Abstract
Comparative studies require knowledge of the evolutionary relationships between taxa. However, neither morphological nor paleontological data have been able to unequivocally resolve the major groups of holometabolous insects so far. Here, we utilize emerging genome projects to assemble and analyze a data set of 185 nuclear genes, resulting in a fully resolved phylogeny of the major insect model species. Contrary to the most widely accepted phylogenetic hypothesis, bees and wasps (Hymenoptera) are basal to the other major holometabolous orders, beetles (Coleoptera), moths (Lepidoptera), and flies (Diptera). We validate our results by meticulous examination of potential confounding factors. Phylogenomic approaches are thus able to resolve long-standing questions about the phylogeny of insects.
The four major orders of holometabolous insects (Hymenoptera, Coleoptera, Lepidoptera, and Diptera) encompass over 45% of all known animal species (Hammond 1992). While analyses based on morphological (Kristensen 1999) or individual molecular markers (such as ribosomal RNA; Whiting 2002b) or mitochondrial DNA sequences (Castro and Dowton 2005) have confirmed the monophyly of these orders, they have been unable to elucidate most of the interordinal relationships with sufficient confidence.
A close relationship between Diptera (flies) and Lepidoptera (moths) within the long-recognized Mecopterida assemblage is generally recovered. However, the affinities of Coleoptera (beetles) and more particularly of Hymenoptera (wasps and bees) (Castro and Dowton 2005) remain elusive. In the most widely accepted phylogenetic hypothesis (Kristensen 1999; Whiting 2002b), a preference is given to a sister-group relationship between Hymenoptera and Mecopterida, while Coleoptera are placed at a more basal position as a sister group to the Neuropterida, another long-recognized assemblage.
To resolve the phylogenetic relationships of the major holometabolous orders, we adopt a phylogenomic approach, utilizing a large number of nuclear genes to maximize phylogenetic signal over noise (Eisen and Fraser 2003; Rokas et al. 2003; Delsuc et al. 2005; DeSalle 2005; Philippe et al. 2005a). Such approaches, based on the simultaneous analysis of a large number of nuclear genes, have already been shown to be a promising route to understand deep metazoan relationships (Dopazo and Dopazo 2005; Philippe et al. 2005b). Here, we demonstrate that these methods are also able to resolve long-standing questions about the phylogeny of insects.
Using EST sequences to obtain phylogenomic data sets has proven fruitful, e.g., in the analysis of Eukaryota (Philippe et al. 2004), Amoebae (Bapteste et al. 2002), and Coleoptera relationships (Hughes et al. 2006). The use of EST sequences in phylogenomic studies of insects was suggested earlier (Theodorides et al. 2002), but sufficient data to answer the questions addressed here has only recently become available.
Our analysis focuses on six holometabolous model species, for which large scale sequencing projects are available or in progress. These encompass two dipterans (the fruit fly Drosophila melanogaster and the mosquito Anopheles gambiae), one lepidopteran (the silk moth Bombyx mori), one coleopteran (the flour beetle Tribolium castaneum), and two hymenopterans (the honey bee Apis mellifera, and the sibling parasitic wasp species Nasonia vitripennis and Nasonia giraulti). We further include one orthopteran (the grasshopper Locusta migratoria) and one hemipteran (the pea aphid Acyrthosiphon pisum), both of which are uncon-tested outgroups to the holometabolous insects based on morphological and molecular markers (Boudreaux 1979; Hennig 1981; Kristensen 1991; Wheeler et al. 2001).
Results
Candidate orthologous clusters were assembled from known or predicted genes based on a stringent sequence similarity criterion, and were then manually curated to ensure orthology (see Methods). After removing ambiguously aligned regions, we assembled the remaining sequences into a concatenated alignment of 33,809 amino acid positions from 185 nuclear genes. As expected, most genes included here perform housekeeping functions (see Table S1 of the Supplemental information for a list of genes).
