had hyphae that were more frequently branched, phenotypes similar to that observed for the wild type treated with DPI (Figures 7A, 7B, and 7I). To test whether the spatial pattern of ROS production was altered in these transformants, colonies were stained with NBT and the ROS distribution pattern within the hyphae examined. For the wild type and the noxR mutant, ∼90% of the growing hyphal tips stained with NBT, indicating ROS production was very localized (Figures 7D, 7E, and 7H) (Tanaka et al., 2006). By contrast, just 20% of the hyphal tips examined in the Ptef-noxR transformants stained with NBT (Figures 7F, 7G and 7H), even though the intensity of NBT staining in the compact colony appeared to be comparable to the wild type (Figure 7C). Instead, some hyphae showed localized ROS production in the middle of the hyphal cell (Figure 7G). As with the DPI treatment, the branching positions in the Ptef-noxR transformants were more randomly distributed (Figure 7J). These results indicate that noxR overexpression compromised localized ROS production in culture-grown hyphae, suggesting that NoxR is involved in the spatial production of ROS, which in turn controls hyphal branching.
Figure 7.
Axenic Culture and Symbiotic Phenotype of a noxR-Overexpressing Strain of E. festucae.
(A) and (B) Light micrograph images captured with DIC of E. festucae wild type (A) and E. festucae Ptef:noxR strain PN2490 (B) growing on PD agar medium for 5 d. Bars = 20 μm.
(C) NBT staining and colony morphology of E. festucae wild type and E. festucae Ptef:noxR on PD agar medium after 7 d.
(D) to (G) Light micrographs showing localized production of ROS (superoxide), as detected by NBT staining, in the wild type (D), noxR mutant strain PN2497 (E), and E. festucae Ptef:noxR strain PN2490 ([F] and [G]). Bars = 20 μm.
(H) Frequency of ROS production at growing tip of hyphae from the wild type, noxR mutant, and noxR-overexpressing strains grown on PD agar medium for 7 d. Values are given as means with sd (n = 3 experiments, at least 84 hyphal tip/genotypes were analyzed for each experiment).
(I) Light micrograph image captured with DIC of E. festucae Ptef:noxR hyphae (highlighted in red) growing on PD agar medium after 7 d. Bar = 50 μm.
(J) Distribution of different branching types of E. festucae wild type, noxR mutant, and noxR-overexpressing strain grown on PD agar medium for 7 d.
(K) Phenotype of perennial ryegrass plants infected with E. festucae wild type and noxR-overexpressing strain (Ptef:noxR) 9 weeks after inoculation.
Plants infected with the Ptef-noxR transformants had the similar stunted phenotype observed for the noxR mutant (Figure 7K), and the hyphae within the leaves of these plants retained the hyperbranched phenotype observed for hyphae in culture. The similarity in plant phenotype observed for the noxR overexpression strain and the loss of function mutant may be due to a compromised localized production of ROS in both cases.
NoxA Activation in Planta Requires Interaction between NoxR and RacA
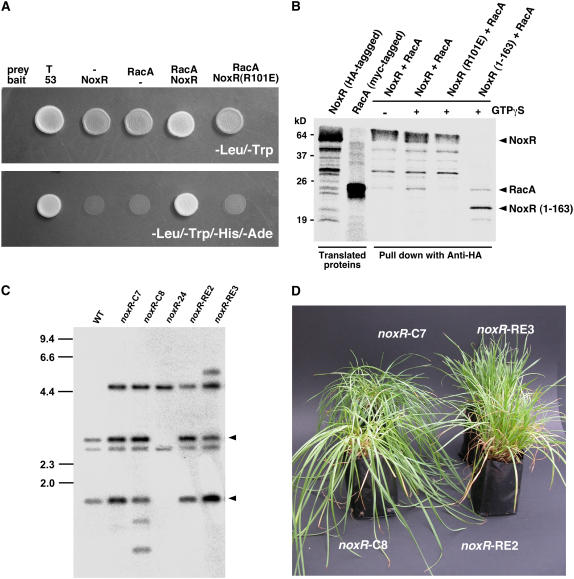
By analogy with the requirement in mammalian cells for p67phox to interact with the small GTP binding protein, Rac2, to activate gp91phox (Koga et al., 1999; Diebold and Bokoch, 2001), we isolated a Rac2 homolog from E. festucae, which we have designated racA (see Supplemental Figure 2 online). Using the yeast two-hybrid assay, an interaction between RacA and wild-type NoxR was demonstrated (Figure 8A). A single amino acid substitution in the predicted RacA binding site of NoxR (R101E) abolished this interaction, a result consistent with the requirement of Arg-102 of the human p67phox for interaction with human Rac2 (Koga et al., 1999). To confirm a physical interaction between NoxR and RacA, constructs were prepared in which these proteins were tagged with HA and Myc, respectively, and expressed in an in vitro translation system (Figure 8B). The multiple lower molecular weight bands observed for the NoxR in vitro translation product could not be inhibited by proteinase inhibitors and correlated with the presence of the C-terminal portion of this protein, suggesting that this portion of the protein is intrinsically unstable in an in vitro translation system. A protein of the same molecular mass as RacA was coimmunoprecipitated with either wild-type NoxR or a C-terminal truncation of NoxR (1-163) using an anti-HA antibody (Figure 8B). The interaction between RacA and NoxR was GTP dependent and was abolished by the NoxR R101E mutation.
Figure 8.
NoxR Requires RacA for Functional Activity.
