Abstract
Circadian gating of light signaling limits the timing of maximum responsiveness to light to specific times of day. The fhy3 (for far-red elongated hypocotyl3) mutant of Arabidopsis thaliana is involved in independently gating signaling from a group of photoreceptors to an individual response. fhy3 shows an enhanced response to red light during seedling deetiolation. Analysis of two independent fhy3 alleles links enhanced inhibition of hypocotyl elongation in response to red light with an arrhythmic pattern of hypocotyl elongation. Both alleles also show disrupted rhythmicity of central-clock and clock-output gene expression in constant red light. fhy3 exhibits aberrant phase advances under red light pulses during the subjective day. Release-from-light experiments demonstrate clock disruption in fhy3 during the early part of the subjective day in constant red light, suggesting that FHY3 is important in gating red light signaling for clock resetting. The FHY3 gating function appears crucial in the early part of the day for the maintenance of rhythmicity under these conditions. However, unlike previously described Arabidopsis gating mutants that gate all light signaling, gating of direct red light–induced gene expression in fhy3 is unaffected. FHY3 appears to be a novel gating factor, specifically in gating red light signaling to the clock during daytime.
INTRODUCTION
Circadian rhythms control a wide range of physiological and biochemical processes in plants in tune with the day/night cycle. An endogenous oscillator based on a transcriptional feedback loop involving the proteins CIRCADIAN CLOCK ASSOCIATED1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), and TIMING OF CAB1 (TOC1) not only ensures that daily processes occur at the optimum time of day but also coordinates metabolic processes to peak in an efficient sequence throughout the day (Harmer et al., 2000; Alabadi et al., 2001). This endogenous oscillator can be entrained to the day/night cycle by environmental cues but nonetheless is capable of sustained oscillation even in the absence of any external signals. However, although such sustained oscillation in constant conditions is robust in its periodicity, it rarely runs with an exactly 24-h period, meaning that daily resetting of the clock is essential. A capacity for resetting is, in fact, very beneficial in a constantly changing daylength, and appropriately, resetting mechanisms appear to be a complex and integral part of clock mechanisms in all organisms studied (Devlin, 2002). Indeed, the effectiveness of this capacity for resetting perhaps explains why evolution has not favored an exactly 24-h rhythm. The most effective of the environmental cues responsible for resetting the clock are the changes in light and temperature, which are both associated with dawn and dusk transitions. In plants, little is known about the mechanism behind temperature resetting, but the way in which light resetting is accomplished is beginning to be understood (Millar, 2004).
One key facet of the response of the circadian clock to environmental cues is a phenomenon known as gating. The most crucial times for the correct resetting of the clock in response to the environment are at dawn and dusk. Consequently, the clock is most sensitive to resetting light stimuli at these times. During the day, it would be extremely disadvantageous for a plant to be continually resetting in response to light, so at this time the clock displays reduced sensitivity (Devlin and Kay, 2001). However, gating is not limited to light input to the circadian clock. Many responses to the light environment have been demonstrated to be subject to gating of photoreceptor action in plants, including direct light-mediated induction of gene expression and light-mediated inhibition of hypocotyl elongation. A range of responses show different times of optimal sensitivity to light (Devlin, 2006). Recently, two components of the pathway gating photoreceptor action, EARLY FLOWERING3 (ELF3) and TIME FOR COFFEE (TIC), have been identified in the model plant Arabidopsis thaliana (McWatters et al., 2000; Covington et al., 2001; Hall et al., 2003). Loss of either component leads to a loss of circadian gating of the effects of pulses of red, blue, or white light both for clock resetting and for direct induction of light-regulated gene expression.
Arabidopsis uses an array of photoreceptors in clock resetting and photomorphogenic responses. These are sensitive to a range of light intensities and cover both ends of the visible spectrum. They fall into two main classes, the red- and far-red-absorbing phytochromes and the blue/UV-A-absorbing cryptochromes (Nagy and Schafer, 2002; Lin and Shalitin, 2003). Five phytochromes, phyA to phyE, and three cryptochromes, cry1 to cry3, exist in Arabidopsis, each group the products of a related gene family. These show discrete and overlapping roles in the control of development throughout the life history of Arabidopsis (Cashmore, 2005; Franklin and Whitelam, 2005).
The identification of a number of mutants in the signal transduction pathways downstream of these photoreceptors has allowed us to begin to dissect these pathways (Fankhauser and Bowler, 2005). The fhy3 (for far-red elongated hypocotyl3) mutant was identified in a screen for phyA mutants deficient in deetiolation responses in constant monochromatic far-red light (Whitelam et al., 1993). Like phyA, fhy3 shows decreased responsiveness to constant far-red light for inhibition of hypocotyl elongation, stimulation of cotyledon opening, and induction of anthocyanin. However, fhy3 possesses wild-type levels of the phyA photoreceptor, implicating it as a component of the phyA signal transduction pathway. PhyA-mediated signaling can be categorized into two distinct modes: the very-low-fluence response, characterized by its requirement for only a small amount of light for saturation, and the high-irradiance response, characterized by its requirement for continuous irradiation and its fluence rate dependence (Whitelam et al., 1993). fhy3 is specifically disrupted in the high-irradiance response (Yanovsky et al., 2000), indicating that it acts in a divergent arm of the phyA signal transduction pathway. In addition, unlike the phyA mutant, the fhy3 mutant is hyperresponsive to red light during seedling establishment, suggesting that it may also affect signaling downstream of other phytochromes (Whitelam et al., 1993). The protein encoded by the FHY3 gene is 839 amino acids in length and forms a member of the FAR family of proteins. It incorporates a coiled-coil motif and a nuclear localization signal and shares homology with type II MuDR family transposons, suggesting that it is a transcription factor (Hudson et al., 2003; Lin and Wang, 2004).
Here, we demonstrate that the enhanced responsivity to continuous red light in fhy3 is associated with the absence of the normal circadian gating of the phytochrome-mediated inhibition of hypocotyl elongation, a result of the conditional arrhythmicity of the circadian clock. We show that the wild-type FHY3 protein plays an important role in specifically gating red light input to the circadian clock during the subjective day to maintain rhythmicity but plays no role in the direct gating of other light responses.
RESULTS
fhy3 Results in Increased Red Light Signaling in the Control of Deetiolation
To eliminate the possibility that the enhanced response to red light in the fhy3-1 mutant was the result of either an allele-specific or ecotype-specific phenomenon, we examined one further allele of fhy3, isolated from the Feldmann T-DNA collection (Forsthofel et al., 1992). Both alleles result in significant disruption to the FHY3 gene. The original fhy3-1 mutation in the Columbia background results in the introduction of a stop codon at position 91, causing an early truncation of the protein (Wang et al., 2002). The fhy3-3 mutation in the Wassilewskija (Ws) background, also described briefly by Wang et al. (2002), is the result of a missense mutation after position 360, introducing a stop codon at position 377.
