Abstract
Molecular imaging is an emerging technology at the life science/physical science interface which is set to revolutionize our understanding and treatment of disease. The tools of molecular imaging are the imaging modalities and their corresponding contrast agents. These facilitate interaction with a biological target at a molecular level in a number of ways. The diverse nature of molecular imaging requires knowledge from both the life and physical sciences for its successful development and implementation. The aim of this review is to introduce the subject of molecular imaging from both life science and physical science perspectives. However, we will restrict our coverage to the prominent in vivo molecular imaging modalities of magnetic resonance imaging, optical imaging and nuclear imaging. The physical basis of these imaging modalities, the use of contrast agents and the imaging parameters of sensitivity, temporal resolution and spatial resolution are described. Then, the specificity of contrast agents for targeting and sensing molecular events, and some applications of molecular imaging in biology and medicine are given. Finally, the diverse nature of molecular imaging and its reliance on interdisciplinary collaboration is discussed.
Keywords: molecular imaging, imaging techniques, contrast agents
1. Introduction
Advances in sequencing the human genome and the genomes of many other organisms provided a vast amount of DNA sequence data and genomic information that transformed the way we can study living organisms. In the twentieth century, biochemical and molecular analysis led to a reductionist view of living processes, where the description of components of living systems (structures and mechanisms) was the primary aim. In contrast, twenty-first century biology is aimed at understanding how component parts collaborate to create complex biological systems. This integrative approach requires the observation of molecular functions in whole organisms. We need to know how components of living cells are organized, how they interact, how they are formed and eliminated at various stages of the life cycle of the organism and how they move around within the cell. In multicellular organisms, cells receive external signals, they communicate with one another, they can migrate within a tissue and, during the development of the organism, they undergo some complex changes from the initial embryonic state to reach the mature adult form. All of these processes can now be visualized at various levels of resolution by in vivo imaging techniques.
The term ‘molecular imaging’ is used in a variety of ways and includes imaging of specific molecules which are present in the living system, the use of externally-added targeted or activatable reporter agents that sense specific molecular targets or cellular processes, the use of labelled or even natural substrates to follow particular pathways, and the introduction of genes that are expressed in the cell into a protein product that can be detected directly or indirectly.
In addition to the study of fundamental biological processes, developments in molecular imaging are aimed at furthering our understanding of disease since most disease processes have a molecular basis. It is hoped that the new imaging methods will lead to better differentiation in diagnosis, earlier detection of disease and objective monitoring of therapies using imaging biomarkers.
The rapid increase in molecular imaging is in part a result of new technologies, but also in part, a rebadging of long-established methods such as the use of 18F-deoxyglucose in positron-emission tomography (PET) imaging. Molecular imaging is a major tool in cell biology and many of the novel optical probes and techniques were developed in studies of cell culture systems. The extension of these techniques to intact multicellular organisms, including to a limited extent to humans, is just beginning. Magnetic resonance techniques, on the other hand, tended to be developed in animal experiments and then extended to human studies. PET was largely developed initially for human application and only with the later introduction of micro-PET instrumentation did animal experiments begin.
In this review, we restrict our discussion to animal and human studies, but will refer to some approaches that have been introduced in isolated cell work which are potentially applicable to the study of higher organisms.
Developments in molecular imaging can be considered under three headings: (i) the physics and engineering of imaging techniques and instruments; (ii) the chemical and materials science needs for designing imaging probes and contrast agents, and the chemical and molecular biology tools required to use these agents in a targeted way; (iii) the biological questions to be answered.
Much of medical imaging to date is based on the invention and development of a particular imaging modality and was, therefore, physics led. In contrast, molecular imaging should be driven primarily by the biological questions. The selection of the imaging technique and the kind of probe to be used then follows.
2. The physics and engineering of imaging techniques and instruments
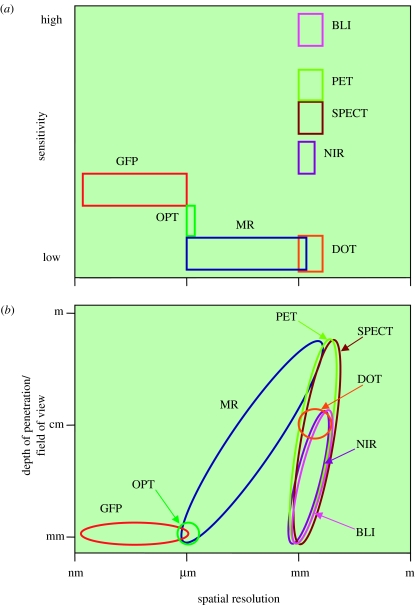
Molecular imaging techniques span the electromagnetic spectrum from ultrasonic to gamma ray and X-ray frequencies. Within this spectrum, magnetic resonance imaging (MRI), optical imaging and nuclear imaging are emerging as key molecular imaging techniques. In general, these are in vivo techniques, with the exception of optical imaging using optical projection tomography (OPT), where ‘fixed’ ex vivo preparations are required (§2.2.2). To gain an understanding of these techniques, and the issues that affect their performance, we will describe their physical basis, instrumentation and the use of contrast mechanisms and agents in some detail. A simplified comparison of their key physical performance parameters is given in figure 1.
Figure 1.
(a) Comparison of sensitivity versus spatial resolution and (b) depth of penetration/field of view versus spatial resolution for the different molecular imaging modalities considered in this review.
2.1 Magnetic resonance imaging
MRI is based on the detection of molecules that contain nuclei that possess the property of nuclear spin (Gadian 1995). When spinning nuclei are exposed to a strong static magnetic field they precess at the Larmor frequency, and for a spin one half system they have either spin-up or spin-down orientations. These orientations correspond to two different energy states, with more spinning nuclei in the lower energy state than in the higher one. The application of an oscillating radio frequency (RF) magnetic field at the Larmor frequency causes some of the spinning nuclei from the lower energy state to move to the higher one. Spins that have moved to the higher energy state will eventually return to the lower energy state, with the emission of a RF signal at the Larmor frequency. This is the detected signal in MRI (detected using an RF coil) and corresponds to the energy difference between the two energy states. The detected signal eventually decays (relaxes) owing to energy exchange between the spin system and the surrounding thermal reservoir, and energy exchange within the spin system itself. These two energy-loss mechanisms have time constants T1 and T2, respectively, which are sample/tissue and static-field strength dependent (Callaghan 1991). MRI relies on the deliberate perturbation of the static magnetic field by the introduction of switched magnetic field gradients, which produce nuclear spins with a spread of Larmor frequencies, and thus spatial encoding of the MRI signal, which can be decoded by an appropriate algorithm to produce an image with a field-of-view (FOV) ranging from sub-millimetre to sub-metre scales.
The sensitivity of MRI is low (mM concentrations) and is dependent on the volume of the sample and the magnitude of the oscillating RF field, the static magnetic field strength, the bandwidth of acquisition, the effective resistance of the RF coil and the number of acquisitions (Hoult & Richards 1976). The number of acquisitions per unit time is limited by T1 and T2 relaxation processes, where T1 is generally longer than T2 for biological tissue, so that signal-to-noise ratio (SNR) per unit time is limited by T1.