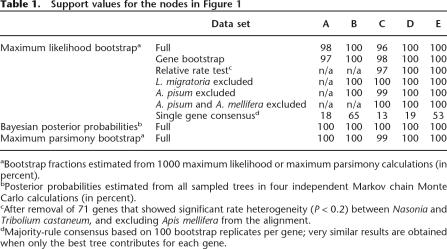
This data set supported the topology in Figure 1 regardless of the phylogenetic methodology (maximum likelihood [Guindon and Gascuel 2003], Bayesian [Yang and Rannala 1997], or maximum parsimony [Felsenstein 2004]), with nearly 100% bootstrap support or 100% posterior probabilities in each case (Table 1). The previously recognized close relationship of Diptera and Lepidoptera is recovered and thus substantiated. However, Hymenoptera and not Coleoptera is the most basal of the four major holometabolous orders. To ensure that our results were not influenced by an unusually evolving subset of sequences (Gadagkar et al. 2005), we utilized a bootstrap strategy based on the resampling of genes (Nei et al. 2001), again resulting in strong support for the topology in Figure 1 (Table 1).
Figure 1.
Holometabolous phylogenetic relationships, showing the monophyly of Coleoptera (beetles), Lepidoptera (moths), and Diptera (flies), to the exclusion of Hymenoptera (bees and wasps). Branch lengths are from maximum likelihood. Numbers report maximum likelihood bootstrap support (in percent); Bayesian posterior probabilities and maximum parsimony bootstrap support are ≥ 99% for each branch (Table 1).
Table 1.
Support values for the nodes in Figure 1
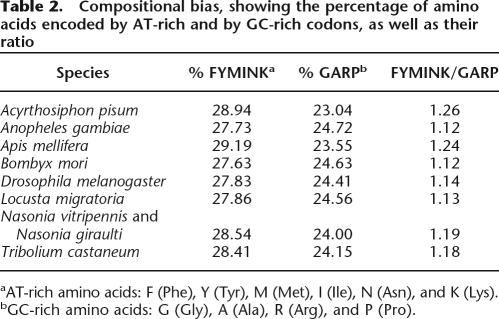
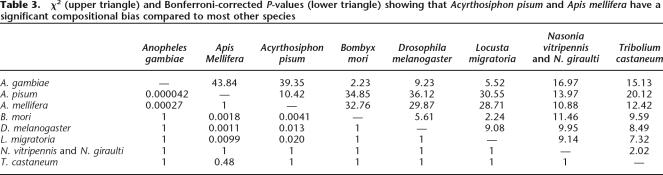
Two common sources of error in phylogenetic reconstructions are compositional biases (Foster and Hickey 1999) and long-branch attraction (Felsenstein 1978). Two of our species, the pea aphid Acyrthosiphon pisum and the honey bee Apis mellifera, have a strongly AT-biased genome. This is reflected in an overrepresentation of amino acids encoded by AT-rich codons (Table 2), confirmed by statistical analysis (pairwise χ2-tests, Table 3). The removal of the outlier species Acyrtho-siphon pisum and Apis mellifera from the data set resulted in the same well supported topology (Table 1).
Table 2.
Compositional bias, showing the percentage of amino acids encoded by AT-rich and by GC-rich codons, as well as their ratio settings, where all sequences flanking a
Table 3.
χ2 (upper triangle) and Bonferroni-corrected P-values (lower triangle) showing that Acyrthosiphon pisum and Apis mellifera have a significant compositional bias compared to most other species
Because long branches in the in-group are restricted to Diptera and Lepidoptera, whose relative positions are un-contested (Kristensen 1999; Whiting 2002b), long-branch attraction is unlikely to have influenced the topology. However, even though the branches of hymenopterans and Tribolium are short, it is still conceivable that genes with particular substitution rates in these species may have biased the phylogeny. Exclusion of such genes on the basis of a relative rate test (Tajima 1993) did not change the tree (Table 1).
Improper outgroup choice can potentially influence the inferred rooting of the ingroup. While our two outgroup species differ profoundly in evolutionary rate and amino acid composition, the results remained unchanged when using either Locusta migratoria or Acyrthosiphon pisum individually as the outgroup (Table 1).
Discussion
The interordinal relationships among holometabolous insect orders had previously proven to be notoriously difficult to resolve. However, most researchers assumed a basal split between two super-orders, the Coleoptera–Neuropterida (including the beetles) and the Hymenoptera–Mecopterida (including wasps, flies, and moths) (Kristensen 1999; Whiting 2002b). The tree presented in Figure 1 necessitates a re-evaluation of this consensual view. The Mecopterida, encompassing Diptera (flies) and Lepidoptera (moths), seem now more closely related to Coleoptera (beetles) than to Hymenoptera (wasps and bees).
