Abstract
The current insect genome sequencing projects provide an opportunity to extend studies of the evolution of developmental genes and pathways in insects. In this paper we examine the conservation and divergence of genes and developmental processes between Drosophila and the honey bee; two holometabolous insects whose lineages separated ∼300 million years ago, by comparing the presence or absence of 308 Drosophila developmental genes in the honey bee. Through examination of the presence or absence of genes involved in conserved pathways (cell signaling, axis formation, segmentation and homeobox transcription factors), we find that the vast majority of genes are conserved. Some genes involved in these processes are, however, missing in the honey bee. We have also examined the orthology of Drosophila genes involved in processes that differ between the honey bee and Drosophila. Many of these genes are preserved in the honey bee despite the process in which they act in Drosophila being different or absent in the honey bee. Many of the missing genes in both situations appear to have arisen recently in the Drosophila lineage, have single known functions in Drosophila, and act early in developmental pathways, while those that are preserved have pleiotropic functions. An evolutionary interpretation of these data is that either genes with multiple functions in a common ancestor are more likely to be preserved in both insect lineages, or genes that are preserved throughout evolution are more likely to co-opt additional functions.
Comparisons of the sequenced genomes of Drosophila, Caenorhabditis, and vertebrates have revealed that many developmental genes and pathways are conserved among animals. These studies tell us little, however, about the novel and rapidly evolving developmental pathways and genes that are likely to encode the evolutionary novelties that are unique in each species. In insects this problem has been severe as, until the recent sequencing of the honey bee (Apis mellifera), Tribolium, Bombyx, and Drosophila species’ genomes, the only two sequenced genomes were Drosophila melanogaster (Adams et al. 2000) and Anopheles gambiae (Holt et al. 2002), both Dipterans. Honey bees diverged from Dipterans ∼300 million years ago (Hennig 1981), and recent phylogenetic evidence implies that the Hymenoptera are the most distant group of holometabolous insects from Drosophila (Whiting 2002; Krauss et al. 2005).
Despite honey bees being an economically important species, few studies of their development have been published. Morphological and classical manipulative studies have been carried out, but studies of gene function or expression are rare. Those studies published have concentrated on genes associated with segmentation (Fleig 1990; Binner and Sander 1997; Osborne and Dearden 2005a), Hox genes (Walldorf et al. 1989, 2000; Fleig et al. 1992), and sex-determining genes (Beye et al. 2003). Recently techniques for studying honey bee development have become available, including robust methods for studying gene expression patterns (Osborne and Dearden 2005b), RNAi to study gene function in both embryos and adults (Beye et al. 2002; Amdam et al. 2003), and cell culture protocols (e.g., see Bergem et al. 2006). These, combined with the genome sequence, will facilitate developmental studies in this genetically, socially, and economically important insect.
Superficially honey bee development is similar to that of Drosophila, in that it is a holometabolous, long-germ-band insect. However, honey bees are different in their development from the Diptera in a number of ways. Honey bee embryos have no pole plasm or morphologically distinct early-segregating germ cells (Nelson 1915; DuPraw 1967; Fleig and Sander 1986; Dearden 2006); they do not undergo germ band extension or head involution, and anterior–posterior progression of patterning during segmentation plays a much larger role than in Diptera (Osborne and Dearden 2005b). Furthermore, honey bees use haplodiploidy to determine sex, a process different from that of sex determination in Drosophila. The adults have several novel evolutionary innovations not present in Dipterans, including poison organs and wax glands. Most important are the polyphenisms associated with the social nature of the honey bee.
In this paper we present initial characterization of developmental genes in the honey bee genome by homology searches and, in some cases, expression studies. The genes we present here are either involved in fundamental and conserved biological processes among insects such as axis formation or signaling pathways, or are associated with derived biological processes such as sex determination, male meiosis, dosage compensation, and germ cell segregation. These studies reveal different levels of conservation in genes underlying both conserved and divergent biological processes.
Results
Genes and pathways of developmental processes that are expected to be conserved
Many of the fundamental pathways and genes that regulate development in insects are expected to be conserved in the honey bee as they are involved in formation of the basic body plan. Pathways and genes such as ancient cell signaling cascades, axis formation, segmentation, and the Hox genes might be expected to be as conserved in the honey bee as they are in other insects (Baron 2003; Logan and Nusse 2004; Bejsovec 2005; Hooper and Scott 2005; Pearson et al. 2005; Peel et al. 2005). To test this, we have examined the presence or absence of genes involved in these processes, and in some cases examined their expression.
Developmental signaling pathways
Several cell signaling pathways have been found in both vertebrates and invertebrates. The components of these pathways are highly conserved, and the pathways themselves are deployed at numerous times and places during development (Pires-daSilva and Sommer 2003). We have examined components of Wnt, Hedgehog (Hh), Notch, and Decapentaplegic (Dpp) signaling.
Wnt signaling is involved in embryogenesis and imaginal disc development in flies, while in vertebrates it is implicated in gastrulation, mesoderm development, cancer, and other developmental processes (Logan and Nusse 2004).
While the honey bee and Drosophila genomes have the same number of Wnt genes, these genes are from different subfamilies. The honey bee and the fruit fly share Wnt1, 5, 6, 7, and 10. In addition, the fly genome contains Wnt9 and D. The honey bee, however, also has Wnt4 and 11. Ancestrally, bilaterian animals had 11 Wnts (Kusserow et al. 2005), implying that Wnt genes have been lost differentially from flies and honey bees. The honey bee genome encodes fewer Wnt receptors than Drosophila. Flies have four Frizzled and three Derailed receptors, while honey bees have two Frizzled and two Derailed receptors. This implies that multiple Wnts function as ligands for a single receptor in honey bees.
Despite these differences, many features of Wnt signaling are conserved between honey bees and fruit flies. The chromosomally-linked, evolutionarily-conserved cluster of Wnt1-Wnt6-Wnt10 (Nusse 2001) is found in the honey bee genome. Furthermore, all downstream components of canonical and noncanonical Wnt signaling (Bejsovec 2005) are conserved in the honey bee (Supplemental Table 1).
In Drosophila and vertebrates, Notch signaling is required for a wide range of processes including neurogenesis and, in vertebrates, somitogenesis (Baron 2003). All of the genes of the Notch signaling cascade are present in the honey bee. The only differences in Notch signaling components between honey bees and Drosophila are in the E(spl) complex, a complex of genes that act as effectors of active Notch signaling. The number of genes in the honey bee complex is reduced relative to those of Dipterans as described by Schlatter and Maier (2005).
Hh and Dpp (the Drosophila homolog of vertebrate BMP2 and BMP4) signaling are also involved in a number of developmental processes. Hh signaling regulates embryogenesis and imaginal disc development in flies and the development of a wide range of tissues in vertebrates (Hooper and Scott 2005). Dpp signaling regulates imaginal disc development in Drosophila and dorso–ventral patterning in both flies and vertebrates (Raftery and Sutherland 1999). The genes of both the Hh and Dpp signal transduction machineries are conserved in honey bees (Supplemental Table 1).
Axis formation
In Drosophila the terminal patterning system acts to determine the anterior-most and posterior-most cells of the embryo (Furriols et al. 1998). In the ovary, Torso-like acts on a ligand, trunk (Trk), to allow it to interact with its receptor, torso (Tor) (Casali and Casanova 2001). Torso is a receptor tyrosine kinase (RTK) that signals to the nucleus to de-repress tailless (Liaw et al. 1995) in anterior and posterior regions of the embryo. This pathway appears to be conserved in the beetle Tribolium, where it acts to regulate the posterior growth zone (Schroder et al. 2000; Schoppmeier and Schroder 2005).