The spatial resolution of MRI is extremely good (μm) and is not limited by excitation wavelength or the diffraction limit of photons. Instead, MRI resolution is limited by a combination of T2 relaxation, diffusion of spinning nuclei, magnetic field susceptibility and in-homogeneity, the magnitude of the switched magnetic field gradients and the available SNR.
The temporal resolution of MRI is good (ms), and a typical image acquisition can be performed in seconds to minutes, although for high spatial-resolution three-dimensional acquisitions, the examination time increases dramatically to the order of several hours (Schneider et al. 2003), owing to the inverse sixth power dependence on spatial resolution (Mansfield & Morris 1982). Temporal resolution and examination time are ultimately governed by sensitivity, which can be modified by the use of contrast agents.
MRI has a number of contrast agent mechanisms for molecular imaging which modify either T1 or T2 relaxation time constants, proton density, or nuclear polarization, for improved sensitivity and specificity.
T1 contrast agents increase the rate of energy exchange between the nuclear spin system and the thermal reservoir, and hence reduce T1 and increase the number of spins that can absorb energy from the next RF pulse (Jacobs et al. 2003). Thus, T1 contrast agents produce a positive signal enhancement on MRI images (Artemov 2003) because the SNR per unit time increases. T1 contrast agents are usually based on paramagnetic ions or stable free radical molecules (Potter 2002), such as the gadolinium III complexes (Aime et al. 2002) with a short circulating half-life (minutes).
T2 contrast agents increase the rate of energy loss within the nuclear spin system by introducing magnetic field perturbations, which are manifested by a reduction in T2 and decreases in the local signal intensity. Thus, T2 contrast agents produce a negative signal enhancement on T2 weighted images. T2 contrast agents are based on ferromagnetic or superparamagnetic centres. These may take the form of monocrystalline, polycrystalline, cross-linked iron oxide magnetic cores (5–30 nm) embedded in a polymer coating with a total particle diameter of 17–50 nm, with the degree of relaxation generally dependent on the molecular size of the carrier, with larger molecules having a higher relaxivity. However, the large molecular size of these contrast agents can cause problems for their biodistribution and circulating half-life (typically 6 min), and thus, ultrasmall superparamagnetic iron oxide nanoparticles have been introduced to overcome these problems with their smaller size (<10 nm), longer half-life (81 min) and improved biodistribution (Weissleder et al. 1990).
Proton density contrast agents introduce contrast to an MRI image by altering the total water signal. This can be achieved using magnetization transfer (MT; Wolff & Balaban 1989), chemical exchange saturation transfer (CEST; Ward et al. 2000), or paramagnetic chemical exchange saturation transfer (PARACEST; Zhang et al. 2003). MT is based on the exchange of magnetization between tissue-bound water and the larger bulk water pool, which results in a decrease in proton density. CEST is based on the slow exchange of magnetization between low molecular weight diamagnetic compounds, which introduces proton density contrast that can be turned on and off. PARACEST is based on the unusually slow water exchange of paramagnetic lanthanide complexes that increases the utility of CEST and has the potential to be used as biologically responsive agents capable of sensing molecular exchange phenomena in tissue (Zhang et al. 2003).
Nuclear polarization contrast agents greatly increase (hyperpolarize) the weak nuclear polarization of atomic nuclei from parts per million (ppm) towards unity. The inherently weak nuclear polarization at room temperature is responsible for the low sensitivity of MRI as compared with other imaging methods. Hyperpolarization can be achieved using a variety of methods, but recent advances in dynamic nuclear polarization (DNP) have shown tremendous potential for molecular imaging. DNP is based on polarizing the nuclear spins of a contrast medium in the solid-state by partly transferring the high electron spin polarization to the nuclear spins by microwave irradiation, and quickly returning the contrast medium to the liquid state. The contrast medium is then administered and MRI acquisitions performed within a short time frame (typically 6 s transfer, MRI within 1 min), before the hyperpolarization enhancement is lost owing to thermal equilibrium, i.e. T1 relaxation. DNP provides novel contrast to MRI by massively increasing the SNR (greater than 10 000 times) and enables molecular imaging of low gyromagnetic ratio nuclei (Golman et al. 2003), within a short acquisition window (seconds to minutes; Ardenkjaer-Larsen et al. 2003).
2.2 Optical imaging
Optical imaging is based on detecting the transmission of light (photons) through biological tissue. Photon transport in biological tissue is severely affected by the processes of absorption and scattering. Absorption occurs when the energy associated with the photon frequency matches an energy transition state within the tissue, and hence the photon energy is absorbed by the tissue and thus reduces the total photon energy. Scattering occurs when light is incident upon the atoms of the tissue. This causes the atom's electrons to accelerate and radiate in various directions, and causes light to deviate from its original path.
The propagation of light through biological tissue experiences both absorption and scattering simultaneously. Absorption and scattering are wavelength and penetration-depth dependent. Absorption is large in the ultraviolet (UV), near visible and infrared (IR), but low in red and near-infrared (NIR; 650–1000 nm; Venugopalan 2004).
For animal and human in vivo and in vitro molecular imaging, the optical imaging technologies of diffuse optical tomography (DOT), OPT, NIR fluorescence imaging, fluorescence protein imaging and bioluminescence imaging (BLI) are emerging as powerful tools for measuring dynamic metabolic processes and probing protease, protein and enzymatic activity.
2.2.1 Diffuse optical tomography.
DOT is based on the transmission and detection of NIR light using an array of sources and detectors distributed over a large region. Three source-detector configurations are generally used: transmissive, in which the sources are on one surface of the tissue and the detectors on the other side; reflective, in which both sources and detectors are interspersed on the same tissue surface; and annular, in which the sources and detectors form one or more rings around the tissue. Either scalar- or vector-based measurement methods are used. Scalar methods measure the optical flux exiting the tissue and the photon path length, whereas vector methods measure both magnitude and the average propagation delay using time-domain or frequency-domain techniques. Time-domain systems use picosecond-wide optical pulses, time-gated photon counting detectors and time-to-amplitude converters; and frequency-domain systems use an RF modulated light source, photomultiplier tubes (PMTs) or fast photodiodes feeding tuned RF amplifiers and an RF inphase/quadrature phase detector followed by post-detection filters (Siegel et al. 1999). Frequency-domain systems use simpler, lower-cost components and have the advantage of greater SNR. The SNR gains are due to their single RF frequency and narrow acquisition bandwidth which results in a lower detected noise level.
The SNR of DOT can be maximized by increasing the source power and decreasing the detected noise level. The source power can be increased up to the safe exposure level of the tissue, and the detected noise level can be decreased by increasing the total number of photons collected by increasing the acquisition time (Siegel et al. 1999). The spatial resolution of DOT is comparable to, and determined by, the grid spacing of the source-detector array (Boas et al. 2004), whereas the depth of penetration is determined by the light transport characteristics of the tissue, with DOT being performed to depths of 10 cm for human brain and breast imaging (Franceschini et al. 1997; Chance et al. 1998; Ntziachristos et al. 2000).