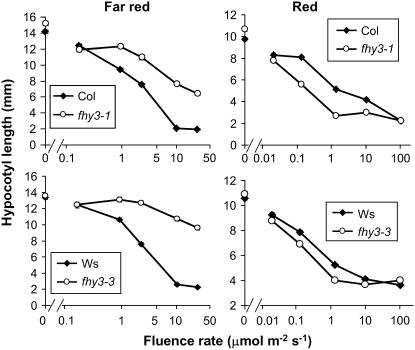
The responses of both alleles to a range of fluence rates of constant monochromatic red light (Rc) or far-red light (FRc) were analyzed. After 1 d of growth in darkness, each of the fhy3 alleles with their respective wild types were transferred to Rc or FRc for 3 d. Each of the fhy3 alleles displayed a long hypocotyl compared with wild-type seedlings, particularly apparent at high fluence rates of FRc, confirming that phyA signaling was indeed impaired in these mutants. As was observed previously for fhy3-1, a slight inhibition of hypocotyl elongation with increasing fluence rate was exhibited by both fhy3 alleles, indicative of some residual phyA action (Figure 1). Both wild-type and fhy3 seedlings showed strong responses to Rc for inhibition of hypocotyl elongation. However, each of the fhy3 alleles analyzed displayed an enhanced response to Rc for inhibition of hypocotyl elongation relative to that observed in their respective wild-type seedlings. This was most apparent at intermediate fluence rates (Figure 1).
Figure 1.
Hypocotyl Length in Wild-Type and fhy3 Seedlings in Red and Far-Red Light.
Seedlings were germinated in darkness for 1 d before transfer to continuous red or far-red light of the fluence rate indicated for a further 3 d. Values shown are means ± se from 30 seedlings. Col, Columbia.
Inhibition of Hypocotyl Elongation under Constant Red Light Displays a Circadian Rhythm in Wild-Type but Not fhy3 Seedlings
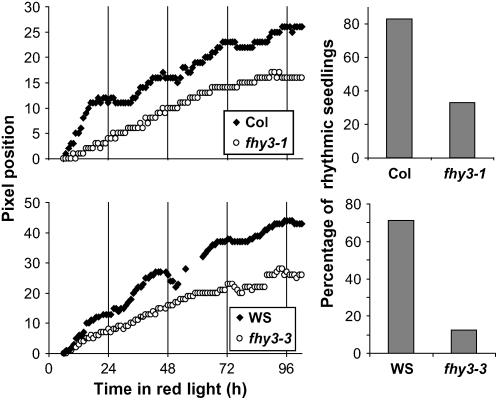
Video imaging was used to follow the kinetics of hypocotyl elongation under Rc in seedlings of the wild type and fhy3. The inhibition of hypocotyl elongation in constant white light (Wc) in Arabidopsis has been demonstrated to be gated by the circadian clock (Dowson-Day and Millar, 1999), resulting in a rhythmic pattern of elongation growth in these conditions. We specifically looked for any such rhythm under Rc in wild-type and fhy3 seedlings. To ensure that any rhythm was entrained to the same phase in all seedlings before measuring the kinetics of the growth, wild-type and fhy3 seedlings were germinated for 2 d in 12-h white light/12-h dark [LD(12:12)] cycles. By this stage, the radicals were just emerging from the seed. The seedlings were then transferred to Rc, and hypocotyl elongation was recorded by following the movement of the hypocotyl apex. Wild-type seedlings displayed a clear circadian rhythm of hypocotyl elongation in Rc for both ecotypes (Figure 2), indicating that light-mediated inhibition of hypocotyl elongation in Rc is gated by the circadian clock in wild-type seedlings as it is in Wc. By contrast, both alleles of fhy3 displayed severely impaired rhythmicity of hypocotyl elongation in Rc. This was confirmed via an analysis of the percentage of rhythmic seedlings in the population (Figure 2). The majority of fhy3 seedlings displayed no apparent rhythmicity under Rc, suggesting that phytochrome-mediated signaling in this response is not gated in these seedlings. Therefore, the enhanced response to Rc observed for the inhibition of hypocotyl elongation in the fhy3 mutant appears to be attributable to the loss of clock-mediated gating of phytochrome signaling.
Figure 2.
Hypocotyl Elongation in Constant Red Light.
Wild-type and fhy3 seedlings were germinated in LD(12:12) for 3 d before transfer to Rc. Left, representative traces produced by individual seedlings recording the vertical pixel position of the apical meristem. Position 0 represents the pixel position at the start of recording. Right, percentage of traces found to be rhythmic by rhythm analysis software. Data shown are from between 6 and 17 seedlings.
fhy3 Causes Arrhythmicity of CAB2 Gene Expression in Continuous Red Light but Not in Continuous Blue Light
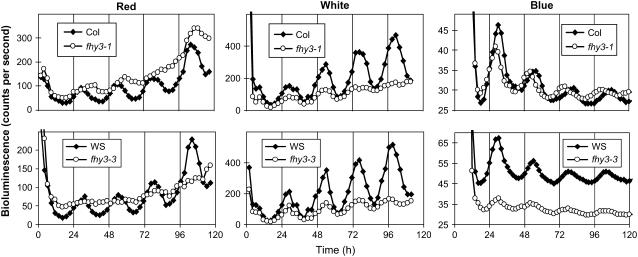
To determine whether the arrhythmicity displayed by fhy3 seedlings during hypocotyl elongation in Rc was more pleiotropic, we examined the pattern of transcription of the CAB2:LUCIFERASE (CAB2:LUC) reporter construct under the same conditions. Transcription from the CAB2:LUC reporter construct has been demonstrated to be under the control of the circadian clock under all conditions tested, including Rc (Millar et al., 1995). Both alleles of the fhy3 mutation were examined using this construct. In each case, the CAB2:LUC construct was introduced into the fhy3 mutant and its respective wild type by crossing these with plants of the appropriate ecotype into which the CAB2:LUC construct had been previously introgressed by repeated backcrossing. Wild-type and fhy3 seedlings containing the CAB2:LUC reporter construct were grown for 6 d in LD(12:12) before being transferred to Rc, Wc, or continuous monochromatic blue light (Bc). Bioluminescence levels, reflecting the activity of the CAB2 promoter, were subsequently monitored at 2-h intervals thereafter. In Rc, wild-type seedlings displayed a clear circadian rhythm of expression of the CAB2:LUC reporter. Conversely, both alleles of fhy3 tested showed a severe disruption of the circadian rhythm of CAB2:LUC expression (Figure 3). This phenotype was most apparent in the fhy3-3 allele, which displayed a completely arrhythmic phenotype (11% of fhy3-1 seedlings were scored as rhythmic as opposed to 100% of corresponding Columbia wild-type seedlings; 0% of fhy3-3 seedlings were scored as rhythmic as opposed to 94% of corresponding Ws wild-type seedlings). In Wc, wild-type seedlings again displayed a clear circadian rhythm of CAB2:LUC expression. As in Rc, both alleles of fhy3 showed a disrupted expression pattern of CAB2:LUC relative to the wild type in the same conditions (Figure 3). The fhy3-1 allele again showed a nearly arrhythmic phenotype after three cycles. A clear circadian rhythm was apparent in the fhy3-3 allele, but this was much less robust than the rhythm seen in wild-type seedlings. In Bc, a clear circadian rhythm of CAB2:LUC expression was again observed in wild-type seedlings. Some disruption of the pattern of expression of CAB2:LUC was observed in fhy3 mutant seedlings, but this disruption was much less dramatic under Bc than that observed under Rc or Wc (Figure 3); seedlings of fhy3-1 and fhy3-3 both demonstrated rhythmicity of CAB2:LUC expression similar to that in wild-type seedlings. However, seedlings of fhy3-1 showed a slightly shorter period length than their respective wild-type seedlings under Bc (variance-weighted mean period ± se for Columbia wild type and fhy3-1 were 25.38 ± 0.14 and 23.56 ± 0.17 h, respectively). Seedlings of fhy3-3 also showed a reduced mean expression level (mean bioluminescence ± se for Ws wild type and fhy3-3 were 50.17 ± 0.70 and 32.20 ± 0.28 counts per second, respectively).