(A) Yeast two-hybrid assay of the interaction between E. festucae NoxR and RacA. Yeast strain AH109 was transformed with prey and bait vectors as indicated and plated on SD medium lacking Leu and Trp (top panel; -Leu/-Trp) or lacking Leu, Trp, His, and adenine (bottom panel; -Leu/-Trp/-His/-Ade). Growth on the latter indicates an interaction between bait and prey. T, SV40 large T-antigen; 53, murine p53.
(B) In vitro pull-down assay showing interaction between E. festucae NoxR and RacA. Autoradiograph of [35S]-labeled in vitro cell-free translation products using noxR and racA plasmids as templates and proteins pulled down with anti-HA, with or without 100 μM GTPγS, following incubation of NoxR and RacA in vitro translation products. The molecular mass of protein standards, in kilodaltons, is shown at the left of the figure.
(C) Autoradiograph of DNA gel blot of BglII-EcoRI genomic digest (2 μg) of E. festucae wild type, noxR mutant complemented with wild-type noxR plasmid pPN109, strains PN2483 (noxR-C7) and PN2484 (noxR-C8), noxR deletion strain PN2497 (noxR-24), noxR mutant not complemented with plasmid pPN110 (noxR with the R101E mutation), and strains PN2485 (noxR-RE2) and PN2486 (noxR-RE3) probed with [32P]-labeled noxR replacement insert amplified from pPN108 with primers pII99-2 and pII99-3. Arrowheads identify DNA fragments derived from the noxR complementation construct. Sizes in kilobases of HindIII-digested λ DNA markers are indicated at left.
(D) Phenotype of perennial ryegrass plants infected with E. festucae noxR mutant transformed with either a wild-type copy of noxR, strains PN2483 (noxR-C7) and PN2484 (noxR-C8), or with noxR containing an R101E mutation, strains PN2485 (noxR-RE3) and PN2486 (noxR-RE2).
To test the biological significance of RacA binding for NoxR to function in vivo, complementation tests were performed by introducing noxR mutant (R101E) and wild-type constructs into the noxR mutant background and inoculating the resulting transformants into perennial ryegrass. DNA gel blot analysis of genomic digests of these transformants probed with the noxR replacement construct confirmed that the appropriate plasmids had integrated into the genome of the noxR mutant (Figure 8C). All five transformants tested that contained the wild-type gene were restored to the wild-type plant interaction phenotype, whereas the four transformants tested that contained the R101E construct had a symbiotic phenotype identical to the noxR mutant (Figures 2 and 8D). These results indicate that RacA binding to NoxR is necessary for NoxR to function in the host plant. These data indicate both NoxR and RacA are required to activate NoxA in the host plant, in an analogous fashion to the requirement for p67phox and Rac2 for activation of the human gp91phox in neutrophils (Diebold and Bokoch, 2001).
DISCUSSION
In mammalian phagocytic cells, activation of gp91phox (NOX2) is triggered by cellular signaling events involving lipid metabolism and protein phosphorylation that result in the recruitment and assembly of cytosolic p40phox, p47phox, p67phox, and Rac-GTP with the membrane-associated gp91phox and p22phox to form a ROS-producing complex (Vignais, 2002; Lambeth, 2004). We report here that filamentous fungi have a single homolog of p67phox, designated NoxR, which in the E. festucae–perennial ryegrass mutualistic interaction has been recruited to control hyphal branching and growth in plant meristematic tissue. Disruption of the noxR leads to a breakdown in the mutualistic interaction between E. festucae and perennial ryegrass, leading to severe stunting and premature senescence of the host, a phenotype similar to that recently reported for loss-of-function mutations in the NoxA isoform of the Nox complex (Tanaka et al., 2006).
The N terminus of the fungal NoxR is similar to mammalian p67phox, containing putative domains for interaction with fungal homologs of the human Rac2 and gp91phox, respectively (Han et al., 1998). By contrast, the C terminus of NoxR is not conserved with p67phox and lacks the protein–protein interaction domains required for interaction with p40phox and p47phox (Lambeth, 2004). The absence of these domains is consistent with the inability to detect homologs of these proteins in fungal genome databases. However, NoxR does possess an alternative type of PB1 protein–protein interaction domain, suggesting that fungal NoxR may have distinct regulatory partners for the activation of the Nox from those found in vertebrates.
While vertebrates generally have two p67phox family members, p67phox and NOXA1, each with different Nox enzyme specificity (Lambeth, 2004), just a single copy of NoxR was identified in each of the filamentous fungal genomes surveyed, including the basidiomycetes Phanerochaete chrysosporium and Coprinus cinereus and the ascomycetes N. crassa, A. nidulans, M. grisea, F. graminearum, and E. festucae. A single copy of a p67phox homolog is also found in the genome of D. discoideum (Lardy et al., 2005), an organism that uses superoxide signaling for multicellular development (Bloomfield and Pears, 2003). With the exception of A. nidulans, at least two isoforms of Nox are found in all these organisms (Lalucque and Silar, 2003; Aguirre et al., 2005). Homologs of p67phox or NoxR are absent from the genomes of the basidiomycetes Cryptococcus neoformans and Ustilago maydis and the hemiascomycete yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, fungi that lack Nox proteins (Lalucque and Silar, 2003; Aguirre et al., 2005). The absence of NoxR from these fungi provides further support for the theory that the Nox complex has evolved in organisms that differentiate multicellular structures (Lalucque and Silar, 2003). No homologs of NoxR or p67phox were found in the genomes of insects (Drosophila melanogaster and Anopheles gambiae), nematodes (Caenorhabditis elegans), and plants (Arabidopsis thaliana and Oryza sativa), organisms whose Nox proteins contain calcium binding domains (EF-hand motif) in the N terminus of the protein, implying a different mechanism of activation.