In the honey bee, two key components of this system, trk and tor, are missing. TBLASTX searches using Drosophila trk indicate that no similar sequences are present in the honey bee genome. A neighbor-joining cladogram of all predicted honey bee RTKs with Drosophila RTKs also indicates that no ortholog for tor is present (Supplemental Fig. 1). All the other components of the terminal system are present. To determine if the lack of trk and tor is honey bee specific, we searched the still incomplete Bombyx mori genome for homologs. Neither of these genes could be found. The absence of trk and tor implies that honey bees, and perhaps all Hymenoptera, use a different pathway to regulate terminal patterning from that of Drosophila and Tribolium.
In Drosophila dorso–ventral (D/V) patterning, gurken (grk) RNA is localized in anterior-ventral regions of the oocyte and when translated, activates the EGF receptor Torpedo in overlying follicle cells (Schüpbach and Roth 1994). These cells then signal back to the oocyte through Spätzle (Spz) and its receptor Toll (Tl), activating the transcription factor Dorsal. grk is not present in the honey bee, Tribolium, or Bombyx genomes, suggesting that grk may be an invention of the Diptera. Tl, EGF-R, and spz are encoded in the honey bee genome. In the honey bee, Tl RNA is expressed in a pattern consistent with a role in D/V patterning in honey bees (Fig. 1) but seems to be limited to an anterior ventral domain, a much smaller region of expression than in Drosophila, where it is ubiquitous (Gay and Keith 1992), or Tribolium, where it is expressed in a ventral-to-dorsal gradient in blastoderm-stage embryos (Maxton Küchenmeister et al. 1999). These differences imply a slightly different mode of dorso–ventral patterning in the honey bee, perhaps one in which the activity of Tl is limited to only anterior trunk regions of the embryo. Despite these differences in the initial parts of the cascade, gene expression patterns regulated by the D/V cascade (e.g., snail) are similar to those in Drosophila (Fig. 1). It seems likely that grk has been co-opted into the Drosophila dorso–ventral patterning system that is constructed of genes present in the honey bee and other insects.
Figure 1.
Expression patterns of genes involved in dorso–ventral patterning. Embryos are oriented with anterior-left, dorsal-up. Scale bars, 100 μm. (A) Expression of honey bee Tl RNA in a stage 2 embryo. Honey bee Tl is expressed only in anterior/ventral regions of the embryo. (B) Expression of honey bee snail RNA in a stage 3 embryo.
In Drosophila, RNA localization in the oocyte plays a key role in specifying the anterior and posterior axes. In the anterior, bicoid (bcd) RNA is localized via the activity of exuperantia (exu), swallow (swa), and staufen (stau) gene products and determines anterior regions of the embryo (MacDonald et al. 1995). bcd is missing from the honey bee genome, a not unexpected finding given previous studies indicating that this gene evolved its anterior role in Diptera (Stauber et al. 2000, 2002). Its absence in the honey bee implies that other genes must carry out its function, possibly orthodenticle and Hunchback as has been shown in Tribolium (Schroder 2003) and Nasonia (Pultz et al. 2005). Both of these genes are present in the honey bee genome. Of the genes involved in bcd RNA localization, swa is missing, but exu and stau are present.
In Drosophila, the posterior localization of proteins and RNA is required for posterior patterning and germ-cell development. These components are localized through interactions with Oskar protein, which acts as a “pole plasm anchor” (Lehmann and Nusslein-Volhard 1986). Oskar (osk) has no homolog in the Apis, Tribolium, or Bombyx genomes. The honey bee genome does, however, contain homologs of all the proteins and RNAs that require Oskar for their localization (Supplemental Table 1). The genomic data imply that osk evolved in the Diptera to regulate pole-plasm assembly.
Segmentation
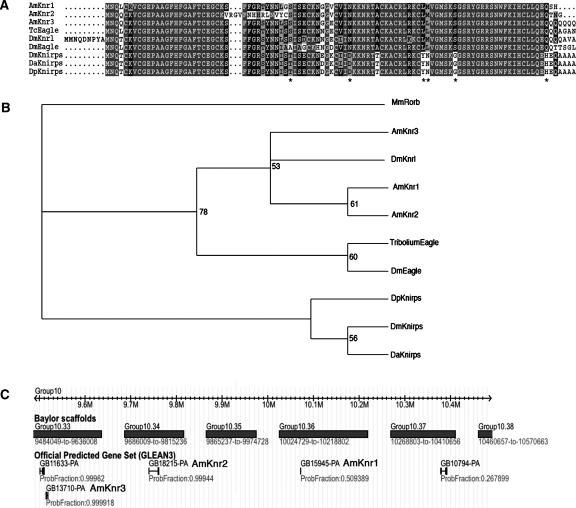
Apart from the genes that act maternally in Drosophila (discussed above), the honey bee genome has orthologs of almost all the major genes in the Drosophila segmentation pathway including gap, pair-rule, and segment polarity genes (Supplemental Table1). The only difference is in a gap gene, knirps (kni). Drosophila kni encodes a zinc finger transcriptional repressor (Nauber et al. 1988). The Drosophila genome encodes two additional genes, knirps-related (knrl) and eagle (egon), but only kni appears to play a role in segmentation (Gonzalez-Gaitan et al. 1994). The honey bee genome contains three knirps-like sequences (Fig. 2), but phylogenetic analyses imply that all three genes encode proteins that are more closely related to knrl and egon than to kni. Both the Drosophila (Rothe et al. 1989) and honey bee genes are clustered in the genome (Fig. 2C). Preliminary in situ hybridization data indicate that none of these honey bee genes are expressed during segmentation (M.J. Wilson and P.K. Dearden, unpubl.). These data imply that knirps-like genes have duplicated in various insect lineages and that one of these, kni has been co-opted into segmentation in the Diptera.
Figure 2.
The honey bee genome contains three knirps-related genes. (A) Amino acids sequence comparison of the Zn finger domain from Knirps-related proteins including those predicted from the honey bee genome sequence. Asterisks indicate residues that are conserved in kni but not egon proteins; (Dp) Drosophila pseudoobscura. (B) Phylogenetic analysis of the Cys–Cys finger domains from Knirps-related proteins. kni from D. melanogaster forms a clade with kni proteins from other Drosophila species. (C) Genomic location of honey bee knirps-related genes. The three predicted knirps-related genes (knr1, knr2, and knr3) are clustered on Baylor scaffold group 10.
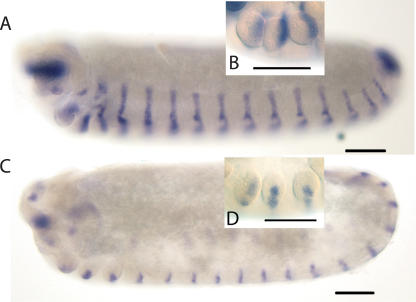
The final stage in the establishment of the parasegments requires the segment polarity genes that encode components of the Wnt and Hh cell signaling pathways and the transcription factor engrailed (en), of which the honey bee has two (e30 and e60) (Walldorf et al. 1989; Fleig 1990). In Drosophila, these genes initiate and maintain parasegment boundaries with wg being expressed just anterior to the parasegment boundary and en and hh just posterior. These expression patterns are conserved in the honey bee (Fig. 3). These data, combined with depletion of en by RNAi (Beye et al. 2002), imply that parasegment boundaries are conserved in the honey bee, consistent with analyses of more basal arthropods (Damen 2002; Dearden et al. 2002).
Figure 3.