The spatial resolution of DOT for human applications is approximately 5 mm (Ntziachristos et al. 2000), but recent advances have increased this by up to a factor of 2 (Boas et al. 2004). For small animal applications, a sub-millimetre spatial resolution is used (Siegel et al. 1999).
DOT has a temporal resolution of a few seconds, and a typical image acquisition can be performed in minutes. Thus, the advantage of DOT over other clinical imaging modalities is that it can simultaneously quantify the tissue concentration of both oxy- and deoxy-haemoglobin (Siegel et al. 1999), and can provide information about blood dynamics, cytochromes, lipids, water, tissue metabolism and blood transport phenomenon with good temporal resolution (Pogue 1999), as well as potentially measure fast scattering changes associated with neuronal activity (Boas et al. 2001).
2.2.2 Optical projection tomography.
OPT is based on the transmission and detection of visible light (400–700 nm) through a 360° step-rotated ex vivo specimen. The ex vivo specimen is bathed in an organic solvent during image capture to reduce its opacity (Sharpe 2003). At each step-rotated position, data are collected, and once the specimen has been rotated through 360°, a three-dimensional image is reconstructed using a back-projection algorithm. OPT has the advantage over confocal microscopy and optical coherence tomography of increased penetration depth and can produce high-resolution (5–10 μm) three-dimensional images of both fluorescent and non-fluorescent biological specimens with a thickness of up to 15 mm (Sharpe et al. 2002).
2.2.3 Near-infrared fluorescence imaging.
NIR fluorescence imaging is based on the absorption and emission of light with a wavelength between 700 and 1000 nm. The absorbed light excites a fluorescent molecule within the biological tissue which emits light (fluoresces) at a wavelength longer than the transmitted light (i.e. it undergoes a Stokes shift). In the NIR range biological tissue is not very fluorescent, but some tissues do exhibit a degree of autofluorescence, which can reduce the signal-to-background ratio (SBR) of an NIR fluorescence image. However, autofluorescence can be removed by the appropriate choice of NIR filtering (Frangioni 2003).
For improved SBR and targeting of specific biological activity, NIR fluorescence imaging relies on the excitation and detection of fluorescence from an exogenous contrast agent. Exogenous NIR contrast agents can be divided into two categories: (i) organic fluorophores and (ii) inorganic fluorescent semiconductor nanocrystals or quantum dots.
Organic fluorophores have been used in NIR applications for many years. They are typically 1200 or less Daltons in size, varying in toxicity, with widely different biodistibution and pharmacokinetics, and are cleared from the circulation by renal filtration in minutes (Frangioni 2003). However, they suffer from problems with controlling their excitation and emission wavelengths, hydrophobia, molecular stability, low quantum yields (efficiency of emission) and are susceptible to photobleaching (loss of fluorescence) which limits their sensitivity to detection (Frangioni 2003).
Quantum dots have recently been introduced to the field of NIR and have the potential to solve many of the problems associated with organic fluorophores. Quantum dots comprise an inorganic core and shell of metal and an outer organic coating. The inorganic core and shell enables tuning of fluorescence with a narrow bandwidth (25–35 nm), thus enabling multi-coloured quantum dots for multiplexed detection of molecular targets (Smith et al. 2004). In addition, quantum dots are resistant to photobleaching, with an emission time constant (tone-half) of approximately 16 min (Chan & Nie 1998), and they have a high quantum yield of close to 90% (Michalet et al. 2005).
However, quantum dots are typically large structures of 3–20 nm hydrodynamic diameter, and hence are difficult to clear from circulation by renal filtration, which results in a high background signal. Also, the issue of in vivo toxicity is as yet unknown, and needs to be addressed (Frangioni 2003).
Near-infra red fluorescent contrast agents can be tailored to target specific biological events. These so-called ‘stealth’ probes are initially dormant when injected, but become active and fluoresce once they reach their biological target. Thus, there is an extremely low background at non-targeted sites and fluorescent signal (‘amplification’) at targeted sites, resulting in good specificity and up to a 1000-fold increase in sensitivity (SBR; Weissleder et al. 1999).
The performance of NIR fluorescence imaging is extremely good with sensitivity being linearly related to the administered contrast agent concentration. An SNR of 3–173 for a 30 s acquisition using 3–200 pM, respectively, and a spatial resolution of 60–300 μm over 15–450 cm2, respectively, with an acquisition time of the order of 10 s has been reported (Mahmood et al. 1999). With these impressive imaging parameters, real-time NIR fluorescence imaging with sub-second temporal resolution to depths of 6–10 cm will soon be possible (Kim et al. 2004).
2.2.4 Green fluorescent protein (GFP) imaging.
GFP imaging is based on the absorption and emission of visible light, where absorption and scattering in tissue have limited the technique to a depth of 1–2 mm, and thus only structures close to the surface can be imaged (Weissleder & Mahmood 2001).
Green fluorescent proteins are native to a few marine organisms, but can be introduced into the cells of other organisms by gene transfer. These transferred GFPs act as a chemical fluorescent contrast agent for protein activity inside the cell (Whitaker 2000). Unlike NIR fluorescent imaging, GFP imaging does not use exogenous fluorochromes.
The original GFP was cloned from the bioluminescent Aequorea victoria jelly fish (Prasher et al. 1992), and had an absorption and excitation wavelengths in the green part of the spectrum from 489 to 508 nm, respectively. Subsequent cloning of ‘GFP-like’ proteins from fluorescent, but non-bioluminescent Anthozoa coral has extended the available palette for GFP imaging from blue to red (440–610 nm; Whitaker 2000). However, significant gaps still occur in the GFP palette with clustering around cyan-green and orange-red parts of the spectrum (Matz et al. 2002).
The extended GFP palette has permitted the imaging of several different proteins simultaneously (Zaccolo 2004), and has provided new perspectives for fluorescence resonance energy transfer (FRET) based techniques (Matz et al. 2002). FRET is based on the distance-dependent energy transfer form neighbouring donor fluorophore and acceptor fluorophore pairs, which are appropriately orientated and have sufficient overlap between their emission and excitation spectra (Zaccolo 2004). FRET causes a decrease in donor fluorescence intensity and lifetime, and an increase in acceptor emission intensity. FRET efficiency is governed by an inverse sixth dependence on distance and is able to resolve proteins to within 2–6 nm, which is well below the 200 nm resolution of optical microscopes (Zaccolo 2004). Thus, FRET provides a unique and very sensitive tool for studying protein–protein and protein–substrate interactions in situ (Whitaker 2000; Matz et al. 2002).