Inactivation of E. festucae noxR changes the interaction of the endophyte with its host from mutualistic to antagonistic. In particular, there is a dramatic increase in the frequency of hyphal branching in meristematic tissues of noxR mutant associations compared with wild-type associations. This leads to an increase in biomass in these tissues and increased fungal colonization of the pseudostem and leaf blade tissues. Microscopy observations suggest that epichloë endophytes grow predominantly by tip growth within the intercellular spaces of the leaf and axillary bud primordia (Tan et al., 2001). After colonization of the leaf expansion zone, the hyphae appear to become physically attached to the cell walls of the expanding leaf cells where they further elongate by intercalary extension (Christensen et al., 2002) (M.J. Christensen, unpublished data). This model predicts that hyphal cells that leave the leaf expansion zone will be fully mature and that the number of hyphae found in the pseudostem and leaf blade tissue will reflect the pattern of endophyte growth in the host meristematic tissue. Therefore, signaling within the meristematic tissue will be vital in determining the outcome of the interaction between epichloë endophytes and their grass hosts. Of interest is the recent finding that plant sesquiterpenes of the strigolactone group induce hyphal branching of arbuscular mycorrhizal fungi during host recognition and colonization (Akiyama et al., 2005). Using cerium chloride to detect H2O2 accumulation, reactive deposits were found in the endophyte ECM and at the interface of the ECM and the host cell walls of meristematic tissue infected with the wild-type strain but were absent from tissues infected with the noxR mutant. A similar observation was made for associations containing the noxA mutant (Tanaka et al., 2006), suggesting that the steady state levels of ROS in the meristematic tissue is an important determinant of the pattern of endophyte growth in the host and the nature of the interaction.
While NoxR is critical for controlling ROS production and hyphal growth in planta, in contrast with NoxA, which is required for both in planta growth and aerial hyphal development (Tanaka et al., 2006), NoxR appears to be dispensable in culture. This suggests that NoxA and other Nox enzyme isoforms have alternative activation mechanisms under these physiological conditions. A precedent for this is the activation of gp91phox in a semirecombinant cell-free system by the calcium binding myeloid-related proteins MRP8 and MRP14, without the need for p67phox, a reaction that involves a Ca2+-dependent specific interaction of these proteins with p22phox (Berthier et al., 2003). The activation of human NOX5, which contains calcium binding EF-hand motifs, also occurs independently of the heterotrimeric cytosolic phox proteins (Kamiguti et al., 2005). In plants, the respiratory burst oxidase homologs require Rac-like GTPases for ROS production (Kawasaki et al., 1999; Park et al., 2004), but the absence of p67phox-like proteins and other regulatory components of the neutrophil Nox imply a different mechanism of activation. The presence of distinctive Ca2+ binding EF-hand motifs in all these proteins suggests that the activation is Ca2+ dependent. Taken together, these results highlight a diverse range of mechanisms for activating Nox complexes, indicating that fungal Nox complexes could be activated in culture, independently of a p67phox-like regulator.
The requirement for NoxR in planta but its dispensability in culture strongly suggests that NoxR is a symbiosis-specific regulator of ROS production. The preferential expression of noxR in planta supports this conclusion. By contrast, noxA is required both in culture and in planta, even though it is preferentially expressed in planta (Tanaka et al., 2006). In the mammalian cell-free system, ROS production by gp91phox requires both p67phox and Rac2 (Diebold and Bokoch, 2001). The demonstration here that E. festucae RacA is necessary for NoxR activation in a plant symbiosis indicates that fungi have evolved a mechanism for activation of the Nox complex, analogous to the gp91phox-p67phox-Rac2 system that has evolved in professional phagocytes.
To gain further insight into the role of NoxA and NoxR in regulating hyphal branching in planta, two experiments were performed in culture that recapitulated the hyperbranching phenotype observed for the noxR mutant in the plant. Depletion of Nox-induced ROS production by growing cultures of the wild-type strain in the presence of DPI disrupted the highly organized pattern of hyphal branch initiation and induced hyperbranching of the hyphae. The same developmental defects were observed when noxR, under the control of a TEF promoter, was overexpressed in a wild-type background. Taken together, these results suggest that the wild-type pattern of hyphal branch initiation is dependent on the spatial production of ROS, which is controlled by NoxR.
While the process of branch formation in filamentous fungi is relatively poorly understood at the molecular level, it is likely that branch formation involves polarity establishment and maintenance steps similar to those used in germ tube emergence (Momany, 2002). AspB, a septin, and SepA, a formin, have both been implicated in branch initiation in A. nidulans (Sharpless and Harris, 2002; Westfall and Momany, 2002). Regulators involved in polar tip growth have also been shown to play important roles in branch formation (Harris and Momany, 2004). The results described here would suggest that bursts of ROS are important in maintaining normal polar tip growth and branch initiation in E. festucae, but how ROS is involved in the release of subapical cells from cell cycle arrest remains to be determined.