Figure 3.
Mean CAB2:LUC Bioluminescence in Constant Light.
Wild-type and fhy3 seedlings expressing the CAB2:LUC reporter construct were entrained in LD(12:12) for 6 d before transfer to constant monochromatic red, white, or monochromatic blue light. Each data point represents the mean of between 16 and 29 seedlings.
Thus, the fhy3 mutant appears to be specifically disrupted in red light signaling. Under Rc, fhy3 mutant seedlings exhibited a severely disrupted rhythm of expression of CAB2:LUC, whereas only minor phenotypic differences between wild-type and fhy3 seedlings were observed under Bc. Under Wc, fhy3 displayed a partial loss of circadian rhythmicity. The loss of CAB2:LUC expression in fhy3 seedlings in Rc supports the proposal that the defect in fhy3 seedlings may be attributable to a disruption of the gating of light signals. However, in contrast with the two previously described gating-deficient mutants, elf3 and tic (Hicks et al., 1996; Hall et al., 2003), in which the light-dependent arrhythmicity of CAB2:LUC expression is apparent under all wavelengths of constant light, this arrhythmicity in fhy3 is specific to red light.
fhy3 Disrupts the Rhythmicity of a Light-Unresponsive Gene in Continuous Red Light but Not in Darkness
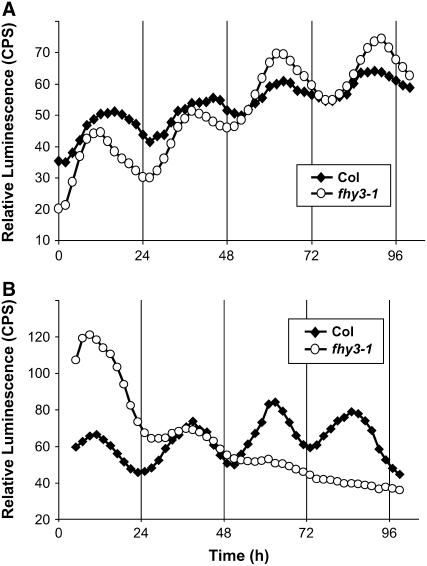
We predicted that if the fhy3 phenotype were specific to red light, then fhy3 mutant seedlings should show normal rhythmicity in darkness. To confirm the light-dependent nature of the arrhythmicity displayed by the fhy3 mutant, the line containing the fhy3-1 allele was crossed with a line containing the CCR2:LUC transgene. CCR2 encodes the COLD AND CIRCADIAN REGULATED2 gene, also known as GLYCINE RICH PROTEIN7 (GRP7). Unlike CAB2, the expression of which is rapidly decreased in constant darkness (DD), the expression of CCR2 retains a strong circadian rhythmicity in both constant light and DD. This offers the opportunity to examine circadian output in DD. Wild-type and fhy3-1 seedlings expressing CCR2:LUC were germinated in LD(12:12) as described above and transferred to DD or to Rc. Both wild-type and fhy3-1 seedlings showed a strong circadian rhythm of CCR2:LUC expression in DD (Figure 4). Little difference in the pattern of expression was observable between the wild type and fhy3-1, indicating that the fhy3 mutation does not disrupt the central circadian oscillator in darkness. In Rc, the disruption of circadian rhythmicity attributable to fhy3 was also apparent for CCR2:LUC expression, further confirming the pleiotropic nature of the mutation (Figure 4).
Figure 4.
Normalized Mean CCR2:LUC Bioluminescence in DD and in Constant Red Light.
Wild-type and fhy3 seedlings expressing the CCR2:LUC reporter construct were entrained in LD(12:12) for 6 d before transfer to DD (A) or to constant red light (B). Each data point represents the normalized mean of between 17 and 59 seedlings. CPS, counts per second.
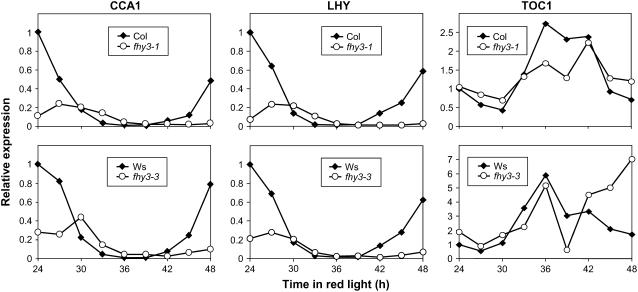
fhy3 Disrupts the Rhythmicity of Central Oscillator Genes in Continuous Red Light
We then analyzed the expression of CCA1, LHY, and TOC1 in Rc in wild-type and mutant seedlings to confirm that the central oscillator itself is disrupted in the fhy3 mutant in Rc. Seedlings were germinated and grown for 6 d in LD(12:12) before being transferred to Rc. After 24 h in Rc, batches of seedlings were harvested every 3 h for a further 24 h. CCA1, LHY, and TOC1 showed a pronounced circadian pattern of expression in wild-type seedlings. Expression of CCA1 and LHY decreased from a high level at subjective dawn to a clear trough corresponding to the middle of the subjective night before increasing again toward subjective dawn (Figure 5), whereas expression of TOC1 increased from a low level in the early subjective day to a peak at subjective dusk before declining again to a low level by the next subjective dawn, in agreement with previous findings. The period of the rhythm appears slightly longer than 24 h, because no clear peak of CCA1/LHY expression was included in this time frame. Again, this is in agreement with previous findings for seedlings grown under these conditions (Devlin and Kay, 2000). Seedlings of both alleles of fhy3 displayed a severely disrupted pattern of expression of CCA1, LHY, and TOC1. In the fhy3 mutants, a reduced-amplitude peak of expression of CCA1 and LHY was observed at the beginning of the window of analysis. By the end of the window, the expression level of CCA1 and LHY effectively remained at trough level in fhy3-1 or showed a very slight increase in fhy3-3. The expression pattern of these genes in fhy3 suggests a delayed phase, and the low level of CCA1 and LHY expression at the end of the window may be a result of this. However, both the delayed phase and the low amplitude in the early part of the window are in exact agreement with the pattern of expression of CAB2:LUC after the same duration in Rc. It is notable, however, that subsequent to this, CAB2 expression becomes arrhythmic. Curiously, the pattern of TOC1 expression in the fhy3 mutants did not show evidence of a delayed phase or reduced amplitude during the early part of the window of analysis. Expression of TOC1 in the fhy3 mutant seedlings appeared to show a trough in the early part of the subjective day before increasing to a peak at subjective dusk, following the pattern observed in wild-type seedlings. However, TOC1 expression in fhy3 then declined rapidly to a sharp second trough at Zeitgeber Time 39 before increasing again sharply and maintaining a higher level of expression than that observed in wild-type seedlings for the remainder of the time course.
Figure 5.
Expression Patterns of CCA1, LHY, and TOC1 in Constant Red Light.
Wild-type and fhy3 seedlings were entrained in LD(12:12) for 6 d before transfer to Rc. After 24 h in red light, individual batches of seedlings were harvested at 3-h intervals for a further 24 h. Expression of CCA1 (left), LHY (middle), and TOC1 (right), relative to the wild type at time 0, was measured using quantitative PCR.