Expression of honey bee hh and wingless (wg) in embryos. Embryos are oriented anterior-left, dorsal-up. Scale bars, 100 μm. (A) hh RNA expression in a stage 10 embryo. Honey bee hh is expressed in a stripe of cells in the posterior of each segment, a pattern similar to the expression of engrailed-like proteins (Fleig 1990) and e30 and e60 mRNA (Osborne and Dearden 2005b). (B) Magnification of the gnathal segments of a stage 10 embryo stained for honey bee hh RNA. hh RNA is located in the posterior of each limb bud. (C) Honey bee wg RNA expression in a stage 9 embryo. wg RNA is expressed in a stripe of cells in each segment. (D) Magnification of the gnathal segments of a stage 9 embryo stained for wg RNA showing expression in a stripe of cells running across the middle of each limb bud.
Homeobox-containing genes
Manually annotated homeobox-containing genes are listed in Supplemental Table 2. We found 74 genes that are predicted to encode proteins containing homeodomains, either alone or in combination with PAX, POU, LIM, and other domains. Two additional Pax family genes (Pax2/5/8 and Pax1/9) lacked homeoboxes, but were included in this analysis because they derive from homeobox genes (Miller et al. 2000).
The 78 homeobox and Pax genes include representatives of all major homeodomain classes. We identify 35 genes within the ANTP class, 19 within the PRD class (including seven Pax genes), four POU genes, eight LIM genes, two CUT genes, three TALE genes, two PROS genes, three SINE genes, and two ZFH genes. Two of these genes, assigned to the LIM and ZFH classes, contain more than one homeobox sequence.
The Hox complex
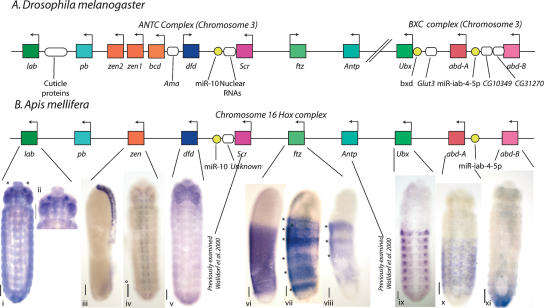
The honey bee Hox gene cluster is located on chromosome 16 and covers ∼1.37 Mb. The honey bee Hox cluster has the characteristics expected of a canonical insect Hox gene cluster. First, similar to Tribolium (Brown et al. 2002), the honey bee complex is not split in two as in Drosophila and Bombyx (Lewis et al. 2003; Yasukochi et al. 2004). Second, it contains all 10 of the expected Hox genes (Fig. 4). Third, all the genes are transcribed from the same strand, indicating lack of inversions in the cluster.
Figure 4.
Organization of the honey bee Hox complex and expression of Hox genes. Illustration (not to scale) of Hox complexes from (A) D. melanogaster and (B) honey bee. Homeobox genes are shown as colored rectangles with arrows indicating the direction of transcription. Non-homeobox genes are shown as ovals, non-protein-coding genes as yellow circles. (B, i–x) In situ hybridization to honey bee embryos using Hox gene probes. Embryos are oriented with anterior up. Scale bars, 100 μm. (i and ii) Honey bee labial (lab) expression in a stage 10 embryo. lab is expressed in small, paired subsets of cells (asterisks) in the intercalary segment (magnified in ii). (iii and iv) Honey bee zen expression in a stage 3 embryo (iii, lateral view) and a stage 10 embryo (iv, ventral view). In early development, zen is expressed in the dorsal extraembryonic membranes. This persists until stage 9. Expression is then seen in cells of the CNS (iv). (v) Honey bee dfd expression in a stage 9 embryo (ventral view). Dfd is expressed in the mandibular and maxillary segments, especially in the limb buds. (vi and viii) Expression of honey bee ftz in stages 4 (vi and vii) and 5 (viii) (lateral views). Cells in a broad trunk domain express honey bee ftz. Expression in this domain fades, forming broad stripes (vii) at a spacing consistent with double segment periodicity (Osborne and Dearden 2005a). In viii, three broad stripes are visible (asterisks), but the embryo is damaged at the posterior end. (ix) Expression of honey bee Ultrabithorax (Ubx) RNA. Ubx is expressed in cells from A1 to A8. (x) Expression of honey bee abd-A RNA in a stage 10 embryo. abd-A is expressed in segments A2–A8 more broadly than Ubx. (xi) Expression of honey bee Abd-B RNA in a stage 9 embryo. Abd-B is expressed in segments A9–A10, but in later stages comes to be expressed in the posterior edges of A7 and A8 (data not shown).
The major difference between the honey bee Hox complex and the Hox complexes of other insects is its size (1.37 Mb). Hox clusters from other insects range in size from Drosophila (0.66: ANT-C [∼0.34 Mb] + BX-C [∼0.32 Mb], respectively) (Drysdale and Crosby 2005), through Tribolium (0.7 Mb) (http://www.bioinformatics.ksu.edu/beetlebase) and Schistocerca (>0.7 Mb) (Ferrier and Akam 1996), to Anopheles at 1.18 Mb (Anobase, 2006; http://www.anobase.org/). The large honey bee complex is not explained by genes inserted into the complex as only a single gene (novel, nonhomeobox) with EST expression evidence, found between Deformed (Dfd) and Sex combs reduced (Scr) and transcribed on the same strand, has been detected. Insertion of transposable elements also does not explain the complex size as only three have been found, all of which are mariner elements, two in an intron of proboscipedia (pb) and one 5′ of pb. The size of the complex can be explained by larger intergenic regions relative to those in other Hox clusters. Excluding gene duplications and splits in complexes, the average sizes of intergenic regions are D. melanogaster, 47,699 bp; Anopheles, 99,897 bp; Tribolium, 84,141 bp; and Apis, 115,890 bp.
Phylogenetic analysis of the predicted genes in the honey bee Hox complex with those from the Drosophila ANTC and BXC indicates a 1:1 orthology between honey bee and Drosophila Hox cluster genes (Supplemental Fig. 2). Hox complexes of insects also encode two microRNAs (Pearson et al. 2005): mir10, which lies between Dfd and Scr, and miR-iab-4-5p, between abdominal-A (abd-A) and Abdominal-B (Abd-B). Both of these sequences and their locations are conserved in the honey bee.
To determine if the collinear expression of Hox genes is conserved in the honey bee, we examined the RNA expression pattern of the honey bee Hox genes that have not been published previously (Fig. 4). All the genes are expressed in patterns consistent with their expression in other insects. Only a homolog of pb has not been examined, because of technical difficulties.
Two Hox genes, referred to as the rogue Hox genes (Hughes and Kaufman 2002), have evolved non-homeotic functions in insects, but still lie within the Hox complex. The first is Hox3/zen, which has evolved extraembryonic membrane functions in insects (Falciani et al. 1996; Dearden et al. 2000) and has also generated the anterior patterning gene bcd by duplication in Diptera (Stauber et al. 1999). The honey bee has only one Hox3/zen gene, probably representing an ancestral gene that was duplicated independently in Tribolium to give two genes (Brown et al. 2002) and in Diptera (to give bcd, zen, and zen2). Honey bee zen is expressed in the extraembryonic membranes of the early honey bee embryo (Fig. 4) as found in other insects, and in the CNS in late honey bee embryos. CNS expression is not reported for zen genes in Tribolium (Falciani et al. 1996; van der Zee et al. 2005), Diptera (Doyle et al. 1989), or Schistocerca (Dearden et al. 2000), implying a novel function in the honey bee.
The second rogue Hox gene, fushi tarazu (ftz), acts in Drosophila in segmentation and nervous system patterning. ftz has been reported to be absent from the honey bee on the basis of library screening (Walldorf et al. 1989). Despite this, a gene with similarity to ftz is present. The RNA expression pattern of the honey bee ftz homolog is consistent with a putative pair-rule segmentation function (Fig. 4), but this needs to be confirmed by costaining with either a segmental marker or a gene with an unambiguous pair-rule expression pattern.