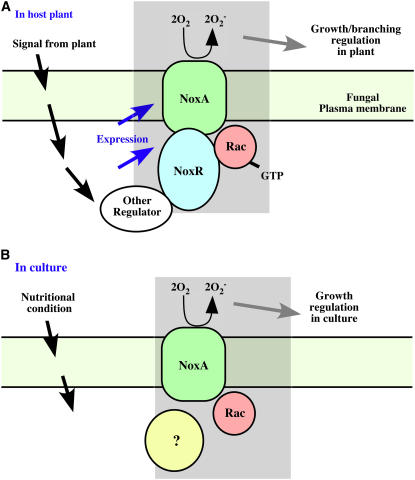
Maintenance of normal polar tip growth and branching of E. festucae in culture is proposed to involve bursts of ROS catalyzed by Nox following recruitment of RacA and other proposed Nox regulators in response to nutritional cues (Figure 9B). Growth of E. festucae in the symbiosis is proposed to involve recruitment of NoxR and RacA to the plasma membrane and activation of NoxA in response to plant signals (Figure 9A). The presence of a unique protein–protein interaction domain in the C terminus of NoxR would suggest that unique fungal regulators of NoxR remain to be identified.
Figure 9.
Models for Regulation of ROS Production by Nox Complex in Planta and in Culture.
(A) Model for regulation of NoxA-catalyzed ROS production in planta by recruitment of RacA and NoxR to the fungal plasma membrane in response to a signal from the host. NoxR is proposed to interact with RacA via the TPR domains and with as yet unidentified cytosolic regulatory components via the PB1 domain.
(B) Model for regulation of NoxA-catalyzed ROS production in culture by recruitment of RacA and other unidentified proteins to the fungal plasma membrane in response to nutritional signals.
In summary, we have shown here that NoxR is a key regulator of NoxA in the symbiosis, where it acts together with RacA to spatially regulate ROS production and control hyphal growth and patterning. That NoxR is dispensable for hyphal branching and growth in axenic culture suggests that an alternative regulator(s) of NoxA is required under these physiological conditions, highlighting how a single enzyme can be adapted for different growth purposes, depending on the physiological environment of the fungus. The use of NoxR as a regulator of ROS production in planta to restrict hyphal growth is a unique adaptation of the epichloë endophytic fungi to maintain the mutualistic state. It will be of considerable interest to determine how widespread this system has been adopted to control growth and development of other symbiotic fungi, including the biotrophic pathogens, and what role it plays in saprophytic fungi.
METHODS
Biological Material and Growth Conditions
Cultures of Epichloë festucae (see Supplemental Table 1 online) were grown on 2.4% PD agar or in 2.4% PD broth at 22°C. Inoculation of epichloë endophyte-free seedlings of perennial ryegrass (Lolium perenne) with E. festucae was performed by the method of Latch and Christensen (1985), and inoculated plants were grown as previously described (Tanaka et al., 2005).
DNA Preparations, Hybridizations, and PCR
Fungal genomic DNA was isolated from mycelium as previously described (Byrd et al., 1990) or using an Extract-N-Amp plant PCR kit (Sigma-Aldrich) according to the manufacturer's instructions. Genomic digests were transferred to positively charged nylon membranes (Roche) by capillary transfer and fixed by UV cross-linking in a UV cross-linker (Cex-800; Ultra-Lum) at 254 nm for 2 min. Filters were probed with [α-32P]dCTP-labeled probes (3000 Ci/mmol; Amersham Biosciences). Probe labeling and purification and hybridization conditions were performed as previously described (Young et al., 1998). Standard PCR amplifications of genomic and plasmid DNA templates were performed as previously described (Tanaka et al., 2005). Sequences of PCR primers used in this study are provided in Supplemental Table 2 online.
Preparation of Deletion, Complementation, and Overexpression Constructs
Cosmids pPN105 (noxR) and pPN91 (racA) were isolated from an Fl1 cosmid library (Tanaka et al., 2005) using probes amplified from genomic DNA with degenerate primer sets P67-f3 and P67-r3, and FRac1f and FRac1r, respectively. Primers were designed to conserved DNA regions identified from an alignment of noxR or racA (rac1) sequences from Fusarium graminearum (FG04123.1 and FG08857.1), Magnaporthe grisea (MG05280.4 and MG02731.4), Neurospora crassa (NCU07850.1 and NCU02160.1), and Aspergillus nidulans (AN6046.2 and AN4743.2). Since pPN91 contained a partial open reading frame of racA, the 3′ end of the racA cDNA was amplified by 3′ rapid amplification of cDNA ends with primer sets EfRac1e and oligo(dT) for primary amplification and EfRac1e and 3′UP for nested PCR. Plasmids pPN106 and pPN107, which contain the noxR open reading frame, were prepared by cloning 6.6-kb EcoRI and 6.7-kb XhoI fragments from pPN105 into pBlueScript II KS+. The noxR replacement vector pPN108 was prepared by sequentially ligating a 1.5-kb BglII-SacI noxR 5′ fragment from pPN106 and a 2.5-kb XbaI-EcoRV noxR 3′ fragment from pPN107 into pSF15. The noxR complementation construct pPN109 was prepared by ligating a 2.0-kb BamHI-SphI fragment and a 3.2-kb SphI-EcoRI fragment from pPN106 into pII99 digested with BglII and EcoRI. The noxR construct pPN110 containing the R101E mutation was prepared by sequentially ligating into pII99, 0.3-kb BglII-XhoI and 1.1-kb XhoI-SalI fragments, followed by ligation of 1.4-kb BamHI-PstI, 1.0-kb PstI-BglII, and 1.4-kb SalI-EcoRI fragments isolated from pPN106. The first two fragments were prepared by digesting PCR products amplified with primer sets Efp67-F7 and Efp67RE-MR2, and XhI-Efp67RE-MF4 and p67-R7, respectively. The noxR overexpression construct pPN114 was prepared by ligating a BamHI-EcoRI digest of a PCR product amplified from pPN112 using primer set BI-p67-F and Efp67-EI-R into pPN94. Plasmid pSF14.10 (3.9 kb) was prepared by cloning a 1.4-kb HindIII fragment containing hph under the control of the trpC promoter from pCB1004 (Carroll et al., 1994) into the HpaI site of pSP72 (Promega). Plasmid pPN94 (5.2 kb) was prepared by sequentially ligating a 0.8-kb SalI-XbaI fragment of TEF promoter from pTEFEGFP (Vanden Wymelenberg et al., 1997) and a 0.6-kb EcoRI-BglII fragment of trpC terminator from pII99 (Namiki et al., 2001) into pSF14.10. The TEF promoter fragment was prepared by digesting a PCR product amplified with primer set TEFsl2 and TEFxb2 using pTEFEGFP as template. The fragment of the trpC terminator was prepared by digesting a PCR product amplified with primer set TnotI and TBg using pII99 as template.