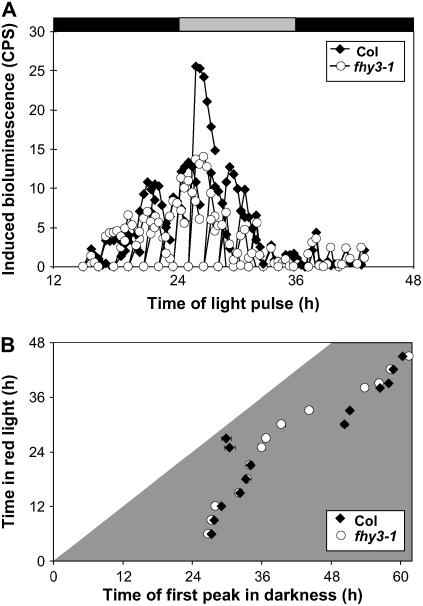
Gating of CAB2:LUC Induction by Pulses of Red Light Is Unaffected in fhy3
The red light specificity of the phenotype conferred by fhy3 was strongly suggestive of a disruption of light input to the clock. As the clock is able to run in the absence of light, we proposed that the defect may be the result of a fault in the gating of red light signaling by the circadian clock rather than a loss of the red light signal per se. A defect in gating would result in light-resetting stimuli affecting the clock at times when the clock is normally immune to such stimuli, potentially causing arrhythmicity. We examined two red light responses known to exhibit pronounced gating. As well as being under the control of the clock, CAB2 expression is directly induced by red light. This induction is gated by the clock so as to be minimal during the subjective night. To examine whether gating of direct red light induction of CAB2:LUC gene expression by red light pulses is disrupted in the fhy3 mutant, wild-type and fhy3 seedlings were subjected to the classical gating experiment of Millar and Kay (1996). Wild-type and fhy3-1 seedlings expressing CAB2:LUC were germinated in LD(12:12) for 6 d and then transferred to DD. After 12 h in DD, successive batches of seedlings were subjected to a single 20-min pulse of red light (Rp) at 2-h intervals for the next 40 h. Acute light-mediated induction of CAB2:LUC expression was followed by imaging luminescence in each batch immediately before the pulse and for 3 h after the pulse. Both wild-type and fhy3-1 seedlings showed a clear circadian gating of acute light-mediated induction of CAB2:LUC expression, with maximal induction occurring around subjective dawn, 28 h after transfer to DD, and a clear inhibition of acute induction being observed during the first and second subjective nights (Figure 6A). Induction of CAB2:LUC expression by Rp was slightly lower in the fhy3 mutant than in the wild type during the subjective day. This is possibly a result of reduced phyA signaling in fhy3. However, key to this investigation, gating of responsiveness to Rp during the subjective night was just as efficient in fhy3 as in the wild type, demonstrating that circadian gating of the acute light-mediated induction of CAB2:LUC expression was unaffected by the fhy3 mutation.
Figure 6.
Effect of Red Light on Direct Induction of CAB2:LUC Expression and on Clock Control of CAB2:LUC Expression.
(A) Gating of the induction of CAB2:LUC expression by a pulse of red light. Wild-type and fhy3 seedlings expressing the CAB2:LUC reporter construct were entrained in LD(12:12) before transfer to darkness at subjective dawn. CAB2:LUC bioluminescence was recorded at the indicated times immediately before seedlings were given a pulse of red light. After return to darkness, CAB2:LUC bioluminescence was recorded for a further 2 h. The mean induction of CAB2:LUC shown was calculated by subtracting basal luminescence before light treatment. Gray and black shading represent subjective day and subjective night, respectively. Each data point represents the mean of between 11 and 24 seedlings. CPS, counts per second.
(B) Release from red light. Wild-type and fhy3 seedlings expressing the CAB2:LUC reporter construct were entrained in LD(12:12) for 6 d and then transferred to Rc at subjective dawn for the duration indicated before transfer to darkness, during which the timing of the first peak of CAB2:LUC bioluminescence was recorded. Each data point represents the mean ± se of between 11 and 28 seedlings.
fhy3 Disrupts the Gating of Clock Resetting in Red Light
The normal gating of CAB2:LUC induction by pulses of red light in fhy3 suggests that the mutation does not result in a general disruption of the gating of red light signaling. To test whether the gating of clock resetting in Rc is disrupted in fhy3, a release-from-light experiment was performed (McWatters et al., 2000). Wild-type and fhy3-1 seedlings expressing the CAB2:LUC transgene were germinated in LD(12:12) for 6 d and then transferred to Rc. After 6 h in Rc, successive batches of seedlings were subsequently transferred to DD at 2-h intervals for the next 40 h. After transfer to DD, luciferase imaging was performed to determine the timing of the first peak of CAB2:LUC expression after transfer. In wild-type seedlings, in which the gating of resetting allows the clock to continue oscillating in Rc, the timing of the first peak after transfer to DD was independent of the time spent in Rc (and of the time of release from Rc). Peaks occurred at times close to those at which they would be expected to occur based on the phase of the rhythm in the preceding LD(12:12) entrainment conditions (Figure 6B). In fhy3-1 seedlings transferred to DD after between 6 and 21 h in Rc, the average timing of the first peak of luminescence corresponded to that observed in wild-type seedlings, indicating that the clock was running normally over this period in Rc. However, in fhy3-1 seedlings transferred to DD between 25 and 33 h in Rc, the average timing of the first peak of luminescence differed greatly from that in the wild type. Between these times in Rc, the average timing of the first peak of luminescence occurred at a fixed time after transfer to DD, indicating that the clock had stopped during the previous Rc treatment in the majority of seedlings, only to start again after transfer to DD (Figure 6B). Thus, the gating of clock resetting by red light appears to be defective in fhy3 during the early part of the subjective day in Rc, the oscillation of the clock being prevented by the occurrence of a constant resetting signal.
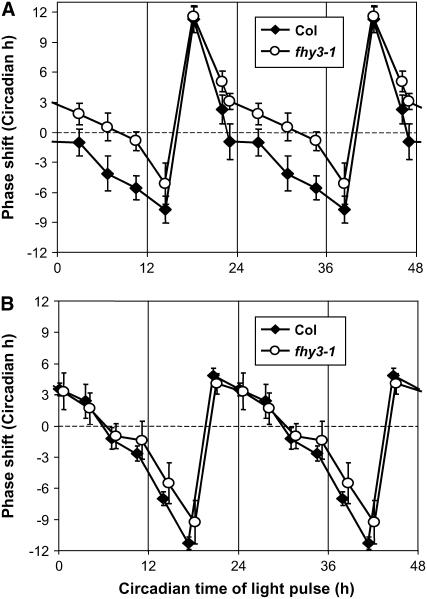
fhy3 Shows Aberrant Clock Resetting in Response to Red Light Pulses
The loss of gating of red light input to the clock during the subjective day in fhy3 was further investigated by generating a phase-response curve for wild-type and mutant seedlings. This examines the magnitude of the shift in the phase of the clock in response to light pulses given through the course of one circadian cycle in darkness. According to the method of Covington et al. (2001), seedlings of the wild type and fhy3 carrying the CCR2:LUC reporter were germinated and grown in LD(12:12) for 6 d before transfer to DD at subjective dawn for a further 24 h. At 4-h intervals throughout the subsequent 24-h period, successive batches of seedlings were subjected to a 1-h pulse of red or blue light before being transferred back to DD. Control seedlings were maintained in DD. Video imaging of CCR2:LUC expression was used to follow the phase of the circadian rhythm. Wild-type seedlings showed a classical phase-response curve in response to red light pulses, demonstrating strong phase advances just before subjective dawn and strong phase delays just after subjective dusk (Figure 7A). During the early part of the subjective day, wild-type seedlings showed little response to red light pulses, whereas during the late part of the subjective day, small phase delays were observed. fhy3 mutant seedlings also showed strong phase advances before subjective dawn and strong phase delays just after subjective dusk in response to red light pulses. However, contrary to the pattern observed for wild-type seedlings, small phase advances were observed throughout the greater part of the subjective day in fhy3 mutant seedlings (Figure 7A). This finding agrees with the proposal that red light input to the clock is disrupted during this part of the day.