In the present framework, the position of Neuropterida could not be assessed. Neither previous molecular phylogenies nor morphological characters allow settlement of this issue; in particular, wing structure features have been argued to support a sister-group relationship of Neuropterida with either Coleoptera (Hornschemeyer 2002) or with Mecopterida (Kukalová-Peck and Lawrence 2004). The morphological characters used to support the traditional Holometabola phylogeny should certainly be reanalyzed in the light of the relationships presented here.
Why was the basal position of hymenopterans not discovered in previous molecular phylogenetic studies? A plausible explanation is the lack of resolution power of single molecules when radiations are old or compressed in time (Rokas et al. 2005). Because the phylogenetic split in question occurred at least 275 million years ago (Mya) (Ponomarenko 2002; Rasnitsyn 2002), analyses based on a single molecule (e.g., 18S rRNA) did not provide sufficient resolution (Whiting 2002a). While 60% of the 185 protein alignments analyzed here were better explained by the tree in Figure 1 than by the previously assumed tree (based on likelihood comparisons), only two proteins supported the basal position of Hymenoptera with a bootstrap support >50% when analyzed individually (removing these proteins does not influence the tree topology, data not shown). Accordingly, a consensus tree based on single gene tree reconstructions yielded the same topology as in Figure 1, but without strong bootstrap support (Table 1). The present analysis thus supports the notion that concatenated sequence trees provide more resolution than consensus gene trees (Rokas et al. 2003; Gadagkar et al. 2005): Combined analysis of a large number of sequences was necessary to resolve the deep evolutionary relationships among holometabolous orders.
Previous studies based on the simultaneous analysis of many proteins also failed to recover the topology in Figure 1. Philippe et al. (2004) analyzed 129 proteins in a maximum likelihood framework, resulting in a strongly supported sister-group relationship between Apis mellifera and Bombyx mori, with Drosophila melanogaster located more basal. This topology is inconsistent with the monophyly of Mecopterida, which is well supported by morphological evidence (Kristensen 1999). It is most likely a long-branch attraction artifact, brought about by the insufficient number of holometabolous species and the use of a very divergent lineage (a tick) as the outgroup (Philippe and Laurent 1998). This study highlights the importance of selecting close outgroup species, and the necessity to test the influence of an alternative outgroup choice. A second study (Philippe et al. 2005b) recently analyzed 146 proteins, failing to resolve the relative positions of Coleoptera and Hymenoptera. Here, the potentially large amount of missing data between the insect species considered might be responsible for the lack of resolution.
The interordinal relationships presented in Figure 1 are in fact also suggested by examination of the fossil record (Rohdendorf and Rasnitsyn 1980), and are supported by phylogenetic analysis of intron positions (Krauss et al. 2005) as well as wing characters (Ross 1965; Kukalová-Peck and Lawrence 2004). Furthermore, we note that phylogenies based on 18S rRNA sequences also yielded a basal Hymenoptera among holometabolous insects, although not with a credible level of support (Whiting 2002b). The sum of evidence hence supports the present phylogenomic analysis as a reliable foundation for comparative analyses of the insect model organisms.
Methods
Sequence data
Drosophila melanogaster (Adams et al. 2000), Anopheles gambiae (Holt et al. 2002), and Apis mellifera peptides were obtained from Ensembl (www.ensembl.org). Bombyx mori (Mita et al. 2003), Locusta migratoria (Kang et al. 2004), Tribolium castaneum, and Acyrthosiphon pisum mRNA sequences were downloaded from NCBI (ftp.ncbi.nlm.nih.gov). Nasonia vitripennis and Nasonia giraulti EST data were generated by authors J.H.W. and H.T. All nucleotide data sets were cleaned of vector, mitochondrial and bacterial contaminations using SeqClean (available from www.tigr.org/tdb/tgi/software/) before being assembled into nonredundant contigs with cap3 using default settings (Huang and Madan 1999). All nucleotide data sets were then searched against all Drosophila melanogaster proteins using BLASTx. The reading frame from the best hit was assumed to be the correct reading frame. We then chose the longest run of peptides uninterrupted by a stop codon as the peptide corresponding to each nucleotide contig.