Originally, GFP and blue fluorescent protein (BFP) pairs were used for FRET applications, but BFPs suffered from low emission intensity, were susceptible to photobleaching and required UV excitation which increased autofluorescence and scattering and therefore reduced sensitivity. Cyan fluorescent protein and yellow fluorescent protein pairs were introduced to overcome these difficulties (Zaccolo 2004). Recently, red fluorescent proteins have been developed with potentially greater tissue penetration and minimized autofluorescence background properties (Campbell et al. 2002), but their application to FRET is at an early stage and requires further developments (Zaccolo 2004).
Associated with FRET is fluorescence lifetime imaging microscopy (FLIM), which produces an image of the fluorescence lifetimes across a sample rather than an intensity map (Whitaker 2000). Combining FLIM and FRET techniques enables populations of interacting proteins to be determined on a point-by-point basis at high spatial (nm) and temporal (ns) resolution with an acquisition time of a few minutes (Peter & Ameer-Beg 2004).
2.2.5 Bioluminescence imaging.
BLI is based on the self-emission of light from yellow to green wavelengths owing to catalysis of luciferase enzymes in the presence of luciferin substrates (Weissleder & Mahmood 2001; Wu et al. 2001).
Luciferase genes are present in a large number of organisms including bacteria, Photinus pyralis firefly, Renilla coral, Aequorea victoria jelly fish and Gonyaulax dinoflagellates (Weissleder & Mahmood 2001). Cloning and transfer of these genes to non-bioluminescent organisms is possible and provides a useful contrast agent for the non-invasive BLI of gene expression in vivo. These transferred luciferase enzymes are ‘probed’ by externally-administered luciferin substrates. For in vivo BLI the firefly luciferase (FL) has been widely used.
The combination of luciferase genes from Renilla coral and GFPs enables bioluminescence resonance energy transfer (BRET) based techniques to be applied to cultured cells. However, the application of BRET in vivo is limited due to the high absorption of emissions in the blue-green region of the spectrum by haemoglobin (Contag & Ross 2002).
Mammalian tissues do not autoluminesce, and because bioluminescence only occurs when luciferase enzymes interact with luciferin substrates, BLI has minimal background signal and an excellent SNR. The spatial resolution of BLI is generally low, but the fast FL enzyme turnover (tone-half=3 h) in the presence of luciferin substrates allows for real-time measurements (Wu et al. 2001), with acquisition times ranging from a few seconds to several minutes (Doyle et al. 2004). Peak FL signal intensity occurs a few days after the injection of luciferin substrate and gradually disappears over several months (Wu et al. 2001), thus serial imaging of the same subject is possible and enables cell tracking in vivo (Tang et al. 2003).
The relationship between FL enzyme concentration and the peak height of emitted light is highly linear (Wu et al. 2001), and it has been shown that it is possible to use BLI to a tissue depth of 2 cm with approximately 106 bioluminescent cells (Contag & Ross 2002). Therefore, BLI is suited to small animals, where improvements to sensitivity can be made by reducing light absorption and scattering by shaving fur and removing skin tissues, although for nude animals this is not necessary (Wu et al. 2001).
The detection of reporter gene expression in vivo often requires reporter genes to be expressed at very high levels which are not always possible. A recently introduced two-step amplification process employing the luciferase gene and a synthetic promoter has produced artificially high levels of reporter gene expression with a 50-fold signal amplification, while maintaining tissue specificity (Contag & Ross 2002).
However, BLI is not a tomographic imaging modality and therefore is unable to localize to the exact site of gene expression (Wu et al. 2002). The repeated administration of FL gene expression causes an immune response lasting 2–3 days that significantly inhibits secondary adenoviral-mediated transduction. This may be overcome by using less immunogenic adenoviruses and alternating the use of different vectors in conjunction with immuno-suppressants (Jooss et al. 1996).
2.3 Nuclear imaging
Nuclear imaging is based on the administration and detection of decaying radioisotopes in vivo. The radioisotopes are combined with biologically active compounds to form radiopharmaceuticals which target specific biochemical events. The decay of a radioisotope emits a positron or a gamma ray which produces either two or a single high-energy photon. The detection of these photons is performed using PET and single photon-emission computed tomography (SPECT), respectively.
Both PET and SPECT have been used for molecular imaging for many years because of their superb sensitivity, availability and specificity of contrast agents, excellent FOV ranging from centimetres to sub-metre scales, good temporal resolution (seconds to minutes) and fast examination times (seconds to minutes). However, both PET and SPECT suffer from low spatial resolution because of the difficulty of determining the exact origin of emission in tissue.
2.3.1 Positron-emission tomography.
PET is based on the decay of a radioisotope, which emits a positron and annihilates with an electron to produce two high-energy (511 keV) photons that propagate in nearly opposite directions. Detection of these propagating photons is performed using a cylindrical ring of scintillation crystals, e.g. bismuth–germinate (BGO), arranged into a block (array) and coupled to PMTs and associated electronics.
When the high-energy photons travel through a crystal they interact with electrons in the valence band and cause them to move to the conduction band. These electrons return to the valence band at impurities in the crystal and emit light, i.e. scintillate, which is collected by a PMT and its associated electronics. The scintillated light decays exponentially at a rate characteristic of each individual crystal in the array. Thus, the output of a PMT can be decoded to reveal in which crystal in the array scintillation occurred (Ollinger & Fessler 1997).
For PET imaging, a positron emission event is recorded every time two photons are detected within a short timing window (ca. 10 ns) by opposing crystals in the cylindrical ring, i.e. the photons are coincident. The detection of coincident photons obviates the need for collimation, and is performed electronically. Image reconstruction is performed using successive summation of detected positron emission events in either two or three dimensions depending on the design of the detector (Ollinger & Fessler 1997).
PET is inherently a contrast-agent (tracer) based imaging method which uses a number of positron emitting isotopes including 11C, 13N, 15O, 18F, 64Cu, 68Ga, 76Br and 94mTc (Phelps 1992; Sharma et al. 2002), which emit two photons upon annihilation. PET contrast agents have a short half-life (tone-half minutes to hours) which requires PET scanners to be geographically close to a cyclotron.
The sensitivity of PET is of the order of 10−11–10−12 M (Mandl et al. 2002), and is determined by the number of counts collected during the scan, which in turn is dependent on the dosage of administered radiopharmaceutical, the duration of the scan, the sensitivity of the detector and the count-rate capability of the scanner (Ollinger & Fessler 1997). The recent introduction of lutetium-oxyorthosilicate (LSO) crystals, which scintillate faster than BGO crystals, has alleviated count-rate limitations of earlier PET scanners and increased the SNR (Miller et al. 2003).
The spatial resolution of a PET image is dependent on the crystal size, decoding process, annihilation photon acollinearity, positron range (distance the positron travels before annihilation with an electron) and reconstruction algorithm (Moses et al. 1997). For clinical PET applications a spatial resolution of 5 mm is typical. The introduction of LSO crystals with fibre optic readout has enabled one-to-one coupling to multiplexed PMTs, and thus has significantly reduced crystal size and improved spatial resolution to less than 2 mm, and therefore has enabled small animal microPET applications (Chatziioannou et al. 1999). Finally, the development of HIDAC-PET cameras (HIDAC: high density avalanche chamber) has further improved spatial resolution to less than 1.3 mm (Jeavons et al. 1999).