Figure 7.
Phase-Response Curves for Red and Blue Light Resetting Pulses.
Wild-type and fhy3 seedlings expressing the CCR2:LUC reporter construct were entrained in LD(12:12) for 6 d before transfer to DD at subjective dawn. After a further 24 h in darkness, individual batches of seedlings were given a 1-h pulse of red light (A) or blue light (B) at 4-h intervals over the next 24 h before being returned to darkness. Data represent the mean phase shifts (±pooled se) plotted against the circadian time of the pulse. Phase advances are plotted as positive values, and delays are plotted as negative values. The data are double plotted to clarify the shape of the phase-response curve. Each data point represents the mean of between 7 and 28 seedlings.
In response to blue light pulses, wild-type and fhy3 seedlings displayed an almost identical phase-response curve (Figure 7B), confirming the red light specificity of the defect in fhy3. In agreement with the findings of Covington et al. (2001), blue light pulses resulted in reduced phase advances and greater phase delays in wild-type seedlings relative to those seen in response to red light pulses.
DISCUSSION
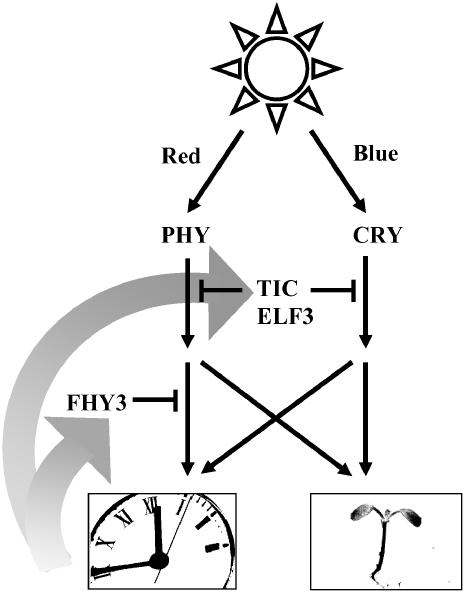
The regulation of light signaling to the plant circadian clock is crucial to the maintenance of an endogenous rhythm during prolonged light periods characteristic of the normal day/night cycle. The clock must be extremely sensitive to light signals at dawn and dusk, as these give important information to set the clock to the right time, but during the rest of the day the clock must avoid being constantly reset by light. The plant clock continues to orchestrate a number of sequential or overlapping physiological and metabolic processes during the day so that they occur in the correct order and so must continue to oscillate. The previously described components ELF3 and TIC act to gate light input to the clock during the early and late parts, respectively, of the subjective night (McWatters et al., 2000; Covington et al., 2001; Hall et al., 2003), and both prevent excessive sensitivity to resetting during the night and even around the dawn and dusk transitions. ELF3 and TIC appear to generally gate light signaling, both to the clock and to other light responses, and they appear to gate light signals originating from both red and blue photoreceptors. Here, we have demonstrated that FHY3 acts to regulate light signaling during the subjective day. Moreover, FHY3 acts to specifically regulate the response to light signals, carrying information to the clock and specifically the response to signals from the phytochrome receptor (Figure 8).
Figure 8.
Scheme Demonstrating the Possible Sites of Action of FHY3, TIC, and ELF3 in the Gating of Signals from the Phytochrome (PHY) and Cryptochrome (CRY) Photoreceptors.
FHY3 acts to gate signals on a branch of the red light pathway mediating light input to the clock. TIC and ELF3 affect all light-signaling pathways tested. TIC and ELF3 together may act to gate both the red and blue light input pathways upstream of a divergence of a branch mediating photomorphogenic responses (light inhibition of hypocotyl elongation and direct induction of CAB2 gene expression) and a branch mediating light input to the clock.
The fhy3 mutant displays an enhanced response to red light for inhibition of hypocotyl elongation. In white light, wild-type Arabidopsis seedlings display a periodic gating of light signaling in the inhibition of hypocotyl elongation during the subjective night, with the result that a period of rapid elongation growth occurs at this time (Dowson-Day and Millar, 1999). We demonstrate here that wild-type Arabidopsis also displays a circadian rhythm of hypocotyl elongation in Rc. In the fhy3 mutant, by contrast, the majority of individuals displayed no such rhythm under these conditions. Therefore, it was proposed that the enhanced inhibition of hypocotyl elongation in Rc in fhy3 was attributable to the lack of such a circadian rhythm of sensitivity to light. This proposal allowed for two possibilities. Either the central clock mechanism itself could have been disrupted in fhy3, with the result that the gate was always open to light signaling, or the lesion in fhy3 could have specifically disrupted the pathway by which a functional clock gates light signaling. The latter possibility, of course, could apply equally to the gating of light input to regulate clock resetting in response to light and to the gating of the inhibition of hypocotyl elongation.
Analysis of the oscillation of transcription from the CAB2 gene promoter revealed that the fhy3 mutant also displayed a severe disruption of the rhythm of expression of CAB2 in constant red light. This finding demonstrated fhy3 to be pleiotropic in its effects, affecting the rhythm of two distinct circadian-regulated markers. However, a relatively normal oscillation of CAB2 expression was observed in blue light, suggesting that the clock itself was capable of functioning normally in fhy3. This conclusion was further supported by the analysis of CCR2 expression. The expression of CCR2 is not decreased in darkness and consequently reports the state of the clock in darkness. A circadian rhythm of expression of CCR2 is observed in wild-type seedlings in both red light and darkness. A circadian rhythm of expression of CCR2 was observed in fhy3 mutant seedlings in darkness. However, CCR2 expression was arrhythmic in fhy3 mutant seedlings in red light. This confirmed that the phenotype of fhy3 is specific to red light. These findings strongly suggested that the lesion did not lie in the clock itself but in the gating of the light input pathway mechanism.
In constant white light, the fhy3-1 allele becomes arrhythmic as in red light, presumably reflecting the lack of gating of the red component of white light. Some rhythmicity is apparent in fhy3-3, although this is of greatly reduced amplitude. It is possible that the poor red content of the cool-white fluorescent light source may explain the less dramatic disruption of rhythmicity in Wc.
Analysis of the pattern of expression of CCA1, LHY, and TOC1 demonstrates that oscillation of the central clock itself is also disrupted in Rc. CCA1 and LHY transcripts showed a greatly reduced amplitude and delayed phase over the early part of the period of analysis before appearing to decrease to a steady trough level. TOC1 transcript showed an apparently wild-type expression pattern over the early part of the period of analysis before displaying a sharp trough in expression at Zeitgeber Time 39, then increasing to a high level at the time when wild-type TOC1 expression returned to a trough level. The timing of the more severe disruption of the pattern of expression of the central clock components in Rc is consistent with the onset of full arrhythmicity in CAB2 expression.