Preparation of RNA and RT-PCR Analysis
Total RNA was isolated from frozen mycelium or perennial ryegrass pseudostem tissue using TRIzol reagent (Invitrogen) and reverse transcribed as previously described (Tanaka et al., 2005). Gene-specific amplifications of cDNA were performed under the same conditions as previously described (Tanaka et al., 2005). The gene-specific primer (see Supplemental Table 2 online) combinations used for expression analysis were as follows: T1.1 and T1.2 (tubB), noxAf2 and noxAr (noxA), nox2e and nox2j (noxB), Efrac1f and Efrac1r (racA), and Efp67-F2 and Efp67-R2 (noxR).
Yeast Two-Hybrid and Pull-Down Assays
The coding sequences of noxR and racA were amplified with primer sets EI-Efp67-F and Efp67-BI-R, and EI-EfRac1-F and EfRac1-BI-R, using cDNA derived from RNA isolated from E. festucae mycelium and ligated into pGADT7 and pGBKT7 (Clontech), respectively, to generate pPN112 and pPN111. To introduce the R101E mutation into NoxR, 5′ and 3′ portions of noxR were amplified from pPN112 with primer sets EI-Efp67-F and Efp67RE-MR2, and XhI-Efp67RE-MF4 and Efp67-BI-R digested with EcoRI-XhoI and XhoI-BamHI, and ligated into pGADT7 to generate pPN113. Plasmid pPN115, expressing a truncated NoxR protein (1-165), was prepared by ligating an EcoRI-BamHI digest of a PCR product amplified from pPN112 using the primer set EI-Efp67-F and Efp67-BI-MR4 into pGADT7. Transformation of yeast (Saccharomyces cerevisiae strain AH109) and yeast two-hybrid assays were performed according to the manufacturer's instructions (Matchmaker Two-Hybrid System3; Clontech).
Myc-tagged RacA, HA-tagged NoxR, and NoxR derivatives were translated in vitro using TNT coupled reticulocyte lysate systems (Promega) with Redivue l-[35S]methionine (Amersham Biosciences) and plasmids pPN111 (RacA), pPN113 (NoxR), pPN113 (NoxR R101E), or pPN115 (NoxR 1-165) according to the manufacturer's instructions. Ten microliters of each of the translated radiolabeled myc-RacA, HA-NoxR, or their derivatives in rabbit reticulocyte lysate were mixed with or without 100 μM GTPγS and subjected to pull-down assay according to the manufacturer's instructions (Matchmaker Co-IP kit; Clontech). After separation of the immunoprecipitated proteins by SDS-PAGE, the gel was fixed with gel fixation solution (10% acetic acid and 20% methanol) for 40 min, treated with Amplify fluorographic reagent (Amersham Biosciences) for 20 min, dried, and exposed to an imaging plate overnight (Fuji). Signals were detected with an image scanner (FLA-5000; Fuji).
E. festucae Transformation
Protoplasts of E. festucae were prepared as previously described (Young et al., 1998, 2005). Protoplasts were transformed with 5 μg of either circular (pPN109, pPN110, and pPN114) or linear PCR product amplified from pPN108 with primer set pII99-2 and pII99-3 or cotransformed with 5 μg of circular pII99 and 10 μg of pPN83 (Tanaka et al., 2006) using a previously described method (Itoh et al., 1994). Putative noxR replacements were initially screened by PCR using primers Efp67-F1 and Efp67-R1, which are within the deleted noxR region. Transformants were selected on YPS medium containing either hygromycin (150 μg/mL) or geneticin (200 μg/mL) (Tanaka et al., 2006). Transformants expressing enhanced GFP were selected under UV light using a BX51 stereomicroscope (Olympus).
DNA Sequencing and Bioinformatics
DNA fragments were sequenced by the dideoxynucleotide chain termination method using Big-Dye (version 3) chemistry (Applied Biosystems). Products were separated on an ABI 3730 analyzer (Applied Biosystems).
Sequence data were assembled into contigs with SEQUENCHER version 4.1 (Gene Codes) and analyzed and annotated as previously described (Tanaka et al., 2005).