2.3.2 Single photon emission tomography.
SPECT is similar to PET, but the radioisotopes used for SPECT emit only a single high-energy (gamma) photon, and hence SPECT does not require the detection of coincidence, and, therefore, a different detector architecture is required.
SPECT detects gamma photons using a gamma camera which is step-rotated around the subject and forms an image using a back-projection algorithm. The gamma camera comprises of a collimator, detector crystals and PMTs. The collimator is a pattern of holes in a gamma-ray-absorbing material such as lead or tungsten, which ensures that all detected photons have propagated along parallel paths so that the origin of emission can be discerned. Thallium-activated sodium iodide (NaI(TI)) detector crystals are generally used in SPECT gamma cameras to convert high-energy photons to visible light.
SPECT is also inherently a contrast agent (tracer) based imaging method which uses a number of gamma emitting isotopes including 133Xe, 99mTc and 123I. These are heavy radioisotopes, they produce a single photon upon decay, and are more readily available and have longer decay times (tone-half hours to days) than those used in PET (Mandl et al. 2002). SPECT tracers are also available in an array of different energy levels, thus enabling simultaneous two-tracer imaging (Labbe 2003). However, SPECT tracers can illuminate other areas rather than their target area, and may require co-registration with another imaging modality such as computer tomography for confirmation of target site (Labbe 2003).
The sensitivity of SPECT is lower than PET because it uses a mechanical collimator which absorbs many photons, whereas in PET, collimation is done electronically. The sensitivity of SPECT is one to two orders of magnitude less than PET (approximately 10−10 M; Mandl et al. 2002).
The spatial resolution of SPECT is affected by collimation errors, although SPECT does not rely on paired photon annihilation, and therefore does not suffer from acollinearity. For clinical applications, the spatial resolution of SPECT is lower than PET. However, for animal microSPECT applications, where pixellated crystals which minimize the spread of scintillated light and pinhole collimators which minimize the barrel distortion at edges (Labbe 2003) are employed, a spatial resolution of 1.25 mm has been reported (MacDonald et al. 2001). However, this spatial resolution decreased with increasing distance from the detector to source, whereas in the HIDAC-PET system (Jeavons et al. 1999), the spatial resolution remained constant over a similar FOV.
3. Targeting and sensing
Probes for molecular imaging essentially consist of two components: a targeting or affinity component and an imaging or sensing component. The cellular or subcellular target may be a receptor, a cell surface marker, a protein or enzyme, DNA and RNA. Specific high-affinity interactions are essential in the design of ligands which may be small molecules (e.g. enzyme substrates, receptor ligands) or larger macromolecules (e.g. monoclonal antibodies, synthesized protein fragments or naturally occurring high affinity compounds, e.g. biotin/avidin). Sensing is achieved by a radioactive label, an optical or fluorescent probe, or by an agent that gives MRI contrast. In each case the chemistry necessary to develop a molecular imaging probe is different. In radiolabelling, sensing can be achieved by isotopic substitution in the targeting agent (e.g. replacing 12C with 11C) or by introducing a label like 18F or a larger chelate containing other isotopes (e.g. 99mTc-based imaging agents; Dilworth & Parrott 1998). In optical imaging, a fluorochrome or dye is attached as the sensor, while in MRI, contrast is given by paramagnetic agents (e.g. gadolinium). Since the magnitude of the relaxation by a single paramagnetic label is generally too small, functionalized polymerized vesicles or nanoparticles are additionally needed to attach more than one label in each molecular imaging probe (Li & Bednarski 2002). It is often necessary to introduce a spacer between the affinity component and the sensor so as to avoid the bulky label inhibiting the specific binding process. Thus, the design of a good molecular imaging probe requires: (i) understanding the nature of the cellular target (i.e. the biological question to be answered); (ii) the use of imaginative chemistry combined with the tools of molecular biology and (iii) a good appreciation of the advantages and limitations of the measurement in terms of sensitivity and spatial resolution. Advances in drug discovery processes such as combinatorial chemistry and high-throughput testing are likely to have a significant impact on the development of new molecular imaging probes (Weissleder 2001).
Targeted imaging probes have several drawbacks. First, a new probe has to be designed and constructed and characterized for each target, be it a protein, a receptor or DNA. Second, even with a strong affinity probe, non-specific binding and distribution in the tissue gives a large background signal. Ideally, the background tracer needs to be cleared for optimal detection of the bound probe. Clearance kinetics in vivo from the interstitial space or from within the cell may be too slow but can sometimes be speeded up by the use of specific ‘chase’ compounds (Goodwin et al. 1988). Third, the number of targets (receptors, DNA and mRNA) are limited and the sensitivity of the imaging measurement may not be sufficient. Targeting proteins and protein function, on the other hand, is relatively less demanding.
Some of the difficulties in using targeted probes can be overcome by strategies that result in signal activation and amplification. Activatable imaging probes (referred to as smart probes, sensors or beacons) give a signal only when they interact with their target. One example of a molecular beacon is a single DNA strand with a fluorophore attached to one end and a quencher to the other. Unhybridized, the DNA folds into a hairpin shape so that the quencher is close to the fluorophore and stops it fluorescing. When the strand binds to DNA or RNA in the cell, it unwinds, and the fluorescence is no longer quenched and the probe ‘lights up’ (Mitchell 2001). Self-quenched fluorescent probes have also been used to investigate different proteolytic functions, e.g. metalloproteases (Bremer et al. 2001), cathepsins (Bogdanov et al. 2002) and others. The two probes are linked with a synthetic peptide spacer that can be specifically cleaved by the enzyme of interest, resulting in the separation of the fluorophores and enhancement of the fluorescence signal.
Optical sensors based on the introduction of luciferase enzymes and luciferin substrates are further examples of activatable optical probes and are particularly relevant to gene expression studies (see below). The calcium sensitive bioluminescent protein aequorin has been used for many years for the detection of intracellular Ca2+, but the quantum yield for its light emission is very low and the signal is difficult to detect. Thus, its use in whole-organism studies is restricted. A new group of calcium-sensitive bioluminescent reporter genes have been constructed by fusing GFP and aequorin, resulting in much more light being emitted as a result of chemiluminescene resonance energy transfer to GFP (Baubet et al. 2000). This system can be targeted to various subcellular components (Chiesa et al. 2001) and is likely to be sufficiently robust for whole organism studies of calcium and other signalling systems.