As in Drosophila, the ParaHox genes intermediate neuroblasts defective (ind or Gsx) and caudal are unlinked in the honey bee, consistent with the hypothesized breakup of the ParaHox cluster in arthropods and their relatives (Ferrier and Holland 2001).
The NK homeobox complex
Mirroring the situation in other insects (Jagla et al. 2001; Luke et al. 2003), several NK class homeobox genes are also clustered in the honey bee genome. Within 300 kb, we find linkage between Lbx, NK3, NK4, and a pair of Msx genes on chromosome 1. This may not represent the full extent of the NK homeobox gene cluster in the honey bee, as there is a Tlx ortholog on an un-mapped scaffold. The NK1 ortholog is not part of the cluster, and is on chromosome 15. The presence of Msx within the NK homeobox gene cluster mirrors the situation in A. gambiae, but differs from that in D. melanogaster (Luke et al. 2003). We conclude that Msx was ancestrally part of the NK cluster, and that a transposition or inversion event has removed it in Drosophila.
Developmental processes that are missing or novel in the honey bee
A notable difference between the honey bee and Drosophila lies in their modes of reproduction. Honey bee males arise from unfertilized eggs that are haploid, while Drosophila males are derived from fertilized diploid eggs. As a consequence of haplo-diploidy, bees have a divergent system of sex determination, lack sex chromosomes and dosage compensation, and have no meiosis in males (Supplemental Fig. 3). In addition, germ-cell development is different from that of Drosophila (Dearden 2006).
Sex determination
Sex in the bee is determined by the allelic composition of a single locus (complementary sex determination, csd) instead of sex chromosomes (Beye et al. 2003). Eggs develop into males when csd is hemizygous or homozygous, while females arise when csd is heterozygous. In Drosophila, the ratio of sex chromosomes to autosomes determines sex. In the honey bee, despite the lack of sex chromosomes, putative orthologs of the genes in Drosophila that assess the sex chromosome to autosome ratio are present—runt, scute, and deadpan—while Sisterless A is missing. Genes that are involved with the transfer of the sex-specific X:A signal to the next downstream gene, Sex lethal (Sxl), have orthologs (groucho, daughterless) in the bee but no extra macrochaetae or hermaphrodite homologs are present. In Drosophila, Sxl, the direct target of the X:A signal, acts as a switch: on in females, off in males. Sxl controls sexual differentiation and dosage compensation and maintains the male/female state during development in the fly. Despite its central role in fly sex determination, Sxl has no obvious sex-determining function in the bee despite being conserved.
Cofactors, sans fille (snf), virilizer (vir), and female lethal d [fl(2)d], which are not sex regulated but are required for Sxl function, also have orthologs in the honey bee. The direct target of Sxl is transformer (tra). tra is only active in females and controls somatic sex differentiation, but has no ortholog in the honey bee. However, the direct partner of tra, transformer-2, has an ortholog. csd, the initial signal that governs sex determination, is thought to be functionally equivalent at the level of tra (Beye et al. 2003). csd, like tra in the fly, acts as a switch, active in females and inactive in males. Downstream of tra in Drosophila are doublesex (dsx) and intersex, which have orthologs in the honey bee. Honey bee dsx is sex-specifically spliced (Beye et al. 2003), consistent with a conserved sex-determining function. This implies that these divergent pathways functionally converge at the level of the dsx gene, supporting the “bottom-up” hypothesis of sex-determining pathways postulated by Wilkins (1995). Members of the pathway that controls gonad development in the fly also have orthologs in the honey bee (ovarian tumor, Mes-4, ovo, snf).
Dosage compensation
In Drosophila males, the single X chromosome is compensated for by a twofold increase in its transcription, a process under the control of the sex-determining cascade (Cline and Meyer 1996). Sxl, in combination with fl(2)d and vir, controls dosage compensation via male specific lethal-2 (msl-2). msl-2 is not present in the honey bee, although orthologs of some (maleless, males absent on the first, male specific lethal-3, and Trithorax-like) but not all (male specific lethal-1 or roX1 and roX2 RNA encoding genes) dosage compensation genes are present.
Meiosis
Sperm form in the honey bee via mitotic segregation of chromosomes rather than meiosis. Seven genes in D. melanogaster were identified that are involved in male but not female meiosis, only three of which have orthologs in the honey bee (boule, courtless, matotopetli). Whether the four potential gene losses in the honey bee lineage reflect the absence of meiosis in males is questionable. When seven genes specifically involved in female meiosis (a process that is present in both insects) are subjected to the same analysis, only four genes have honey bee orthologs (altered disjunction, bifocal, eIF-4E, Fimbrin).
Germ-cell development
In Drosophila and many other holometabolous insects, a specialized cytoplasm, the pole plasm, present in early eggs is partitioned into the pole cells, which go on to make the germ cells (Mahowald 2001). In some hymenopterans, germ-cell fate is determined by the inheritance of an oosome, a subcellular organ-elle. In the honey bee, however, there is no pole plasm, oosome, or any pole cells present in early embryos (Nelson 1915; DuPraw 1967; Dearden 2006). Genes involved in pole-plasm assembly are all conserved except two, osk (as discussed above) and swa (which plays a role in bcd RNA localization and actin organization). Genes required for pole cell formation are also conserved, with no major genes, except osk and swa, missing.
Discussion
To learn more about evolutionary trends in insects, we have focused our survey of developmental genes in the honey bee on two classes of developmental processes that are well described in Drosophila: (1) developmental processes that encode the basic body plan and appear to be conserved among insects and (2) processes that we know to be different in the honey bee and Drosophila.
We looked first for orthology of genes that function in conserved developmental pathways. This approach identifies genes that are present in the honey bee, and those that are missing, despite functional roles in basic developmental processes in Drosophila. This approach does not, however, identify honey bee-specific genes, or other genes not acting in Drosophila, with roles in these pathways in non-Drosophilid insects.
In an evolutionary context, a gene “missing” from the honey bee genome could be interpreted in two ways: It could be lost (or changed in sequence so significantly to appear absent) during the course of evolution within the honey bee lineage, or it could have newly arisen in the Drosophila lineage.
Our survey of genes involved in apparently conserved cellular and developmental processes revealed that most genes of the cell signaling pathways (wnt, hh, notch, dpp), axis formation (terminal, dorso–ventral, anterior–posterior patterning), and segmentation have conserved counterparts in the honey bee, consistent with the view that these are basic processes that are fundamental to insect or animal development. The organization and expression of the Hox complex, including the rapidly evolving rogue Hox genes, and the complement of homeobox-containing transcription factors indicates a general conservation of the framework of these genes. If these genes and pathways regulate novel developmental events in the bee, then this must be associated with changes in their regulation and their downstream target genes, rather than wholesale gain or loss of genes. Despite this general rule, it is clear that several key genes (bcd, osk, grk, tor, trk, and kni) involved in these supposedly conserved processes in Drosophila are missing.
To explore whether the few cases in which genes are “missing” provide clues about trends of evolution in developmental pathways, we asked whether those genes are more likely to be associated with single or pleiotropic functions in Drosophila or if their absence might be related to their position in a developmental cascade. A survey of attributed functions of genes in FlyBase indicates that genes missing from the honey bee tend to have a single reported function in Drosophila (χ2 = 7.03; P < 0.01) (Supplemental Table 3). This finding is consistent with the idea that many, but not all, of these genes seem to have arisen recently in the Dipteran lineage. The recent emergence of a gene and its single documented function may thus be related.