A disruption in gating of light input to the clock could be the result of either a general disruption of gating of light signaling, both to the clock and to other light responses, or merely a disruption of only one of these aspects. The direct induction of CAB2 expression by light is one example of a light response gated by the clock. When wild-type and fhy3 seedlings were entrained to light/dark cycles and then transferred to darkness, a clear circadian gating of CAB2:LUC induction by subsequent pulses of red light was observed in both the wild type and fhy3. Furthermore, this gating was observed over a period of 72 h in darkness, indicating that the direct gating of the light response is unaffected in fhy3. This conclusion is supported by the CAB2:LUC expression pattern observed in fhy3 seedlings in 16-h red light/8-h dark cycles [RD(16:8)]. Under these conditions, each fhy3 allele behaves exactly like its respective wild type (see Supplemental Figure 1 online). This contrasts with the tic and elf3 mutants, in which the wild-type decline in CAB2:LUC expression during the afternoon in 16-h light/8-h dark cycles is absent. The absence of this decline is attributable to the fact that elf3 and tic are defective in the gating of direct light induction of CAB2 expression.
By contrast, a release-from-light experiment confirmed the disruption of the gating of clock resetting in red light in fhy3. Wild-type seedlings, entrained to light/dark cycles and then transferred to constant red light, maintained the entrained rhythm after subsequent release from light irrespective of the duration of exposure to red light, indicating that red light input to the clock had been successfully gated during this time. Although fhy3 seedlings treated in the same way maintained the entrained rhythm after between 6 and 21 h of exposure to red light, they experienced a cessation of the rhythm after red light exposure of between 25 and 33 h. This finding indicated that between these points the gating of red light input to the clock was no longer fully functional in fhy3. This time point corresponds to the early part of the subjective day, suggesting a role for FHY3 in gating during this period. Some residual rhythmicity can be observed for all outputs tested for a few hours beyond this point in seedlings maintained in Rc. This is also true for the central clock components, but this rhythm is clearly of insufficient robustness not to be reset by the lights-off transition used in this experiment during this crucial period of the early part of the subjective day.
Confirmation of the specific disruption of red light input to the clock is provided by the analysis of the phase-response curve charting the degree of clock resetting resulting from the application of pulses of red or blue light to seedlings otherwise maintained in darkness. The fhy3-1 mutant displayed phase advances in response to red light pulses during the subjective day at a time when wild-type seedlings displayed phase delays. During darkness, the circadian rhythm is unaffected in fhy3, meaning that the difference in the response to red light cannot be a result of any difference in phase of the clock in the fhy3 mutant. Clock resetting primarily occurs as a result of light induction of expression of the central clock components CCA1 and LHY, causing their levels to increase to levels normally associated with a different phase of the rhythm. CCA1 and LHY expression is particularly responsive to light during the subjective night, the period during which the greatest light-induced phase shifts can be observed. During the subjective day, however, little or no induction of CCA1 or LHY expression is observed in response to light (Locke et al., 2005). Another route for light input to the clock acting during the day was recently proposed by Locke et al. (2005). This provides a mechanism by which daylength can be tracked, delaying the phase of the clock in response to a lengthening of the day. This proposal is also consistent with the persistence of a resetting response to light in the cca1 lhy double mutant. The GIGANTEA (GI) protein forms an interlocked loop with TOC1. GI acts to promote TOC1 expression during the subjective day at the same time that LHY is suppressing TOC1 expression. GI expression is also induced by light and appears to provide a mechanism by which plants respond to light throughout the day, allowing any extension of the light period at subjective dusk to delay the clock. Stimulation of GI and, therefore, TOC1 expression beyond the normal peak of TOC1 expression around Zeitgeber Time 11 [just before dusk in a LD(12:12) day] would delay the clock. Conversely, enhanced activation of GI expression by light during the earlier part of the day would result in a premature increase in TOC1 expression and a phase advance, such as that observed in the fhy3 mutant. In addition, modeling of the clock loop predicts that constant activation of TOC1 by light under constant light would cause arrhythmia (Locke et al., 2005). Therefore, the phenotype of fhy3 in Rc is also consistent with the proposal that FHY3 normally acts to gate the response of GI to red light. Further analysis of the expression patterns of GI in the fhy3 mutant under a range of light conditions will be required to test this hypothesis. The mode of action of FHY3 also remains to be determined. The similarity of FHY3 to the type II MuDR family transposons has led to suggestions that it is a transcription factor (Hudson et al., 2003) and so may act directly to regulate gene expression. FHY3 expression does not itself appear to be regulated by light (Lin and Wang, 2004) or the clock (Edwards et al., 2006), so its activity must be determined posttranscriptionally. Most likely, it is the protein activity that is dependent on both red light and the phase of the clock. Tests of misexpression lines would reveal whether it is the presence or the activity of FHY3 that is key.
Therefore, the short-hypocotyl phenotype observed in red light in fhy3 would appear to be, primarily, the result of the cessation of the oscillation of the clock under these conditions. This loss of oscillation, presumably secondarily, results in a loss of gating of the light signal, causing an inhibition of elongation. This would require that the clock be stopped in a position at which the gate is always open in fhy3 in red light.
It is interesting that FHY3, defined previously as a component of the phyA signal transduction pathway, should play a role in gating of light input to the clock. The phyA mutant displays a loss of sensitivity for clock resetting in response to low-fluence-rate light in both the red and blue regions of the spectrum (Somers et al., 1998). In fact, FHY3 has been demonstrated previously to be involved in this phyA signaling to cause clock resetting (Yanovsky et al., 2001). However, the defect displayed by fhy3 in Rc is quite distinct from this role as a phyA signaling component. Despite the reduced response to resetting stimuli, the phyA mutant maintains a clear circadian rhythmicity in Rc of all fluence rates (Somers et al., 1998). Consequently, the defect in fhy3 cannot be the result of the loss of phyA signaling. This indicates that FHY3 plays two distinct roles in the plant: one as a positively acting signal transduction component downstream of phyA, the other as a component of the machinery gating red light input to the circadian clock. Such dual roles have been suggested for a number of other circadian clock–associated components. TOC1 appears to have a dual role in circadian and photomorphogenic responses in Arabidopsis (Más et al., 2003). Similarly, GI was recently demonstrated to play distinct roles in promoting flowering and in regulating circadian rhythms in Arabidopsis (Mizoguchi et al., 2005). This dual role of FHY3 may not be unique among the FAR family members, as a FRS9 RNA interference transgenic line has been shown to have a similar short-hypocotyl phenotype in Rc (Lin and Wang, 2004). It will be interesting to determine whether this also results from a defect in the gating of red light input to the circadian clock.
Red light input to the clock is mediated by at least four of the phytochrome family photoreceptors, with phyA playing the major role in low-fluence-rate red light and phyB playing the major role in high-fluence-rate red light (Somers et al., 1998; Devlin and Kay, 2000). The enhanced inhibition of hypocotyl elongation observed as a result of the fhy3 mutation was particularly observed at intermediate fluence rates, suggesting that both phyA and phyB signaling may be affected. It is proposed that the enhanced inhibition is no longer observed at the highest fluence rates of red light due to the wild-type response becoming increasingly saturated at higher fluence rates, as is particularly evident by the response of the Ws wild type in Figure 1. The enhanced inhibition of hypocotyl elongation observed as a result of the fhy3 mutation was not observed in the absence of either phyA or phyB (see Supplemental Figure 2 online). This result supports the proposal that a combination of signals from each of these phytochromes is necessary in the absence of FHY3 to effect sufficient clock-resetting stimulus to cause arrhythmicity.