Many ‘Smart’ MRI agents that report specific enzymatic activities have also been designed (Josephson et al. 2002; Meade et al. 2003) and used in particular for following gene expression. The use of surface-functionalized nanoparticles provides an alternative means of increasing the sensitivity of detection, particularly in the case of molecular MRI contrast agents. A whole range of nanoparticles have been used including polymerized vesicles (from derivatised polymerizable lipids; Li & Bednarski 2002; Wickline & Lanza 2002), caged magnetic nanoparticles (Schellenberger et al. 2004), nanoparticulate emulsions (Winter et al. 2003). The functional groups attached were aimed at targeting biochemical epitopes (e.g. integrins, apoptotic cells) and to carry multiple groups (normally chelators) that carried the contrast agent (e.g. gadolinium). Functionalized nanoparticle libraries could provide a rapid synthesis and screening for specific target interactions (Schellenberger et al. 2004). Multifunctional nanoparticle probes based on fluorescent quantum dots have been developed by several groups (Mitchell 2001). The quantum dots, for example, were encapsulated with a triblock copolymer and linking this amphiphilic polymer to targeting ligands to specific tumours. Using these quantum dots multicolour imaging of cancer cells in vivo was achieved (Gao et al. 2004). One important advantage of most nanoparticle constructs is that as well as providing specific biomarkers, they can also incorporate drug-delivery functionalities.
Designing and constructing high affinity or activated probes with appropriate sensors for in vivo applications is only one part of the problem. While in studies of cellular preparations the delivery of the probe to the cell (or even inside the cell) is relatively straightforward, for the examination of intact organism and human applications additional steps are needed for efficient and safe use of the molecular probe. For example, polyethylene glycol conjugates of biomolecules decrease immunogenecity and the use of peptide-mediated translocation signals will aid the transport of probes across the cell membrane (Polyakov et al. 2000).
4. Some applications of molecular imaging in biology and medicine
The explosion in the application of molecular imaging in the last 5 years has resulted in many excellent review articles. Some recent articles include those by Massoud & Gambhir (2003), an excellent general and comprehensive review; molecular imaging in the cardiovascular system (Jaffer et al. 2004), in cancer (Meares et al. 2003), of gene expression and protein function using PET and SPECT (Blasberg & Gelovani-Tjuvajev 2002; Sharma et al. 2002), of cellular events in developing embryos using GFP (Yu et al. 2003). A whole special issue of the Journal of Magnetic Resonance Imaging (vol 16, issue 4: 2002) is devoted to molecular imaging and two recent volumes of Methods in Enzymology (vol 385 and 386: 2004) cover many aspects of the subject.
One of the most intensively studied applications is aimed at determining the pattern of gene expression in vivo. This can be directed at genes externally introduced into the system or monitoring the expression of endogenous genes (Phelps 2000; Massoud & Gambhir 2003). PET (Phelps 2000; Sharma et al. 2002) and optical methods have led the field, but some ingenious specific magnetic resonance (MR) approaches have also been developed. A reporter gene codes for a protein that is detectable with an imaging probe. For example, the enzyme thymidine kinase of the herpes simplex virus (HSV) can be detected with PET using the inhibitor 8-(F-18)fluoroganciclovir (an analogue of the drug used for treating HSV; Phelps 2000). Optical imaging based on the luciferase reporter gene, where the detection is bioluminescence requiring the delivery of a luminescent substrate (Bhaumik & Gambhir 2002), is increasingly used for rapid screening of gene delivery in transgenic mice. More precise localization of gene expression can be achieved using OPT (Sharpe et al. 2002), provided the specimen can be made to have a uniform refractive index, and the gene expression product mRNA or protein can be labelled or stained externally. Endogenous genes during development of small organisms such as Caenorhabditis elegans have been studied with great success. For example, de Bono et al. (2002) examined several genes in the nervous system of the worm and were able to relate their presence or absence of the ‘social behaviour’ of different strains.
MRI, on the other hand, does not require optical transparency and a molecular probe that is sensitive to the activity of β-galactosidase (a gadolinium chelate protected by a carbohydrate cap that is cleaved by the enzyme) has been developed (Louie et al. 2000) and was used to examine the developing Xenopus embryo. Magnetic resonance spectroscopy (MRS) has also been used to follow the expression of marker genes. Viral-mediated gene transfer using recombinant adenovirus construct with the gene for the invertebrate enzyme arginine kinase was introduced into the hind limb of neonatal mice and the appearance of the enzyme in muscle was detected by 31P MRS following the formation of phosphoarginine (Walter et al. 2000).
Protein–protein interactions are fundamental to cellular functions. They are involved in the control of gene expression, signal transduction, metabolic pathways, in development and many other processes. At the cellular level, the so-called ‘two-hybrid system’ has been successfully used to map out protein–protein interactions, particularly in yeast cells. The principle of this method is that when a target protein binds to its ‘bait’ in the cell, their interaction brings together two halves of a transcriptional activator which then switches on the expression of the reporter gene. In a cell, this method can be used to screen a whole range of proteins using cDNA libraries to search for interactions. In several proof-of-concept-type studies, specific protein–protein interactions were followed in living animals using PET, fluorescence (GFP) and luminescence reporters (Luker et al. 2002; Ray et al. 2002). More recently, the luciferase complementation-based bioassay was used to study the homodimerization of HSV type 1 thymidine kinase in mammalian cells and living mice (Massoud et al. 2004).
Apoptosis, programmed cell death, is a key biological process in organ development, the removal of damaged cells in tissues and defects in the apoptotic pathways may contribute to several diseases. Specific probes for apoptotic cells were designed on the basis that the protein annexin binds strongly to the lipid phosphatidyl-serine in cell membranes and that at an early stage of apoptosis, these lipid molecules translocate to the outer face of the membrane (Martin et al. 1995). It has been demonstrated that cells undergoing apoptosis can be detected in vivo by imaging radionuclide-labelled annexin in animal models including fulminant hepatitis, cardiac allograft rejection and tumour response to treatment (Blankenberg et al. 1998). Tumour apoptosis was observed in nude mice injected with annexin labelled with a NIR probe (Petrovsky et al. 2003). At a more general level, probes for apoptotic cells, detectable by MR or fluorescence were derived by screening libraries of surface-functionalized nanoparticles (Schellenberger et al. 2004).
Cell migration and cell implantation are important to follow both in relation to immune function in vivo and because of the new cell therapies largely based on the use of stem cells. Cells can be labelled with radionuclides, MRI contrast agents, fluorescent or bioluminescent markers. The last of these methods has been particularly successful in elucidating the spatio-temporal trafficking patterns of lymphocytes in animal models for human disease (Mandl et al. 2002). To study the migration of implanted stem cells, mouse embryonic stem cells were labelled with ultrasmall superparamagnetic iron-oxide particles and injected into the unaffected and healthy contralateral hemisphere of an infracted rat brain. MRI was used to show that during 3 weeks, cells migrated towards the damaged brain tissue on the hemisphere opposite to the implantation sites. These experiments demonstrated the high migrational dynamics of embryonic stem (ES) cells (Hoehn et al. 2002). Several groups have shown, in various animal models of cardiac infarct, that myogenic precursor cells (from different sources) loaded with iron-oxide can be detected in vivo by cardiac MRI (Garot et al. 2003).