Another conclusion is that Drosophila genes “missing” in the honey bee genome tend to act early in a developmental cascade. This is true of the axis formation genes, where it is the early acting members, grk, tor, trk and osk, that are missing, and is consistent with the hypotheses of Wilkins (2002) and Davidson (2001), who postulate that the initial steps in a cascade are likely to have evolved most recently.
We next examined the orthology of genes involved in biological processes in Drosophila that are different in the honey bee. This approach does not identify genes that encode honey bee-specific, or non-Drosophila, biological functions.
Sixty-nine, or 78%, of the Drosophila genes involved in processes that are substantially diverged or absent in the honey bee have orthologs in the honey bee genome. This finding is best illustrated by the genes of the upper part of the sex-determining cascade of Drosophila, where previous studies have shown that several genes have been co-opted recently to their sex-determining function (Schutt and Nothiger 2000) from functions in a wide range of developmental processes. The evolution of the Drosophila neo-X chromosome (Bachtrog 2005) and the absence of dosage compensation in other dipterans (Dubendorfer et al. 2002) imply that the genes involved in dosage compensation in Drosophila have also been recently co-opted. These examples and the large number of preserved genes we have found imply that co-option is a common event in evolution.
The majority of these conserved genes have additional reported biological functions in FlyBase. These additional functions may represent the ancestral functions of these genes, and their function in the honey bee may indicate this. In genes acting in divergent processes between honey bee and Drosophila, we see the same relationship between “missing” genes and pleiotropy. In this case, there is a tendency for genes with multiple functions in Drosophila to be preserved in the honey bee more often than genes that have a single function (χ2 = 19.03; P < 0.001) (Supplemental Table 3).
Possible interpretations of this are that genes that have multiple functions in a common ancestor are more likely to be preserved in both insect lineages, or that genes that are preserved throughout evolution are more likely to co-opt additional functions.
Our data provide evidence that novel biological processes (or function) may arise either with the evolution of new genes (genes that arise by gene duplication/by rapid sequence evolution), or from co-option of existing genes into new biological functions, or, as is most likely, with a combination of both.
Outlook
Comparing the genome sequence of the honey bee to other genomes allows us to identify genes that are missing from the honey bee genome and thus act in rapidly evolving developmental processes. It does not, however, identify genes that have newly evolved or been co-opted into novel developmental pathways in the honey bee. To complete our understanding of conserved and divergent developmental processes, it is necessary to identify these genes in the honey bee. It is these genes that will encode the components of the developmental pathways underlying honey bee-specific biology. Forward genetics, like that employed to find csd (Beye et al. 2003), RNAi-based screens, proteomics, or microarrays will be required to identify these genes. Such approaches will be enhanced by the honey bee genome sequence and will lead to a more complete understanding of how molecular functions and mechanisms differ, how pathways have evolved, and how this led to the rise of the different biology of the fly and the honey bee.
Methods
Genes involved in Drosophila development were identified by GO analysis using FlyBase (Drysdale and Crosby 2005). Using sequences from D. melanogaster, Tribolium castaneum, and A. gambiae (accessed at NCBI), we searched the Apis mellifera genome Assembly 2 for homologous sequences using TBLASTN (Altschul et al. 1990). Orthology calls were made by best reciprocal BLAST matches and CLUSTALW (Thompson et al. 1994) alignments. Where orthology was more difficult to assign, phylogenetic trees were constructed using either Phylip (Felsenstein 2004) or Mr-Bayes (Ronquist and Huelsenbeck 2003). In cases in which orthologous genes could not be found, two possible scenarios were implemented. Either no sequences in the honey bee genome, including reads unable to be assembled, had any homology with the Drosophila gene of interest, indicating that the gene is missing, or homologs were present in the honey bee genome but they gave another gene as the top hit in reciprocal BLAST searches of the Drosophila genome. In this second case, phylogenetic analysis, as described above, was used to test orthologous relationships between genes.
Gene models were manually curated using the Apollo genome annotation browser (Lewis et al. 2002). Evidence included a honey bee consensus gene set and six independently generated gene prediction sets: Ensembl, NCBI, Fgenesh, Fgenesh++, an Evolutionary Conserved Core set, and a Drosophila Ortholog set, as described in The Honey Bee Genome Sequencing Consortium 2006. BLAST similarity searches to UniProt proteins and EST/cDNA were also used. Identifiers from BeeBase (http://racerx00.tamu.edu/bee_resources.html) were assigned to the annotated proteins.
Total RNA was extracted using an RNAeasy kit (Qiagen). Primer sequences are shown in Supplemental Table 4. Probe synthesis and in situ hybridization were carried out as described previously (Osborne and Dearden 2005b). Honey bee embryos were staged according to the scheme of DuPraw (1967).
Acknowledgments
We thank the Baylor College of Medicine Human Genome Sequencing Center for making the Apis mellifera and Tribolium castaneum genome sequences publicly available prior to publication. This work was supported by a Royal Society of New Zealand Marsden Grant (UOO0401) to P.K.D., an NIH grant to S.J.B., and by grants from the Deutsche Forschungsgemeinschaft to M.B.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5108606.
References
- Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Li P.W., Hoskins R.A., Galle R.F., Hoskins R.A., Galle R.F., Galle R.F., et al. The genome sequence of Drosophila melanogaster . Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Altschul S., Gish W., Miller W., Myers E., Lipman D., Gish W., Miller W., Myers E., Lipman D., Miller W., Myers E., Lipman D., Myers E., Lipman D., Lipman D. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amdam G.V., Simoes Z.L.P., Guidugli K.R., Norberg K., Omholt S.W., Simoes Z.L.P., Guidugli K.R., Norberg K., Omholt S.W., Guidugli K.R., Norberg K., Omholt S.W., Norberg K., Omholt S.W., Omholt S.W. Disruption of vitellogenin gene function in adult honeybees by intra-abdominal injection of double stranded RNA. BMC Biotechnol. 2003;3:1. doi: 10.1186/1472-6750-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. Sex chromosome evolution: Molecular aspects of Y-chromosome degeneration in Drosophila . Genome Res. 2005;15:1393–1401. doi: 10.1101/gr.3543605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M. An overview of the Notch signalling pathway. Semin. Cell Dev. Biol. 2003;14:113–119. doi: 10.1016/s1084-9521(02)00179-9. [DOI] [PubMed] [Google Scholar]