Microscopy
Small leaf pieces, 1- to 2-mm thick, from endophyte-infected plant tissues were fixed in 3% glutaraldehyde and 2% formaldehyde in 0.1 M phosphate buffer, pH 7.2, and then prepared for light microscopy and transmission electron microscopy as previously described (Spiers and Hopcroft, 1993). For light microscopy, the sections were stained with toluidine blue as previously described (Christensen et al., 2002). A Philips 201C transmission electron microscope was used to examine hyphal structure.
For H2O2 detection, a modified cytochemical method was used (Briggs et al., 1975; Bestwick et al., 1997). Meristematic or pseudostem tissue (2- to 3-mm thick) was vacuum-infiltrated in 5 mM CeCl3 solution buffered with 50 mM MOPS, pH 7.2, or MOPS at room temperature for 1 min, and incubated at room temperature for 1 h. The samples were then prefixed in 2.5% glutaraldehyde buffered with 0.1 M cacodylate buffer, pH 7.2, at 4°C overnight. After prefixation, the specimens were prepared for transmission electron microscopy as previously described (Shinogi et al., 2001) and observed with a Hitachi 7100 transmission electron microscope.
Production of superoxide was detected with NBT using a method modified from that previously described (Shinogi et al., 2003). Cultures of E. festucae were grown on slides covered with a thin layer of PD agar for 10 d. Mycelia were incubated in 0.05 M sodium phosphate buffer, pH 7.5, containing 0.05% (w/v) NBT (Sigma-Aldrich). After 5 h of incubation with NBT, the cultures were fixed in ethanol to stop the reaction. The stained samples were either photographed or examined with a compound microscope at ×100 and ×400 magnification.
Confocal laser scanning fluorescence images were recorded on a TCS 4D confocal system (Leica Microsystems) with a ×40 numerical aperture, 1.0 oil-immersion lens. A krypton-argon laser was used as the excitation source at 488 nm, and GFP fluorescence was recorded between 515 and 545 nm. Images of GFP fluorescence shown in the figures are a depth series of 28 optical sections taken at 1.5 μm. The images were stored as TIF files and processed with Canvas 10 software (ACD Systems International).
Phylogenetic Analysis
The encoded protein sequences of vertebrate p67phox, NOXA1, and fungal NoxR genes were aligned (see Supplemental Figure 3 online) using the ClustalX program (version 1.8; Thompson et al., 1997) with default settings. Phylogenetic analysis was conducted using the neighbor-joining method (Saitou and Nei, 1987) with the PHYLIP software package (version 3.63; http://evolution.genetics.washington.edu/phylip.html) with 1000 bootstrap trials. The dendrogram was displayed graphically using the TreeView program (version 1.6.6).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL/DDBJ databases under accession numbers AB260937 (E. festucae noxR) and AB260938 (E. festucae racA cDNA).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of Deduced Fungal NoxR Sequences with Vertebrate p67phox and NOXA1 Sequences.
Supplemental Figure 2. Comparison of Amino Acid Sequences of Human, Fungal, and Yeast Rac and Cdc42 Proteins.
Supplemental Figure 3. Alignment Using ClustalX of Fungal NoxR Sequences with Vertebrate p67phox and NOXA1 Sequences That Were Used as Input for Construction of the Phylogenetic Tree Shown in Figure 1.
Supplemental Table 1. Biological Materials.
Supplemental Table 2. Primers Used in This Study.
Supplementary Material
Acknowledgments
This research was supported by Grant MAU0403 from the Royal Society of New Zealand Marsden Fund. We thank Andrea Bryant, Ruth Wrenn, and Elizabeth Nickless (Massey University, Palmerston North, New Zealand), Michael Christensen, Wayne Simpson, and Anouk de Bonth (AgResearch, Palmerston North, New Zealand), and Douglas Hopcroft and Raymond Bennett (HortResearch, Palmerston North, New Zealand) for technical assistance. We also thank Jeremy Hyams, Matthew Nicholson, and Michelle Bryant (Massey University) and Jesus Aguirre (University of Mexico, Mexico City, Mexico) for comments on the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions to Authors (www.plantcell.org) is: Barry Scott (d.b.scott@massey.ac.nz).
Online version contains Web-only data.