Identification of orthologs
We performed BLASTp searches of all proteome pairs. Orthologs were selected based on reciprocal best BLAST hits (Tatusov et al. 1997) using an E-value cut-off of 10−25. A group of sequences with exactly one member in each species (including either one or both Nasonia species) was accepted as a candidate orthologous family if each sequence had each of the other family sequences as the best BLASTp hit in the respective proteome. This requirement of all-against-all reciprocal best hits is very stringent, and thus gives good confidence in the inferred orthology. Multiple sequence alignments were performed with MUSCLE (Edgar 2004) using default settings. Resulting alignments were then manually curated to ensure completeness and consistency. Poorly conserved families or clusters potentially containing paralogous sequences were discarded. For the sibling Nasonia species, when orthologous sequences were available for both species, the longer one was chosen. Alignments were then purged from unreliably aligned positions as well as gaps with Gblocks (Castresana 2000) using highly stringent block had to be conserved and where blocks smaller than 20 amino acids were discarded. We concatenated the final set of 185 nuclear sequences for phylogenetic analysis, resulting in an eight-species alignment of 33,809 amino acid positions. The list of genes included in our analysis is available as Supplemental-Table S1.
Phylogenetic reconstructions
We first analyzed the data in a maximum likelihood framework, using phyML (Guindon and Gascuel 2003) under an empirical model of amino acid substitutions (Jones et al. 1992), allowing for substitution rate variation among sites with a gamma distribution (four rate categories). Branch support values in Table 1 are from analysis of 1000 bootstrap replicates. Using alternative models of amino acid evolution (WAG, Whelan and Goldman 2001; and VT, Bapteste et al. 2002) led to the same well supported tree.
Additionally, we estimated the tree in a Bayesian framework, using MrBayes (Huelsenbeck and Ronquist 2001) and employing the same model of sequence evolution as above. We ran four independent searches, each starting from a random tree and sampling every tenth tree over 100,000 generations. Each run had equilibrated after less than 1000 generations; thus, the first 100 trees were disregarded as burn-in. The independent runs consistently resulted in the same topology; posterior probabilities (Table 1) were calculated from all sampled trees across independent runs. Finally, we also analyzed 1000 bootstrap replicates under a maximum parsimony criterion, using PROTPARS from the PHYLIP package (Felsenstein 2004).
Gene bootstrap resampling
To test if the obtained tree was dominated by one or a few disparate genes, we performed maximum likelihood analyses of 1000 bootstrap data sets obtained from the resampling of complete genes (Nei et al. 2001). For each replicate, we drew 185 gene alignments from the full curated data set described above. Because genes were not removed from the pool after being chosen, each bootstrap data set contained some gene alignments more than once, while others were missing altogether; this is analogous to the widely used bootstrap strategy based on individual amino acid sites. For each replicate, alignments were then concatenated and analyzed with phyML as above.
Relative rate tests
To determine if the relative position of Coleoptera and Hymenoptera was caused by rate variation among these orders, we performed a relative rate test (Tajima 1993) between Nasonia and Tribolium castaneum. We restricted this analysis and the following tree reconstruction to sites that were identical between Locusta migratoria and Acyrthosiphon pisum, and where the ancestral state could thus be inferred reliably. To be conservative, we removed all genes for which ×2 > 1.64, i.e., P < 0.2 (1 degree of freedom) (Kumar and Hedges 1998). Excluding Apis mellifera, we concatenated the remaining 114 genes into an alignment of 16,495 amino acid positions. Maximum likelihood bootstrap analysis was performed as above.
Phylogenetic analysis of individual proteins
We also analyzed each individual protein alignment with the maximum likelihood method as described above. To analyze each protein's support for the tree in Figure 1 compared to the previously assumed tree (with the positions of hymenopterans and Tribolium exchanged), we compared the likelihoods calculated under each topology; statistical support was estimated using the method of Kishino and Hasegawa (1989) as implemented in the PAML package (Yang 1997).
Compositional heterogeneity among species pairs was assessed with a χ2-test, where χmn2=∑i(fmi−fni)2/(fmi+fni) with fmi the total number of amino acids of type i in the concatenated sequence for species m. The values in Table 3 are based on those amino acids that are biased in the GC content of their codons (FYMINK/GARP, see also Table 2; 9 degrees of freedom). Qualitatively very similar results are obtained when using all amino acids (data not shown).
All phylogenomic analyses and tests were implemented in Perl scripts, which are available in the Supplemental material.
Acknowledgments
J.H.W. thanks Wayne Hunter (USDA, ARS) and Phat Dang (USDA, ARS) for construction of the Nasonia EST libraries. We wish to thank Shannon K. McWeeney and Csaba Pal for helpful discussions. J.S. and D.T. acknowledge support through grants from the HFSPO and the DFG. M.J.L. acknowledges financial support from the Royal Society and the DFG. The Nasonia EST project of J.H.W. and H.T. was supported by the 21st Century Research & Technology Fund.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5204306.