- Bejsovec A. Wnt pathway activation: New relations and locations. Cell. 2005;120:11–14. doi: 10.1016/j.cell.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Bergem M., Norberg K., Aamodt R.M., Norberg K., Aamodt R.M., Aamodt R.M. Long-term maintenance of in vitro cultured honeybee (Apis mellifera) embryonic cells. BMC Dev. Biol. 2006;6:17. doi: 10.1186/1471-213X-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beye M., Hartel S., Hagen A., Hasselmann M., Omholt S.W., Hartel S., Hagen A., Hasselmann M., Omholt S.W., Hagen A., Hasselmann M., Omholt S.W., Hasselmann M., Omholt S.W., Omholt S.W. Specific developmental gene silencing in the honey bee using a homeobox motif. Insect Mol. Biol. 2002;11:527–532. doi: 10.1046/j.1365-2583.2002.00361.x. [DOI] [PubMed] [Google Scholar]
- Beye M., Hasselmann M., Fondrk M.K., Page R.E., Omholt S.W., Hasselmann M., Fondrk M.K., Page R.E., Omholt S.W., Fondrk M.K., Page R.E., Omholt S.W., Page R.E., Omholt S.W., Omholt S.W. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell. 2003;114:419–429. doi: 10.1016/s0092-8674(03)00606-8. [DOI] [PubMed] [Google Scholar]
- Binner P., Sander K., Sander K. Pair-rule patterning in the honeybee Apis Mellifera; expression of even-skipped combines traits known from beetles and fruitfly. Dev. Genes Evol. 1997;206:447–454. doi: 10.1007/s004270050074. [DOI] [PubMed] [Google Scholar]
- Brown S.J., Fellers J.P., Shippy T.D., Richardson E.A., Maxwell M., Stuart J.J., Denell R.E., Fellers J.P., Shippy T.D., Richardson E.A., Maxwell M., Stuart J.J., Denell R.E., Shippy T.D., Richardson E.A., Maxwell M., Stuart J.J., Denell R.E., Richardson E.A., Maxwell M., Stuart J.J., Denell R.E., Maxwell M., Stuart J.J., Denell R.E., Stuart J.J., Denell R.E., Denell R.E. Sequence of the Tribolium castaneum homeotic complex: The region corresponding to the Drosophila melanogaster antennapedia complex. Genetics. 2002;160:1067–1074. doi: 10.1093/genetics/160.3.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali A., Casanova J., Casanova J. The spatial control of Torso RTK activation: A C-terminal fragment of the Trunk protein acts as a signal for Torso receptor in the Drosophila embryo. Development. 2001;128:1709–1715. doi: 10.1242/dev.128.9.1709. [DOI] [PubMed] [Google Scholar]
- Cline T.W., Meyer B.J., Meyer B.J. Vive la difference: Males vs females in flies vs. worms. Annu. Rev. Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- Damen W.G. Parasegmental organization of the spider embryo implies that the parasegment is an evolutionarily conserved entity in arthropod embryogenesis. Development. 2002;129:1239–1250. doi: 10.1242/dev.129.5.1239. [DOI] [PubMed] [Google Scholar]
- Davidson E.H. Genomic regulatory systems. Academic Press; London: 2001. [Google Scholar]
- Dearden P.K. Germ cell development in the honeybee (Apis mellifera); vasa and nanos expression. BMC Dev. Biol. 2006;6:6. doi: 10.1186/1471-213X-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearden P., Grbic M., Falciani F., Akam M., Grbic M., Falciani F., Akam M., Falciani F., Akam M., Akam M. Maternal expression and early zygotic expression of the Hox3/zen gene in Schistocerca . Evol. Dev. 2000;2:261–270. doi: 10.1046/j.1525-142x.2000.00065.x. [DOI] [PubMed] [Google Scholar]
- Dearden P., Donly C., Grbic M., Donly C., Grbic M., Grbic M. Expression of pair-rule gene homologues in a chelicerate: Early patterning of the Two-Spotted Spider Mite Tetranychus urticae . Development. 2002;129:5461–5472. doi: 10.1242/dev.00099. [DOI] [PubMed] [Google Scholar]
- Doyle H.J., Kraut R., Levine M., Kraut R., Levine M., Levine M. Spatial regulation of zerknullt: A dorsal–ventral patterning gene in Drosophila . Genes & Dev. 1989;3:1518–1533. doi: 10.1101/gad.3.10.1518. [DOI] [PubMed] [Google Scholar]
- Drysdale R.A., Crosby M.A., Crosby M.A. FlyBase: Genes and gene models. Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubendorfer A., Hediger M., Burghardt G., Bopp D., Hediger M., Burghardt G., Bopp D., Burghardt G., Bopp D., Bopp D. Musca domestica, a window on the evolution of sex-determining mechanisms in insects. Int. J. Dev. Biol. 2002;46:75–79. [PubMed] [Google Scholar]
- DuPraw E.J. The honeybee embryo. In: Wilt F.H., Wessells N.K., Wessells N.K., editors. Methods in developmental biology. Thomas Y; Cromwell, New York: 1967. pp. 183–217. [Google Scholar]
- Falciani F., Hausdorf B., Schroder R., Akam M., Tautz D., Denell R., Brown S., Hausdorf B., Schroder R., Akam M., Tautz D., Denell R., Brown S., Schroder R., Akam M., Tautz D., Denell R., Brown S., Akam M., Tautz D., Denell R., Brown S., Tautz D., Denell R., Brown S., Denell R., Brown S., Brown S. Class 3 Hox genes in insects and the origin of zen. Proc. Natl. Acad. Sci. 1996;93:8479–8484. doi: 10.1073/pnas.93.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genomic Sciences, University of Washington; Seattle, WA: 2004. [Google Scholar]
- Ferrier D.E.K., Akam M., Akam M. Organization of the Hox gene cluster in the grasshopper, Schistocerca gregaria . Proc. Natl. Acad. Sci. 1996;93:13024–13029. doi: 10.1073/pnas.93.23.13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrier D.E.K., Holland P.W.H., Holland P.W.H. Sipunculan ParaHox genes. Evol. Dev. 2001;3:263–270. doi: 10.1046/j.1525-142x.2001.003004263.x. [DOI] [PubMed] [Google Scholar]
- Fleig R. Engrailed expression and body segmentation in the honeybee, Apis mellifera . Rouxs Arch. Dev. Biol. 1990;198:467–473. doi: 10.1007/BF00399057. [DOI] [PubMed] [Google Scholar]
- Fleig R., Sander K., Sander K. Embryogenesis of the honeybee Apis mellifera L. (Hymenoptera: Apidae): An SEM study. Int. J. Insect Morphol. Embryol. 1986;15:449–462. [Google Scholar]
- Fleig R., Walldorf U., Gehring W.J., Sander K., Walldorf U., Gehring W.J., Sander K., Gehring W.J., Sander K., Sander K. Development of the Deformed protein pattern in the embryo of the honeybee Apis mellifera L. (Hymenoptera) Rouxs Arch. Dev. Biol. 1992;201:235–242. doi: 10.1007/BF00188754. [DOI] [PubMed] [Google Scholar]
- Furriols M., Casali A., Casanova J., Casali A., Casanova J., Casanova J. Dissecting the mechanism of torso receptor activation. Mech. Dev. 1998;70:111–118. doi: 10.1016/s0925-4773(97)00181-0. [DOI] [PubMed] [Google Scholar]