References
- Aguirre, J., Rios-Momberg, M., Hewitt, D., and Hansberg, W. (2005). Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13 111–118. [DOI] [PubMed] [Google Scholar]
- Akiyama, K., Matsuzaki, K., and Hayashi, H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435 824–827. [DOI] [PubMed] [Google Scholar]
- Berthier, S., Paclet, M.H., Lerouge, S., Roux, F., Vergnaud, S., Coleman, A.W., and Morel, F. (2003). Changing the conformation state of cytochrome b558 initiates NADPH oxidase activation: MRP8/MRP14 regulation. J. Biol. Chem. 278 25499–25508. [DOI] [PubMed] [Google Scholar]
- Bestwick, C.S., Brown, I.R., Bennett, M.H., and Mansfield, J.W. (1997). Localization of hydrogen peroxide accumulation during the hypersensitive reaction of lettuce cells to Pseudomonas syringae pv phaseolicola. Plant Cell 9 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield, G., and Pears, C. (2003). Superoxide signalling required for multicellular development of Dictyostelium. J. Cell Sci. 116 3387–3397. [DOI] [PubMed] [Google Scholar]
- Boyce, K.J., Hynes, M.J., and Andrianopoulos, A. (2003). Control of morphogenesis and actin localization by the Penicillium marneffei RAC homolog. J. Cell Sci. 116 1249–1260. [DOI] [PubMed] [Google Scholar]
- Briggs, R.T., Drath, D.B., Karnovsky, M.L., and Karnovsky, M.J. (1975). Localization of NADH oxidase on the surface of human polymorphonuclear leukocytes by a new cytochemical method. J. Cell Biol. 67 566–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd, A.D., Schardl, C.L., Songlin, P.J., Mogen, K.L., and Siegel, M.R. (1990). The β-tubulin gene of Epichloë typhina from perennial ryegrass (Lolium perenne). Curr. Genet. 18 347–354. [DOI] [PubMed] [Google Scholar]
- Carol, R.J., Takeda, S., Linstead, P., Durrant, M.C., Kakesova, H., Derbyshire, P., Drea, S., Zarsky, V., and Dolan, L. (2005). A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438 1013–1016. [DOI] [PubMed] [Google Scholar]
- Carroll, A.M., Sweigard, J.A., and Valent, B. (1994). Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl. 41 22. [Google Scholar]
- Chen, C., and Dickman, M.B. (2004). Dominant active Rac and dominant negative Rac revert the dominant active Ras phenotype in Colletotrichum trifolii by distinct signalling pathways. Mol. Microbiol. 51 1493–1507. [DOI] [PubMed] [Google Scholar]
- Christensen, M.J., Bennett, R.J., and Schmid, J. (2002). Growth of Epichloë/Neotyphodium and p-endophytes in leaves of Lolium and Festuca grasses. Mycol. Res. 106 93–106. [Google Scholar]
- Diebold, B.A., and Bokoch, G.M. (2001). Molecular basis for Rac2 regulation of phagocyte NADPH oxidase. Nat. Immunol. 2 211–215. [DOI] [PubMed] [Google Scholar]
- Doke, N. (1983). Involvement of superoxide anion generation in the hypersensitive response of potato tuber tissues to infection with an incompatible race of Phytophthora infestans and to the hyphal wall components. Physiol. Plant Pathol. 23 345–357. [Google Scholar]
- Foreman, J., Demidchik, V., Bothwell, J.H., Mylona, P., Miedema, H., Torres, M.A., Linstead, P., Costa, S., Brownlee, C., Jones, J.D., Davies, J.M., and Dolan, L. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422 442–446. [DOI] [PubMed] [Google Scholar]
- Han, C.H., Freeman, J.L., Lee, T., Motalebi, S.A., and Lambeth, J.D. (1998). Regulation of the neutrophil respiratory burst oxidase. Identification of an activation domain in p67(phox). J. Biol. Chem. 273 16663–16668. [DOI] [PubMed] [Google Scholar]
- Harris, S.D., and Momany, M. (2004). Polarity in filamentous fungi: Moving beyond the yeast paradigm. Fungal Genet. Biol. 41 391–400. [DOI] [PubMed] [Google Scholar]
- Itoh, Y., Johnson, R., and Scott, B. (1994). Integrative transformation of the mycotoxin-producing fungus, Penicillium paxilli. Curr. Genet. 25 508–513. [DOI] [PubMed] [Google Scholar]
- Kamiguti, A.S., Serrander, L., Lin, K., Harris, R.J., Cawley, J.C., Allsup, D.J., Slupsky, J.R., Krause, K.-H., and Zuzel, M. (2005). Expression and activity of NOX5 in the circulating malignant B cells of hairy cell leukemia. J. Immunol. 175 8424–8430. [DOI] [PubMed] [Google Scholar]
- Kawasaki, T., Henmi, K., Ono, E., Hatakeyama, S., Iwano, M., Satoh, H., and Shimamoto, K. (1999). The small GTP-binding protein Rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. USA 96 10922–10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga, H., Terasawa, H., Nunoi, H., Takeshige, K., Inagaki, F., and Sumimoto, H. (1999). Tetratricopeptide repeat (TPR) motifs of p67(phox) participate in interaction with the small GTPase Rac and activation of the phagocyte NADPH oxidase. J. Biol. Chem. 274 25051–25060. [DOI] [PubMed] [Google Scholar]
- Lalucque, H., and Silar, P. (2003). NADPH oxidase: An enzyme for multicellularity? Trends Microbiol. 11 9–12. [DOI] [PubMed] [Google Scholar]
- Lambeth, J.D. (2004). NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 4 181–189. [DOI] [PubMed] [Google Scholar]
- Lambeth, J.D., Cheng, G., Arnold, R.S., and Edens, W.A. (2000). Novel homologs of gp91phox. Trends Biochem. Sci. 25 459–461. [DOI] [PubMed] [Google Scholar]
- Lara-Ortiz, T., Riveros-Rosas, H., and Aguirre, J. (2003). Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol. Microbiol. 50 1241–1255. [DOI] [PubMed] [Google Scholar]
- Lardy, B., Bof, M., Aubry, L., Paclet, M.H., Morel, F., Satre, M., and Klein, G. (2005). NADPH oxidase homologs are required for normal cell differentiation and morphogenesis in Dictyostelium discoideum. Biochim. Biophys. Acta 1744 199–212. [DOI] [PubMed] [Google Scholar]