This discovery of the role of FHY3 in specifically gating red light input to the clock reveals new information about the topology of the phytochrome signaling pathway. Gating of phytochrome signals by FHY3 occurs after a branch point between signals regulating clock resetting and signals regulating other light responses. In fact, evidence for this branch point can be seen in the manner in which Arabidopsis shows a gating of light input to the clock during the subjective day (Covington et al., 2001). The fact that, at this time of day, light signals for the induction of CAB2 gene expression and the inhibition of hypocotyl elongation appear not to be gated is clear evidence that the two are subjected to distinct mechanisms of gating. Indeed, a number of photomorphogenic responses in plants are gated with different phases of maximal responsivity, suggesting that each phase may be the result of the action of a specific gating factor (Devlin, 2006). Gating defects in the elf3 and tic mutants affect both aspects of phytochrome signaling, suggesting that ELF3 and TIC gate the signaling pathway upstream of this branch point. However, the fact that ELF3 and TIC gate both red and blue light signals means that they would have to act independently in both red and blue signaling pathways in this way (Figure 8). The alternative possibility is that ELF3 and TIC could act independently on both clock resetting and photomorphogenic signals after the branch point between the two but downstream of a convergence of red and blue signaling in both pathways.
METHODS
Plant Materials
The fhy3-1 mutant allele of Arabidopsis thaliana in the Columbia ecotype has been described previously (Whitelam et al., 1993). The fhy3-3 mutant allele in the Ws ecotype was isolated from the Feldmann T-DNA collection (Feldmann and Marks, 1987; Forsthofel et al., 1992) in a screen to identify mutants that displayed a long hypocotyl under FRc.
The wild-type CAB2:LUC transgenic lines were generated by introgressing the CAB2:LUC transgene from the original 2CAC transgenic line (C24 ecotype) described by Millar et al. (1992) into the respective ecotypes at least five times (Somers et al., 1998). The CAB2:LUC transgenic lines homozygous for fhy3 were selected after a cross of each allele to the wild-type CAB2:LUC transgenic line in the relevant ecotype. Homozygous fhy3 lines were selected on the basis of hypocotyl length under FRc.
The wild-type CCCR2:LUC transgenic line in the Columbia ecotype was the WT450 transgenic line described previously (Harmer et al., 2000). The CCCR2:LUC transgenic line homozygous for fhy3-1 was selected after a cross of fhy3-1 to the wild-type CCR2:LUC transgenic line. The homozygous fhy3-1 line was selected on the basis of hypocotyl length under FRc.
Seedlings for luminescence, RNA extraction, or leaf-movement analysis were germinated and grown under LD(12:12) for the indicated durations as described previously (Devlin and Kay, 2000).
Light Sources
The white light source for LD(12:12) entrainment and Wc CAB2:LUC luminescence assays (45 μmol·m−2·s−1) and the red light source for Rc, RD(16:8), and Rp CAB2:LUC luminescence assays (22 μmol·m−2·s−1) and for Rc leaf-movement analysis (8 μmol·m−2·s−1) were as described previously (Devlin and Kay, 2000). Rp for the phase-response curve assay (40 μmol·m−2·s−1) was provided by light-emitting diodes at a maximum of 660 nm (Farnell). Bc for the CAB2:LUC luminescence assay (18 μmol·m−2·s−1) and Bp for the phase-response curve assay (25 μmol·m−2·s−1) were provided by light-emitting diodes at a maximum of 450 nm (Farnell). Rc and FRc sources for hypocotyl measurement (variable intensity) and the Rc source for the growth of seedlings for RNA extraction (22 μmol·m−2·s−1) were as described previously (Franklin et al., 2003). Light measurements were performed using either an LI-1800 portable spectroradiometer (Li-Cor) or an EPP2000 fiber-optic spectrometer (Stellarnet). Fluence-response curves for hypocotyl elongation were determined on standard medium without sucrose, as described previously (Franklin et al., 2003). All experiments were performed at a constant 22°C.
Rhythm Analysis
Luminescence levels were measured by ultra-low-light video imaging using a NightOwl cooled CCD camera and Winlight32 imaging software (Berthold Technologies), with the exception of the white light and red light data presented in Figure 3 and the data presented in Supplemental Figure 1 online, which were obtained using a VIM intensified charge-coupled device camera and an ARGUS-100 photon-counting image processor (Hamamatsu Photonics). Seedlings were prepared for imaging as described by Millar et al. (1992). An exposure time of 20 min was used for each image. Data for CCR2:LUC expression in Figure 4 were normalized to wild-type mean luminescence. Hypocotyl elongation rhythms were measured by time-lapse imaging using the Kujata imaging system (Mark Heyett) as described by Dowson-Day and Millar (1999), except that seedlings were germinated and grown under LD(12:12) for 3 d before imaging. In each case, data were imported into the Biological Rhythm Analysis Software System (developed by Paul Brown in the laboratory of Andrew Millar, University of Edinburgh), in which rhythmic traces for individual seedlings were scored by fast Fourier transform nonlinear least-squares analysis (Johnson and Frasier, 1985; Straume et al., 1991; Plautz et al., 1997) as having a circadian period if the strongest period detected was in the 20- to 30-h range. Mean periods and standard errors were variance-weighted as described by Millar et al. (1995). Rhythmic robustness was assessed using the relative amplitude error: a rhythm is described as being robust if its relative amplitude error value is less than the wild-type mean relative amplitude error plus 2 sd (Hall et al., 2003). Analysis of the gating of CAB2:LUC induction by pulses of red light was performed as described by McWatters et al. (2000), except that red light was used to induce CAB2:LUC expression. Release-from-light assays were conducted as described by McWatters et al. (2000), except that seedlings were maintained in Rc before release. All data are representative of two independent experiments. Phase-response curve analysis was performed as described by Covington et al. (2001), except that seedlings were germinated and grown under LD(12:12) for 6 d before transfer to imaging and that light pulses were given a 4-h intervals.
Quantitative PCR
RNA was extracted using the Sigma-Aldrich GenElute Mammalian Total RNA Miniprep kit, followed by Sigma-Aldrich DNaseI treatment. cDNA was synthesized using oligo(dT) primer and Moloney murine leukemia virus reverse transcriptase from Promega. Quantitative PCR was performed on a MJ Research Chromo4 machine, using SYBR Green JumpStart Taq ReadyMix. Data represent means of three technical replicates. Relative expression was calculated according to Pfaffl (2001). The data for Columbia and fhy3-1 are representative of two independent biological repeats.
Accession Numbers
The Arabidopsis Genome Initiative locus numbers for the genes analyzed in this article are as follows: FHY3, At3g22170; CAB2, At1g29920; CCR2, At2g21660; CCA1, At2g46830; LHY, At1g01060; TOC1, At5g61380; and GI, At1g22770.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Mean CAB2:LUC Bioluminescence in 16-h Red Light/8-h Dark Cycles in Wild-Type and fhy3 Seedlings.
Supplemental Figure 2. Inhibition of Hypocotyl Elongation in Red Light in Wild-Type, fhy3, phyB, phyB fhy3, phyA, and phyA fhy3 Seedlings.