References
- Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Li P.W., Hoskins R.A., Galle R.F., Hoskins R.A., Galle R.F., Galle R.F., et al. The genome sequence of Drosophila melanogaster . Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Bapteste E., Brinkmann H., Lee J.A., Moore D.V., Sensen C.W., Gordon P., Durufle L., Gaasterland T., Lopez P., Muller M., Brinkmann H., Lee J.A., Moore D.V., Sensen C.W., Gordon P., Durufle L., Gaasterland T., Lopez P., Muller M., Lee J.A., Moore D.V., Sensen C.W., Gordon P., Durufle L., Gaasterland T., Lopez P., Muller M., Moore D.V., Sensen C.W., Gordon P., Durufle L., Gaasterland T., Lopez P., Muller M., Sensen C.W., Gordon P., Durufle L., Gaasterland T., Lopez P., Muller M., Gordon P., Durufle L., Gaasterland T., Lopez P., Muller M., Durufle L., Gaasterland T., Lopez P., Muller M., Gaasterland T., Lopez P., Muller M., Lopez P., Muller M., Muller M., et al. The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba . Proc. Natl. Acad. Sci. 2002;99:1414–1419. doi: 10.1073/pnas.032662799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreaux H.B. Arthropod phylogeny with special reference to insecs. Wiley; New York: 1979. [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Castro L.R., Dowton M., Dowton M. The position of the Hymenoptera within the Holometabola as inferred from the mitochondrial genome of Perga condei (Hymenoptera: Symphyta: Pergidae) Mol. Phylogenet. Evol. 2005;34:469–479. doi: 10.1016/j.ympev.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Delsuc F., Brinkmann H., Philippe H., Brinkmann H., Philippe H., Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat. Rev. Genet. 2005;6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- DeSalle R. Animal phylogenomics: Multiple interspecific genome comparisons. Methods Enzymol. 2005;395:104–133. doi: 10.1016/S0076-6879(05)95008-8. [DOI] [PubMed] [Google Scholar]
- Dopazo H., Dopazo J., Dopazo J. Genome-scale evidence of the nematode-arthropod clade. Genome Biol. 2005;6:R41. doi: 10.1186/gb-2005-6-5-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J.A., Fraser C.M., Fraser C.M. Phylogenomics: Intersection of evolution and genomics. Science. 2003;300:1706–1707. doi: 10.1126/science.1086292. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 1978;27:401–410. [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 36. Department of Genome Sciences, University of Washington; Seattle, WA: 2004. [Google Scholar]
- Foster P.G., Hickey D.A., Hickey D.A. Compositional bias may affect both DNA-based and protein-based phylogenetic reconstructions. J. Mol. Evol. 1999;48:284–290. doi: 10.1007/pl00006471. [DOI] [PubMed] [Google Scholar]
- Gadagkar S.R., Rosenberg M.S., Kumar S., Rosenberg M.S., Kumar S., Kumar S. Inferring species phylogenies from multiple genes: Concatenated sequence tree versus consensus gene tree. J. Exp. Zool. (B Mol. Dev. Evol.) 2005;304:64–74. doi: 10.1002/jez.b.21026. [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hammond P. Species inventory. In: Groombridge B., editor. Global biodiversiy. Chapman & Hall; London: 1992. pp. 17–39. [Google Scholar]
- Hennig W. Insect phylogey. Wiley; New York: 1981. [Google Scholar]
- Holt R.A., Subramanian G.M., Halpern A., Sutton G.G., Charlab R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Subramanian G.M., Halpern A., Sutton G.G., Charlab R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Halpern A., Sutton G.G., Charlab R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Sutton G.G., Charlab R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Charlab R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Clark A.G., Ribeiro J.M., Wides R., Ribeiro J.M., Wides R., Wides R., et al. The genome sequence of the malaria mosquito Anopheles gambiae . Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Hornschemeyer T. Phylogenetic significance of the wing-base of the Holometabola (Insecta) Zool. Scr. 2002;31:17–29. [Google Scholar]