- Latch, G.C.M., and Christensen, M.J. (1985). Artificial infection of grasses with endophytes. Ann. Appl. Biol. 107 17–24. [Google Scholar]
- Malagnac, F., Lalucque, H., Lepère, G., and Silar, P. (2004). Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 41 982–997. [DOI] [PubMed] [Google Scholar]
- Momany, M. (2002). Polarity in filamentous fungi: Establishment, maintenance and new axes. Curr. Opin. Microbiol. 5 580–585. [DOI] [PubMed] [Google Scholar]
- Namiki, F., Matsunaga, M., Okuda, M., Inoue, I., Nishi, K., Fujita, Y., and Tsuge, T. (2001). Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis. Mol. Plant Microbe Interact. 14 580–584. [DOI] [PubMed] [Google Scholar]
- Noda, Y., Kohjima, M., Izaki, T., Ota, K., Yoshinaga, S., Inagaki, F., Ito, T., and Sumimoto, H. (2003). Molecular recognition in dimerization between PB1 domains. J. Biol. Chem. 278 43516–43524. [DOI] [PubMed] [Google Scholar]
- Park, J., Gu, Y., Lee, Y., Yang, Z., and Lee, Y. (2004). Phosphatidic acid induces leaf cell death in Arabidopsis by activating the Rho-related small G protein GTPase-mediated pathway of reactive oxygen species generation. Plant Physiol. 134 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves, E.P., Lu, H., Jacobs, H.L., Messina, C.G., Bolsover, S., Gabella, G., Potma, E.O., Warley, A., Roes, J., and Segal, A.W. (2002). Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature 416 291–297. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and Nei, N. (1987). A neighbor-joining method: A new method for constructing phylogenetic tree. Mol. Biol. Evol. 44 406–425. [DOI] [PubMed] [Google Scholar]
- Sharpless, K.E., and Harris, S.D. (2002). Functional characterization and localization of the Aspergillus nidulans formin SEPA. Mol. Biol. Cell 13 469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinogi, T., Suzuki, T., Kurihara, T., Narusaka, Y., and Park, P. (2003). Microscopic detection of reactive oxygen species generation in the compatible and incompatible interactions of Alternaria alternata Japanese pear pathotype and host plants. J. Gen. Plant Pathol. 69 7–16. [Google Scholar]
- Shinogi, T., Suzuki, T., Tagashira, M., Yamane, K., Yao, N., Uwo, M., Kawakami, S., Narusaka, Y., and Park, P. (2001). A low viscosity epoxy resin “Quetol-651” as a substitute of Spurr's resin for hard biological materials in transmission electron microscopy. J. Electr. Microsc. Technol. Med. Biol. 16 1–10. [Google Scholar]
- Spiers, A.G., and Hopcroft, D.H. (1993). Black canker and leaf spot of Salix in New Zealand caused by Glomerella miyabeana (Colletotrichum gloeosporioides). Eur. J. Forest Pathol. 23 92–102. [Google Scholar]
- Suh, Y.A., Arnold, R.S., Lassegue, B., Shi, J., Xu, X., Sorescu, D., Chung, A.B., Griendling, K.K., and Lambeth, J.D. (1999). Cell transformation by the superoxide-generating oxidase Mox1. Nature 401 79–82. [DOI] [PubMed] [Google Scholar]
- Tan, Y.Y., Spiering, M.J., Scott, V., Lane, G.A., Christensen, M.J., and Schmid, J. (2001). In planta regulation of extension of an endophytic fungus and maintenance of high metabolic rates in its mycelium in the absence of apical extension. Appl. Environ. Microbiol. 67 5377–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, A., Christensen, M.J., Takemoto, D., Park, P., and Scott, B. (2006). Reactive oxygen species play a role in regulating a fungus-perennial ryegrass mutualistic association. Plant Cell 18 1052–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, A., Tapper, B.A., Popay, A.J., Parker, E.J., and Scott, B. (2005). A symbiosis expressed non-ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Mol. Microbiol. 57 1036–1050. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, M.A., and Dangl, J.L. (2005). Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8 397–403. [DOI] [PubMed] [Google Scholar]
- Vanden Wymelenberg, A.J., Cullen, D., Spear, R.N., Schoenike, B., and Andrews, J.H. (1997). Expression of green fluorescent protein in Aureobasidium pullulans and quantification of the fungus on leaf surfaces. Biotechniques 23 686–690. [DOI] [PubMed] [Google Scholar]
- Vignais, P.V. (2002). The superoxide-generating NADPH oxidase: Structural aspects and activation mechanism. Cell. Mol. Life Sci. 59 1428–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinzierl, G., Leveleki, L., Hassel, A., Kost, G., Wanner, G., and Bolker, M. (2002). Regulation of cell separation in the dimorphic fungus Ustilago maydis. Mol. Microbiol. 45 219–231. [DOI] [PubMed] [Google Scholar]
- Westfall, P.J., and Momany, M. (2002). Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch, and conidiophore development. Mol. Biol. Cell 13 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, C., Itoh, Y., Johnson, R., Garthwaite, I., Miles, C.O., Munday-Finch, S.C., and Scott, B. (1998). Paxilline-negative mutants of Penicillium paxilli generated by heterologous and homologous plasmid integration. Curr. Genet. 33 368–377. [DOI] [PubMed] [Google Scholar]
- Young, C.A., Bryant, M.K., Christensen, M.J., Tapper, B.A., Bryan, G.T., and Scott, B. (2005). Molecular cloning and genetic analysis of a symbiosis-expressed gene cluster for lolitrem biosynthesis from a mutualistic endophyte of perennial ryegrass. Mol. Genet. Genomics 274 13–29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.