Cancer, receptors and angiogenesis have been extensively investigated by the whole range of molecular imaging approaches. In all of these, the key has been the design of imaging probes where the specificity of the binding relied on defining agents that recognized some component of the cancer cell, the receptor or the biological process that was characteristic of the system to be studied. In angiogenesis of tumours, for example, ανβ3-integrin is an attractive biochemical epitope because it is highly expressed in activated neovascular endothelial cells and is essentially absent in mature quiescent cells. Sipkins et al. (1998) constructed paramagnetic (gadolinium labelled) polymerized liposomes conjugated to biotinylated antibodies against ανβ3-integrins and used MRI to detect angiogenic vasculature in a rabbit carcinoma model. More recently, in a similar rabbit model integrin-targeted paramagnetic nanoparticles were shown to detect and spatially characterize neovascularity by MRI at the common clinical field strength of 1.5 T (Winter et al. 2003).
5. Concluding remarks
Molecular imaging seeks to understand the components, processes, dynamics and therapies of disease from a molecular perspective, by using and developing imaging technologies and contrast agents. The key to success in this diverse endeavour is the multi-disciplinary interaction between the life and physical sciences. For example, the disciplines of physics and engineering have provided the hardware; mathematics and computing the software and analysis tools; chemistry, materials science and biology the contrast agents; and biology and medicine have posed the biological and medical questions to be answered.
The tools of molecular imaging are the imaging modalities and their corresponding contrast agents. The imaging modalities of MRI, optical imaging and nuclear imaging are emerging as key modalities for in vivo molecular imaging because of their ability to detect molecular events when combined with contrast agents with sufficient sensitivity, specificity, temporal resolution and spatial resolution. Effective use of the tools of molecular imaging requires knowledge of the basis of detection of the imaging modality, contrast agent mechanism and the biological environment.
Molecular imaging provides an interface between the life and physical sciences at the level of the contrast agent. Recent developments in contrast agent technology have increased their specificity and therapeutic potential. This will revolutionize the manner in which disease is managed and will rely on a multi-modal and multi-disciplinary approach drawn from specialists from the life and physical sciences, who understand each other's language and share the common goal of combating disease.
Acknowledgments
We are grateful to the British Heart Foundation for supporting this work.
References
- Aime S, Cabella C, Colombatto S, Geninatti Crich S, Gianolio E, Maggioni F. Insights into the use of paramagnetic Gd(III) complexes in MR-molecular imaging investigations. J. Magn. Reson. Imaging. 2002;16:394–406. doi: 10.1002/jmri.10180. [DOI] [PubMed] [Google Scholar]
- Ardenkjaer-Larsen J.H, Fridlund B, Gram A, Hansson G, Hansson L, Lerche M.H, Servin R, Thaning M, Golman K. Increase in signal-to-noise ratio of >10,000 times in liquid-state NMR. Proc. Natl Acad. Sci. USA. 2003;100:10 158–10 163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artemov D. Molecular magnetic resonance imaging with targeted contrast agents. J. Cell. Biochem. 2003;90:518–524. doi: 10.1002/jcb.10660. [DOI] [PubMed] [Google Scholar]
- Baubet V, Le Mouellic H, Campbell A.K, Lucas-Meunier E, Fossier P, Brulet P. Chimeric green fluorescent protein-aequorin as bioluminescent Ca2+ reporters at the single-cell level. Proc. Natl Acad. Sci. USA. 2000;97:7260–7265. doi: 10.1073/pnas.97.13.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S, Gambhir S.S. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc. Natl Acad. Sci. USA. 2002;99:377–382. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg F.G. In vivo detection and imaging of phosphatidylserine expression during programmed cell death. Proc. Natl Acad. Sci. USA. 1998;95:6349–6354. doi: 10.1073/pnas.95.11.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasberg R.G, Gelovani-Tjuvajev J. In vivo molecular-genetic imaging. J. Cell. Biochem. Suppl. 2002;39:172–183. doi: 10.1002/jcb.10433. [DOI] [PubMed] [Google Scholar]
- Boas D.A, Brooks D.H, Miller E.L, DiMarzio C.A, Gaudette R.J, Zhang Q. Imaging the body with diffuse optical tomography. IEEE Sig Process Mag. 2001;18:57–75. [Google Scholar]
- Boas D.A, Chen K, Grebert D, Franceschini M.A. Improving the diffuse optical imaging spatial resolution of the cerebral hemodynamic response to brain activation in humans. Opt. Lett. 2004;29:1506–1508. doi: 10.1364/ol.29.001506. [DOI] [PubMed] [Google Scholar]
- Bogdanov A.A, Jr, Lin C.P, Simonova M, Matuszewski L, Weissleder R. Cellular activation of the self-quenched fluorescent reporter probe in tumor microenvironment. Neoplasia. 2002;4:228–236. doi: 10.1038/sj.neo.7900238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer C, Tung C.H, Weissleder R. In vivo molecular target assessment of matrix metalloproteinase inhibition. Nat. Med. 2001;7:743–748. doi: 10.1038/89126. [DOI] [PubMed] [Google Scholar]
- Callaghan P.T. Oxford University Press; Oxford: 1991. Principles of nuclear magnetic resonance microscopy. [Google Scholar]
- Campbell R.E, Tour O, Palmer A.E, Steinbach P.A, Baird G.S, Zacharias D.A, Tsien R.Y. A monomeric red fluorescent protein. Proc. Natl Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W.C, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- Chance B. A novel method for fast imaging of brain function, non-invasively, with light. Opt. Express. 1998;2:411–423. doi: 10.1364/oe.2.000411. [DOI] [PubMed] [Google Scholar]
- Chatziioannou A.F, Cherry S.R, Shao Y, Silverman R.W, Meadors K, Farquhar T.H, Pedarsani M, Phelps M.E. Performance evaluation of microPET: a high-resolution lutetium oxyorthosilicate PET scanner for animal imaging. J. Nucl. Med. 1999;40:1164–1175. [PMC free article] [PubMed] [Google Scholar]
- Chiesa A, Rapizzi E, Tosello V, Pinton P, de Virgilio M, Fogarty K.E, Rizzuto R. Recombinant aequorin and green fluorescent protein as valuable tools in the study of cell signalling. Biochem. J. 2001;355:1–12. doi: 10.1042/0264-6021:3550001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contag C.H, Ross B.D. It's not just about anatomy: in vivo bioluminescence imaging as an eyepiece into biology. J. Magn. Reson. Imaging. 2002;16:378–387. doi: 10.1002/jmri.10178. [DOI] [PubMed] [Google Scholar]