- Huang X., Madan A., Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Ronquist F., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hughes J., Longhorn S.J., Papadopoulou A., Theodorides K., de Riva A., Mejia-Chang M., Foster P.G., Vogler A.P., Longhorn S.J., Papadopoulou A., Theodorides K., de Riva A., Mejia-Chang M., Foster P.G., Vogler A.P., Papadopoulou A., Theodorides K., de Riva A., Mejia-Chang M., Foster P.G., Vogler A.P., Theodorides K., de Riva A., Mejia-Chang M., Foster P.G., Vogler A.P., de Riva A., Mejia-Chang M., Foster P.G., Vogler A.P., Mejia-Chang M., Foster P.G., Vogler A.P., Foster P.G., Vogler A.P., Vogler A.P. Dense taxonomic EST sampling and its applications for molecular systematics of the Coleoptera (beetles) Mol. Biol. Evol. 2006;23:268–278. doi: 10.1093/molbev/msj041. [DOI] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M., Taylor W.R., Thornton J.M., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kang L., Chen X., Zhou Y., Liu B., Zheng W., Li R., Wang J., Yu J., Chen X., Zhou Y., Liu B., Zheng W., Li R., Wang J., Yu J., Zhou Y., Liu B., Zheng W., Li R., Wang J., Yu J., Liu B., Zheng W., Li R., Wang J., Yu J., Zheng W., Li R., Wang J., Yu J., Li R., Wang J., Yu J., Wang J., Yu J., Yu J. The analysis of large-scale gene expression correlated to the phase changes of the migratory locust. Proc. Natl. Acad. Sci. 2004;101:17611–17615. doi: 10.1073/pnas.0407753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino H., Hasegawa M., Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J. Mol. Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- Krauss V., Pecyna M., Kurz K., Sass H., Pecyna M., Kurz K., Sass H., Kurz K., Sass H., Sass H. Phylogenetic mapping of intron positions: A case study of translation initiation factor eIF2γs. Mol. Biol. Evol. 2005;22:74–84. doi: 10.1093/molbev/msh255. [DOI] [PubMed] [Google Scholar]
- Kristensen N.P. The phylogeny of hexapods. In: Naumann I.D., editor. The insects of Australia: A Textbook for students and research workes. Melbourne University Press; Carlton, Australia: 1991. pp. 125–140. [Google Scholar]
- Kristensen N.P. Phylogeny of endopterygote insects, the most successful lineage of living organisms. Eur. J. Entomol. 1999;96:237–253. [Google Scholar]
- Kukalová-Peck J., Lawrence J.F., Lawrence J.F. Relationships among coleopteran suborders and major endoneopteran lineages: Evidence from hind wing characters. Eur. J. Entomol. 2004;101:95–144. [Google Scholar]
- Kumar S., Hedges S.B., Hedges S.B. A molecular timescale for vertebrate evolution. Natur. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Mita K., Morimyo M., Okano K., Koike Y., Nohata J., Kawasaki H., Kadono-Okuda K., Yamamoto K., Suzuki M.G., Shimada T., Morimyo M., Okano K., Koike Y., Nohata J., Kawasaki H., Kadono-Okuda K., Yamamoto K., Suzuki M.G., Shimada T., Okano K., Koike Y., Nohata J., Kawasaki H., Kadono-Okuda K., Yamamoto K., Suzuki M.G., Shimada T., Koike Y., Nohata J., Kawasaki H., Kadono-Okuda K., Yamamoto K., Suzuki M.G., Shimada T., Nohata J., Kawasaki H., Kadono-Okuda K., Yamamoto K., Suzuki M.G., Shimada T., Kawasaki H., Kadono-Okuda K., Yamamoto K., Suzuki M.G., Shimada T., Kadono-Okuda K., Yamamoto K., Suzuki M.G., Shimada T., Yamamoto K., Suzuki M.G., Shimada T., Suzuki M.G., Shimada T., Shimada T., et al. The construction of an EST database for Bombyx mori and its application. Proc. Natl. Acad. Sci. 2003;100:14121–14126. doi: 10.1073/pnas.2234984100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Xu P., Glazko G., Xu P., Glazko G., Glazko G. Estimation of divergence times from multiprotein sequences for a few mammalian species and several distantly related organisms. Proc. Natl. Acad. Sci. 2001;98:2497–2502. doi: 10.1073/pnas.051611498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H., Laurent J., Laurent J. How good are deep phylogenetic trees? Curr. Opin. Genet. Dev. 1998;8:616–623. doi: 10.1016/s0959-437x(98)80028-2. [DOI] [PubMed] [Google Scholar]