Supplementary Material
Acknowledgments
We thank Ceinwen Tilley for technical assistance. We also thank Paloma Más (Consejo Superior de Investigaciones Cientificas, Barcelona, Spain) and Andrew Millar (University of Edinburgh) for helpful discussion during the preparation of the manuscript. This work was supported by grants from the European Molecular Biology Organization (LTF720), the Royal Society (574006.G503/22295/SM), and the University of London Central Research Fund (AR/CRF/B) to P.F.D.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Garry C. Whitelam (gcw1@le.ac.uk) and Paul F. Devlin (paul.devlin@rhul.ac.uk).
Online version contains Web-only data.
References
- Alabadi, D., Oyama, T., Yanovsky, M.J., Harmon, F.G., Más, P., and Kay, S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293 880–883. [DOI] [PubMed] [Google Scholar]
- Cashmore, A.R. (2005). Cryptochromes. In Photomorphogenesis in Plants and Bacteria, E. Schafer and F. Nagy, eds (Dordrecht, The Netherlands: Springer), pp. 379–406.
- Covington, M.F., Panda, S., Liu, X.L., Strayer, C.A., Wagner, D.R., and Kay, S.A. (2001). ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin, P.F. (2002). Signs of the time: Environmental input to the circadian clock. J. Exp. Bot. 53 1535–1550. [DOI] [PubMed] [Google Scholar]
- Devlin, P.F. (2006). Circadian regulation of photomorphogenesis. In Photomorphogenesis in Plants and Bacteria, E. Schafer and F. Nagy, eds (Dordrecht, The Netherlands: Springer), pp. 567–604.
- Devlin, P.F., and Kay, S.A. (2000). Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12 2499–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin, P.F., and Kay, S.A. (2001). Circadian photoperception. Annu. Rev. Physiol. 63 677–694. [DOI] [PubMed] [Google Scholar]
- Dowson-Day, M.J., and Millar, A.J. (1999). Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. 17 63–71. [DOI] [PubMed] [Google Scholar]
- Edwards, K.D., Anderson, P.E., Hall, A., Salathia, N.S., Locke, J.C., Lynn, J.R., Straume, M., Smith, J.Q., and Millar, A.J. (2006). FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser, C., and Bowler, C. (2005). Biochemical and molecular analysis of signaling components. In Photomorphogenesis in Plants and Bacteria, E. Schafer and F. Nagy, eds (Dordrecht, The Netherlands: Springer), pp. 379–406.
- Feldmann, K.A., and Marks, M.D. (1987). Agrobacterium-mediated transformation of germinating seeds of Arabidopsis thaliana: A non-tissue culture approach. Mol. Gen. Genet. 208 1–9. [Google Scholar]
- Forsthofel, N.R., Wu, Y., Schulz, B., Bennett, M.J., and Feldmann, K.A. (1992). T-DNA insertion mutagenesis in Arabidopsis: Prospects and perspectives. Plant Physiol. 19 353–366. [Google Scholar]
- Franklin, K.A., Davis, S.J., Stoddart, W.M., Vierstra, R.D., and Whitelam, G.C. (2003). Mutant analyses define multiple roles for phytochrome C in Arabidopsis photomorphogenesis. Plant Cell 15 1981–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin, K.A., and Whitelam, G.C. (2005). The role of phytochromes in adult plants. In Photomorphogenesis in Plants and Bacteria, E. Schafer and F. Nagy, eds (Dordrecht, The Netherlands: Springer), pp. 475–497.
- Hall, A., Bastow, R.M., Davis, S.J., Hanano, S., McWatters, H.G., Hibberd, V., Doyle, M.R., Sung, S., Halliday, K.J., Amasino, R.M., and Millar, A.J. (2003). The TIME FOR COFFEE (TIC) gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15 2719–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer, S.L., Hogenesch, J.B., Straume, M., Chang, H.S., Han, B., Zhu, T., Wang, X., Kreps, J.A., and Kay, S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290 2110–2113. [DOI] [PubMed] [Google Scholar]
- Hicks, K.A., Millar, A.J., Carré, I.A., Somers, D.E., Straume, M., Meeks-Wagner, R., and Kay, S.A. (1996). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274 790–792. [DOI] [PubMed] [Google Scholar]
- Hudson, M.E., Lisch, D.R., and Quail, P.H. (2003). The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 34 453–471. [DOI] [PubMed] [Google Scholar]
- Johnson, M.L., and Frasier, S.G. (1985). Nonlinear least squares analysis. Methods Enzymol. 117 301–342. [Google Scholar]
- Lin, C., and Shalitin, D. (2003). Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 54 469–496. [DOI] [PubMed] [Google Scholar]
- Lin, R., and Wang, H. (2004). Arabidopsis FHY3/FAR1 gene family and distinct roles of its members in light control of Arabidopsis development. Plant Physiol. 136 4010–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, J.C., Southern, M.M., Kozma-Bognar, L., Hibberd, V., Brown, P.E., Turner, M.S., and Millar, A.J. (June 28, 2005). Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol. Syst. Biol. (online), doi/10.1038/msb4100018. [DOI] [PMC free article] [PubMed]
- Más, P., Alabadi, D., Yanovsky, M.J., Oyama, T., and Kay, S.A. (2003). Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWatters, H.G., Bastow, R.M., Hall, A., and Millar, A.J. (2000). The ELF3 zeitnehmer regulates light signaling to the circadian clock. Nature 408 716–720. [DOI] [PubMed] [Google Scholar]
- Millar, A.J. (2004). Input signals to the plant circadian clock. J. Exp. Bot. 55 277–283. [DOI] [PubMed] [Google Scholar]
- Millar, A.J., and Kay, S.A. (1996). Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc. Natl. Acad. Sci. USA 93 15491–15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.J., Short, S.R., Chua, N.-H., and Kay, S.A. (1992). A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell 4 1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.J., Straume, M., Chory, J., Chua, N.-H., and Kay, S.A. (1995). The regulation of circadian period by phototransduction pathways in Arabidopsis. Science 267 1163–1166. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Wright, L., Fujiwara, S., Cremer, F., Lee, K., Onouchi, H., Mouradov, A., Fowler, S., Kamada, H., Putterill, J., and Coupland, G. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17 2255–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, F., and Schafer, E. (2002). Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu. Rev. Plant Biol. 53 329–355. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz, J.D., Straume, M., Stanewsky, R., Jamison, C.F., Brandes, C., Dowse, H., Hall, J.C., and Kay, S.A. (1997). Quantitative analysis of Drosophila period gene transcription in living animals. J. Biol. Rhythms 12 204–217. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Devlin, P.F., and Kay, S.A. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282 1488–1490. [DOI] [PubMed] [Google Scholar]
- Straume, M., Frasier-Cadoret, S.G., and Johnson, M.L. (1991). Least-Squares Analysis of Fluorescence Data. (New York: Plenum Press), pp. 171–240.
- Wang, H., Ma, L., Habashi, J., Li, J., Zhao, H., and Deng, X.W. (2002). Analysis of far-red light-regulated genome expression profiles of phytochrome A pathway mutants in Arabidopsis. Plant J. 32 723–733. [DOI] [PubMed] [Google Scholar]
- Whitelam, G.C., Johnson, E., Peng, J., Carol, P., Anderson, M.L., Cowl, J.S., and Harberd, N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J., Mazzella, M.A., Whitelam, G.C., and Casal, J.J. (2001). Resetting of the circadian clock by phytochromes and cryptochromes in Arabidopsis. J. Biol. Rhythms 16 523–530. [DOI] [PubMed] [Google Scholar]
- Yanovsky, M.J., Whitelam, G.C., and Casal, J.J. (2000). fhy3-1 retains inductive responses of phytochrome A. Plant Physiol. 123 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.