- Gay N.J., Keith F.J., Keith F.J. Regulation of translation and proteolysis during the development of embryonic dorso–ventral polarity in Drosophila. Homology of easter proteinase with Limulus proclotting enzyme and translational activation of Toll receptor synthesis. Biochim. Biophys. Acta. 1992;1132:290–296. doi: 10.1016/0167-4781(92)90163-t. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Gaitan M., Rothe M., Wimmer E.A., Taubert H., Jackle H., Rothe M., Wimmer E.A., Taubert H., Jackle H., Wimmer E.A., Taubert H., Jackle H., Taubert H., Jackle H., Jackle H. Redundant functions of the genes knirps and knirps-related for the establishment of anterior Drosophila head structures. Proc. Natl. Acad. Sci. 1994;91:8567–8571. doi: 10.1073/pnas.91.18.8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig W. Insect phylogeny. John Wiley; Bath, UK: 1981. [Google Scholar]
- Holt R.A., Subramanian G.M., Halpern A., Sutton G.G., Charlab R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Subramanian G.M., Halpern A., Sutton G.G., Charlab R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Halpern A., Sutton G.G., Charlab R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Sutton G.G., Charlab R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Charlab R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Clark A.G., Ribeiro J.M., Wides R., Ribeiro J.M., Wides R., Wides R., et al. The genome sequence of the malaria mosquito Anopheles gambiae . Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- The Honey Bee Genome Sequencing Consortium. Insights into social insects from the genome of the honey bee Apis mellifera. Nature. 2006;((in press)) [Google Scholar]
- Hooper J.E., Scott M.P., Scott M.P. Communicating with Hedgehogs. Nat. Rev. Mol. Cell Biol. 2005;6:306–317. doi: 10.1038/nrm1622. [DOI] [PubMed] [Google Scholar]
- Hughes C.L., Kaufman T.C., Kaufman T.C. Hox genes and the evolution of the arthropod body plan. Evol. Dev. 2002;4:459–499. doi: 10.1046/j.1525-142x.2002.02034.x. [DOI] [PubMed] [Google Scholar]
- Jagla K., Bellard M., Frasch M., Bellard M., Frasch M., Frasch M. A cluster of Drosophila homeobox genes involved in mesoderm differentiation programs. Bioessays. 2001;23:125–133. doi: 10.1002/1521-1878(200102)23:2<125::AID-BIES1019>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Krauss V., Pecyna M., Kurz K., Sass H., Pecyna M., Kurz K., Sass H., Kurz K., Sass H., Sass H. Phylogenetic mapping of intron positions: A case study of translation initiation factor eIF2γ. Mol. Biol. Evol. 2005;22:74–84. doi: 10.1093/molbev/msh255. [DOI] [PubMed] [Google Scholar]
- Kusserow A., Pang K., Sturm C., Hrouda M., Lentfer J., Schmidt H.A., Technau U., von Haeseler A., Hobmayer B., Martindale M.Q., Pang K., Sturm C., Hrouda M., Lentfer J., Schmidt H.A., Technau U., von Haeseler A., Hobmayer B., Martindale M.Q., Sturm C., Hrouda M., Lentfer J., Schmidt H.A., Technau U., von Haeseler A., Hobmayer B., Martindale M.Q., Hrouda M., Lentfer J., Schmidt H.A., Technau U., von Haeseler A., Hobmayer B., Martindale M.Q., Lentfer J., Schmidt H.A., Technau U., von Haeseler A., Hobmayer B., Martindale M.Q., Schmidt H.A., Technau U., von Haeseler A., Hobmayer B., Martindale M.Q., Technau U., von Haeseler A., Hobmayer B., Martindale M.Q., von Haeseler A., Hobmayer B., Martindale M.Q., Hobmayer B., Martindale M.Q., Martindale M.Q., et al. Unexpected complexity of the Wnt gene family in a sea anemone. Nature. 2005;433:156–160. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- Lehmann R., Nusslein-Volhard C., Nusslein-Volhard C. Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila . Cell. 1986;47:141–152. doi: 10.1016/0092-8674(86)90375-2. [DOI] [PubMed] [Google Scholar]
- Lewis S.E., Searle S.M., Harris N., Gibson M., Iyer V., Richter J., Wiel C., Bayraktaroglir L., Birney E., Crosby M.A., Searle S.M., Harris N., Gibson M., Iyer V., Richter J., Wiel C., Bayraktaroglir L., Birney E., Crosby M.A., Harris N., Gibson M., Iyer V., Richter J., Wiel C., Bayraktaroglir L., Birney E., Crosby M.A., Gibson M., Iyer V., Richter J., Wiel C., Bayraktaroglir L., Birney E., Crosby M.A., Iyer V., Richter J., Wiel C., Bayraktaroglir L., Birney E., Crosby M.A., Richter J., Wiel C., Bayraktaroglir L., Birney E., Crosby M.A., Wiel C., Bayraktaroglir L., Birney E., Crosby M.A., Bayraktaroglir L., Birney E., Crosby M.A., Birney E., Crosby M.A., Crosby M.A., et al. Apollo: A sequence annotation editor. Genome Biol. 2002;3:research0082. doi: 10.1186/gb-2002-3-12-research0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E.B., Pfeiffer B.D., Mathog D.R., Celniker S.E., Pfeiffer B.D., Mathog D.R., Celniker S.E., Mathog D.R., Celniker S.E., Celniker S.E. Evolution of the homeobox complex in the Diptera. Curr. Biol. 2003;13:R587–R588. doi: 10.1016/s0960-9822(03)00520-7. [DOI] [PubMed] [Google Scholar]
- Liaw G.J., Rudolph K.M., Huang J.D., Dubnicoff T., Courey A.J., Lengyel J.A., Rudolph K.M., Huang J.D., Dubnicoff T., Courey A.J., Lengyel J.A., Huang J.D., Dubnicoff T., Courey A.J., Lengyel J.A., Dubnicoff T., Courey A.J., Lengyel J.A., Courey A.J., Lengyel J.A., Lengyel J.A. The torso response element binds GAGA and NTF-1/Elf-1, and regulates tailless by relief of repression. Genes & Dev. 1995;9:3163–3176. doi: 10.1101/gad.9.24.3163. [DOI] [PubMed] [Google Scholar]
- Logan C.Y., Nusse R., Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Luke G.N., Castro L.F., McLay K., Bird C., Coulson A., Holland P.W.H., Castro L.F., McLay K., Bird C., Coulson A., Holland P.W.H., McLay K., Bird C., Coulson A., Holland P.W.H., Bird C., Coulson A., Holland P.W.H., Coulson A., Holland P.W.H., Holland P.W.H. Dispersal of NK homeobox gene clusters in amphioxus and humans. Proc. Natl. Acad. Sci. 2003;100:5292–5295. doi: 10.1073/pnas.0836141100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald P.M., Leask A., Kerr K., Leask A., Kerr K., Kerr K. exl protein specifically binds BLE1, a bicoid mRNA localization element, and is required for one phase of its activity. Proc. Natl. Acad. Sci. 1995;92:10787–10791. doi: 10.1073/pnas.92.23.10787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald A.P. Assembly of the Drosophila germ plasm. Int. Rev. Cytol. 2001;203:187–213. doi: 10.1016/s0074-7696(01)03007-8. [DOI] [PubMed] [Google Scholar]
- Maxton Küchenmeister J., Handel K., Schmidt-Ott U., Roth S., Jäckle H., Handel K., Schmidt-Ott U., Roth S., Jäckle H., Schmidt-Ott U., Roth S., Jäckle H., Roth S., Jäckle H., Jäckle H. Toll homologue expression in the beetle Tribolium suggests a different mode of dorsoventral patterning than in Drosophila embryos. Mech. Dev. 1999;83:107–114. doi: 10.1016/s0925-4773(99)00041-6. [DOI] [PubMed] [Google Scholar]
- Miller D.J., Hayward D.C., Reece-Hoyes J.S., Scholten I., Catmull J., Gehring W.J., Callaerts P., Larsen J.E., Ball E.E., Hayward D.C., Reece-Hoyes J.S., Scholten I., Catmull J., Gehring W.J., Callaerts P., Larsen J.E., Ball E.E., Reece-Hoyes J.S., Scholten I., Catmull J., Gehring W.J., Callaerts P., Larsen J.E., Ball E.E., Scholten I., Catmull J., Gehring W.J., Callaerts P., Larsen J.E., Ball E.E., Catmull J., Gehring W.J., Callaerts P., Larsen J.E., Ball E.E., Gehring W.J., Callaerts P., Larsen J.E., Ball E.E., Callaerts P., Larsen J.E., Ball E.E., Larsen J.E., Ball E.E., Ball E.E. Pax gene diversity in the basal cnidarian Acropora millepora (Cnidaria, Anthozoa): Implications for the evolution of the Pax gene family. Proc. Natl. Acad. Sci. 2000;97:4475–4480. doi: 10.1073/pnas.97.9.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauber U., Pankratz M.J., Kienlin A., Seifert E., Klemm U., Jackle H., Pankratz M.J., Kienlin A., Seifert E., Klemm U., Jackle H., Kienlin A., Seifert E., Klemm U., Jackle H., Seifert E., Klemm U., Jackle H., Klemm U., Jackle H., Jackle H. Abdominal segmentation of the Drosophila embryo requires a hormone receptor-like protein encoded by the gap gene knirps . Nature. 1988;336:489–492. doi: 10.1038/336489a0. [DOI] [PubMed] [Google Scholar]