- Philippe H., Snell E.A., Bapteste E., Lopez P., Holland P.W., Casane D., Snell E.A., Bapteste E., Lopez P., Holland P.W., Casane D., Bapteste E., Lopez P., Holland P.W., Casane D., Lopez P., Holland P.W., Casane D., Holland P.W., Casane D., Casane D. Phylogenomics of eukaryotes: Impact of missing data on large alignments. Mol. Biol. Evol. 2004;21:1740–1752. doi: 10.1093/molbev/msh182. [DOI] [PubMed] [Google Scholar]
- Philippe H., Delsuc F., Brinkmann H., Lartillot N., Delsuc F., Brinkmann H., Lartillot N., Brinkmann H., Lartillot N., Lartillot N. Phylogenomics. Annu. Rev. Ecol. Evol Syst. 2005a;36:541–562. [Google Scholar]
- Philippe H., Lartillot N., Brinkmann H., Lartillot N., Brinkmann H., Brinkmann H. Multigene analyses of bilaterian animals corroborate the monophyly of Ecdysozoa, Lophotrochozoa, and Protostomia. Mol. Biol. Evol. 2005b;22:1246–1253. doi: 10.1093/molbev/msi111. [DOI] [PubMed] [Google Scholar]
- Ponomarenko A.G. Superorder Scarabaeidea Laicharting, 1781. Order Coleoptera Linné, 1758. The Beetles. In: Rasnitsyn A.P., Quicke D.L.J., Quicke D.L.J., editors. History of insecs. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2002. pp. 164–176. [Google Scholar]
- Rasnitsyn A.P. Cohors Scarabaeiformes Laicharting, 1781. The Holometabolans. In: Rasnitsyn A.P., Quicke D.L.J., Quicke D.L.J., editors. History of insects. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2002. pp. 157–159. [Google Scholar]
- Rohdendorf B.B., Rasnitsyn A.P., Rasnitsyn A.P. Historical development of the class insecta. Nauka Press; Moscow: 1980. [Google Scholar]
- Rokas A., Williams B.L., King N., Carroll S.B., Williams B.L., King N., Carroll S.B., King N., Carroll S.B., Carroll S.B. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- Rokas A., Kruger D., Carroll S.B., Kruger D., Carroll S.B., Carroll S.B. Animal evolution and the molecular signature of radiations compressed in time. Science. 2005;310:1933–1938. doi: 10.1126/science.1116759. [DOI] [PubMed] [Google Scholar]
- Ross H.H. A textbook of entomoloy. Wiley; New York: 1965. [Google Scholar]
- Tajima F. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R.L., Koonin E.V., Lipman D.J., Koonin E.V., Lipman D.J., Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Theodorides K., De Riva A., Gomez-Zurita J., Foster P.G., Vogler A.P., De Riva A., Gomez-Zurita J., Foster P.G., Vogler A.P., Gomez-Zurita J., Foster P.G., Vogler A.P., Foster P.G., Vogler A.P., Vogler A.P. Comparison of EST libraries from seven beetle species: Towards a framework for phylogenomics of the Coleoptera. Insect Mol. Biol. 2002;11:467–475. doi: 10.1046/j.1365-2583.2002.00355.x. [DOI] [PubMed] [Google Scholar]
- Wheeler W.C., Whiting M., Wheeler Q.D., Carpenter J.M., Whiting M., Wheeler Q.D., Carpenter J.M., Wheeler Q.D., Carpenter J.M., Carpenter J.M. The phylogeny of the extant hexapod orders. Cladistics. 2001;17:113–169. doi: 10.1111/j.1096-0031.2001.tb00115.x. [DOI] [PubMed] [Google Scholar]
- Whelan S., Goldman N., Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Whiting M.F. Phylogeny of the holometabolous insect orders based on 18S ribosomal data: When bad things happen to good data. In: DeSalle G.G.R., Wheeler W.C., Wheeler W.C., editors. Molecular systematics and evolution:Theory and practice. Birkhäuser Verlag AG; Basel, Switzerland: 2002a. pp. 69–83. [DOI] [PubMed] [Google Scholar]
- Whiting M.F. Phylogeny of the holometabolous insect orders: Molecular evidence. Zool. Scr. 2002b;31:3–15. [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z.H., Rannala B., Rannala B. Bayesian phylogenetic inference using DNA sequences: A Markov Chain Monte Carlo method. Mol. Biol. Evol. 1997;14:717–724. doi: 10.1093/oxfordjournals.molbev.a025811. [DOI] [PubMed] [Google Scholar]