- Nelson J.A. The embryology of the honeybee. Princeton University Press; Princeton, NJ: 1915. [Google Scholar]
- Nusse R. An ancient cluster of Wnt paralogues. Trends Genet. 2001;17:443. doi: 10.1016/s0168-9525(01)02349-6. [DOI] [PubMed] [Google Scholar]
- Osborne P., Dearden P.K., Dearden P.K. Expression of Pax group III genes in the honeybee (Apis mellifera) Dev. Genes Evol. 2005a;215:499–508. doi: 10.1007/s00427-005-0008-9. [DOI] [PubMed] [Google Scholar]
- Osborne P., Dearden P.K., Dearden P.K. Non-radioactive in situ hybridisation to honeybees embryos and ovaries. Apidologie (Celle) 2005b;36:113–118. [Google Scholar]
- Pearson J.C., Lemons D., McGinnis W., Lemons D., McGinnis W., McGinnis W. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- Peel A.D., Chipman A.D., Akam M., Chipman A.D., Akam M., Akam M. Arthropod segmentation: Beyond the Drosophila paradigm. Nat. Rev. Genet. 2005;6:905–916. doi: 10.1038/nrg1724. [DOI] [PubMed] [Google Scholar]
- Pires-daSilva A., Sommer R.J., Sommer R.J. The evolution of signalling pathways in animal development. Nat. Rev. Genet. 2003;4:39–49. doi: 10.1038/nrg977. [DOI] [PubMed] [Google Scholar]
- Pultz M.A., Westendorf L., Gale S.D., Hawkins K., Lynch J., Pitt J.N., Reeves N.L., Yao J.C., Small S., Desplan C., Westendorf L., Gale S.D., Hawkins K., Lynch J., Pitt J.N., Reeves N.L., Yao J.C., Small S., Desplan C., Gale S.D., Hawkins K., Lynch J., Pitt J.N., Reeves N.L., Yao J.C., Small S., Desplan C., Hawkins K., Lynch J., Pitt J.N., Reeves N.L., Yao J.C., Small S., Desplan C., Lynch J., Pitt J.N., Reeves N.L., Yao J.C., Small S., Desplan C., Pitt J.N., Reeves N.L., Yao J.C., Small S., Desplan C., Reeves N.L., Yao J.C., Small S., Desplan C., Yao J.C., Small S., Desplan C., Small S., Desplan C., Desplan C., et al. A major role for zygotic hunchback in patterning the Nasonia embryo. Development. 2005;132:3705–3715. doi: 10.1242/dev.01939. [DOI] [PubMed] [Google Scholar]
- Raftery L.A., Sutherland D.J., Sutherland D.J. TGF-β family signal transduction in Drosophila development: From Mad to Smads. Dev. Biol. 1999;210:251–268. doi: 10.1006/dbio.1999.9282. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rothe M., Nauber U., Jackle H., Nauber U., Jackle H., Jackle H. Three hormone receptor-like Drosophila genes encode an identical DNA-binding finger. EMBO J. 1989;8:3087–3094. doi: 10.1002/j.1460-2075.1989.tb08460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlatter R., Maier D., Maier D. The Enhancer of split and Achaete-Scute complexes of Drosophilids derived from simple ur-complexes preserved in mosquito and honeybee. BMC Evol. Biol. 2005;5:67. doi: 10.1186/1471-2148-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoppmeier M., Schroder R., Schroder R. Maternal torso signaling controls body axis elongation in a short germ insect. Curr. Biol. 2005;15:2131–2136. doi: 10.1016/j.cub.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Schroder R. The genes orthodenticle and hunchback substitute for bicoid in th beetle Tribolium. Nature. 2003;422:621–625. doi: 10.1038/nature01536. [DOI] [PubMed] [Google Scholar]
- Schroder R., Eckert C., Wolff C., Tautz D., Eckert C., Wolff C., Tautz D., Wolff C., Tautz D., Tautz D. Conserved and divergent aspects of terminal patterning in the beetle Tribolium castaneum . Proc. Natl. Acad. Sci. 2000;97:6591–6596. doi: 10.1073/pnas.100005497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T., Roth S., Roth S. Dorsoventral patterning in Drosophila oogenesis. Curr. Opin. Genet. Dev. 1994;4:502–507. doi: 10.1016/0959-437x(94)90064-a. [DOI] [PubMed] [Google Scholar]
- Schutt C., Nothiger R., Nothiger R. Structure, function and evolution of sex-determining systems in Dipteran insects. Development. 2000;127:667–677. doi: 10.1242/dev.127.4.667. [DOI] [PubMed] [Google Scholar]
- Stauber M., Jackle H., Schmidt-Ott U., Jackle H., Schmidt-Ott U., Schmidt-Ott U. The anterior determinant bicoid of Drosophila is a derived Hox class 3 gene. Proc. Natl. Acad. Sci. 1999;96:3786–3789. doi: 10.1073/pnas.96.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber M., Taubert H., Schmidt-Ott U., Taubert H., Schmidt-Ott U., Schmidt-Ott U. Function of bicoid and hunchback homologs in the basal cyclorrhaphan fly Megaselia (Phoridae) Proc. Natl. Acad. Sci. 2000;97:10844–10849. doi: 10.1073/pnas.190095397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber M., Prell A., Schmidt-Ott U., Prell A., Schmidt-Ott U., Schmidt-Ott U. A single Hox3 gene with composite bicoid and zerknullt expression characteristics in non-Cyclorrhaphan flies. Proc. Natl. Acad. Sci. 2002;99:274–279. doi: 10.1073/pnas.012292899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zee M., Berns N., Roth S., Berns N., Roth S., Roth S. Distinct functions of the Tribolium zerknullt genes in serosa specification and dorsal closure. Curr. Biol. 2005;15:624–636. doi: 10.1016/j.cub.2005.02.057. [DOI] [PubMed] [Google Scholar]
- Walldorf U., Fleig R., Gehring W.J., Fleig R., Gehring W.J., Gehring W.J. Comparison of homeobox-containing genes of the honeybee and Drosophila . Proc. Natl. Acad. Sci. 1989;86:9971–9975. doi: 10.1073/pnas.86.24.9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walldorf U., Binner P., Fleig R., Binner P., Fleig R., Fleig R. Hox genes in the honey bee Apis mellifera . Dev. Genes Evol. 2000;210:483–492. doi: 10.1007/s004270000091. [DOI] [PubMed] [Google Scholar]
- Whiting M.F. Phylogeny of the holometabolous insect orders: Molecular evidence. Zool. Scr. 2002;31:3–15. [Google Scholar]
- Wilkins A.S. Moving up the hierarchy: A hypothesis on the evolution of a genetic sex determination pathway. Bioessays. 1995;17:71–77. doi: 10.1002/bies.950170113. [DOI] [PubMed] [Google Scholar]
- Wilkins A.S. The evolution of developmental pathways. Sinauer Associates; Sunderland, MA: 2002. [Google Scholar]
- Yasukochi Y., Ashakumary L.A., Wu C., Yoshido A., Nohata J., Mita K., Sahara K., Ashakumary L.A., Wu C., Yoshido A., Nohata J., Mita K., Sahara K., Wu C., Yoshido A., Nohata J., Mita K., Sahara K., Yoshido A., Nohata J., Mita K., Sahara K., Nohata J., Mita K., Sahara K., Mita K., Sahara K., Sahara K. Organization of the Hox gene cluster of the silkworm, Bombyx mori: A split of the Hox cluster in anon-Drosophila insect. Dev. Genes Evol. 2004;214:606–614. doi: 10.1007/s00427-004-0441-1. [DOI] [PubMed] [Google Scholar]