- de Bono M, Tobin D.M, Davis M.W, Avery L, Bargmann C.I. Social feeding in Caenorhabditis elegans is induced by neurons that detect aversive stimuli. Nature. 2002;419:899–903. doi: 10.1038/nature01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth J.R, Parrott S.J. The biomedical chemistry of technetium and rhenium. Chem. Soc. Rev. 1998;27:43–55. [Google Scholar]
- Doyle T.C, Burns S.M, Contag C.H. In vivo bioluminescence imaging for integrated studies of infection. Cell. Microbiol. 2004;6:303–317. doi: 10.1111/j.1462-5822.2004.00378.x. [DOI] [PubMed] [Google Scholar]
- Franceschini M.A, et al. Frequency-domain techniques enhance optical mammography: initial clinical results. Proc. Natl Acad. Sci. USA. 1997;94:6468–6473. doi: 10.1073/pnas.94.12.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangioni J.V. In vivo near-infrared fluorescence imaging. Curr. Opin. Chem. Biol. 2003;7:626–634. doi: 10.1016/j.cbpa.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Gadian D.G. Oxford University Press; Oxford: 1995. NMR and its application to living systems. [Google Scholar]
- Gao X, Cui Y, Levenson R.M, Chung L.W, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat. Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- Garot J, Unterseeh T, Teiger E, Champagne S, Chazaud B, Gherardi R, Hittinger L, Gueret P, Rahmouni A. Magnetic resonance imaging of targeted catheter-based implantation of myogenic precursor cells into infarcted left ventricular myocardium. J. Am. Coll. Cardiol. 2003;41:1841–1846. doi: 10.1016/s0735-1097(03)00414-5. [DOI] [PubMed] [Google Scholar]
- Golman K, Ardenkjaer-Larsen J.H, Petersson J.S, Mansson S, Leunbach I. Molecular imaging with endogenous substances. Proc. Natl Acad. Sci. USA. 2003;100:10 435–10 439. doi: 10.1073/pnas.1733836100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin D, Meares C, McCall M, McTigue M, Chaovapong W. Pre-targeted immunoscintigraphy of murine tumors with indium-111-labeled bifunctional haptens. J. Nucl. Med. 1988;29:226–234. [PubMed] [Google Scholar]
- Hoehn M, et al. Monitoring of implanted stem cell migration in vivo: a highly resolved in vivo magnetic resonance imaging investigation of experimental stroke in rat. Proc. Natl Acad. Sci. USA. 2002;99:16 267–16 272. doi: 10.1073/pnas.242435499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoult D.I, Richards R.E. The signal-to-noise ratio of the nuclear magnetic resonance experiment. J. Magn. Reson. 1976;24:71–85. doi: 10.1016/j.jmr.2011.09.018. [DOI] [PubMed] [Google Scholar]
- Jacobs R.E, Papan C, Ruffins S, Tyszka J.M, Fraser S.E. MRI: volumetric imaging for vital imaging and atlas construction. Nat. Rev. Mol. Cell. Biol. 2003;(Suppl.):SS10–SS16. [PubMed] [Google Scholar]
- Jaffer F.A, Tung C.H, Wykrzykowska J.J, Ho N.H, Houng A.K, Reed G.L, Weissleder R. Molecular imaging of factor XIIIa activity in thrombosis using a novel, near-infrared fluorescent contrast agent that covalently links to thrombi. Circulation. 2004;110:170–176. doi: 10.1161/01.CIR.0000134484.11052.44. [DOI] [PubMed] [Google Scholar]
- Jeavons A.P, Chandler R.A, Dettmar C.A.R. A 3D HIDAC-PET camera with sub-millimetre resolution for imaging small animals. IEEE Trans. Nucl. Sci. 1999;46:468–473. [Google Scholar]
- Jooss K, Yang Y, Wilson J.M. Cyclophosphamide diminishes inflammation and prolongs transgene expression following delivery of adenoviral vectors to mouse liver and lung. Hum. Gene Ther. 1996;7:1555–1566. doi: 10.1089/hum.1996.7.13-1555. [DOI] [PubMed] [Google Scholar]
- Josephson L, Kircher M.F, Mahmood U, Tang Y, Weissleder R. Near-infrared fluorescent nanoparticles as combined MR/optical imaging probes. Bioconjug. Chem. 2002;13:554–560. doi: 10.1021/bc015555d. [DOI] [PubMed] [Google Scholar]
- Kim S, et al. Near-infrared fluorescent type II quantum dots for sentinel lymph node mapping. Nat. Biotechnol. 2004;22:93–97. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe J.P. SPECT/CT emerges from the shadow of PET/CT. Biophotonics Int. 2003:50–57. [Google Scholar]
- Li K.C, Bednarski M.D. Vascular-targeted molecular imaging using functionalized polymerized vesicles. J. Magn. Reson. Imaging. 2002;16:388–393. doi: 10.1002/jmri.10174. [DOI] [PubMed] [Google Scholar]
- Louie A.Y, Huber M.M, Ahrens E.T, Rothbacher U, Moats R, Jacobs R.E, Fraser S.E, Meade T.J. In vivo visualization of gene expression using magnetic resonance imaging. Nat. Biotechnol. 2000;18:321–325. doi: 10.1038/73780. [DOI] [PubMed] [Google Scholar]
- Luker G.D, Sharma V, Pica C.M, Dahlheimer J.L, Li W, Ochesky J, Ryan C.E, Piwnica-Worms H, Piwnica-Worms D. Noninvasive imaging of protein–protein interactions in living animals. Proc. Natl Acad. Sci. USA. 2002;99:6961–6966. doi: 10.1073/pnas.092022399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald L.R, Patt B.E, Iwanczyk J.S, Tsui B.M.W, Wang Y, Frey E.C, Wessell D.E, Acton P.D, Kung H.F. Pinhole SPECT of mice using the LumaGEM gamma camera. IEEE Trans. Nucl. Sci. 2001;48:830–836. [Google Scholar]
- Mahmood U, Tung C.H, Bogdanov A, Jr, Weissleder R. Near-infrared optical imaging of protease activity for tumor detection. Radiology. 1999;213:866–870. doi: 10.1148/radiology.213.3.r99dc14866. [DOI] [PubMed] [Google Scholar]
- Mandl S, Schimmelpfennig C, Edinger M, Negrin R.S, Contag C.H. Understanding immune cell trafficking patterns via in vivo bioluminescence imaging. J. Cell. Biochem. Suppl. 2002;39:239–248. doi: 10.1002/jcb.10454. [DOI] [PubMed] [Google Scholar]
- Mansfield P, Morris P.G. Academic Press; New York: 1982. NMR imaging in biomedicine. [Google Scholar]
- Martin S.J, Reutelingsperger C.P, McGahon A.J, Rader J.A, van Schie R.C, LaFace D.M, Green D.R. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J. Exp. Med. 1995;182:1545–1556. doi: 10.1084/jem.182.5.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoud T.F, Gambhir S.S. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- Massoud T.F, Paulmurugan R, Gambhir S.S. Molecular imaging of homodimeric protein–protein interactions in living subjects. FASEB J. 2004;18:1105–1107. doi: 10.1096/fj.03-1128fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz M.V, Lukyanov K.A, Lukyanov S.A. Family of the green fluorescent protein: journey to the end of the rainbow. Bioessays. 2002;24:953–959. doi: 10.1002/bies.10154. [DOI] [PubMed] [Google Scholar]
- Meade T.J, Taylor A.K, Bull S.R. New magnetic resonance contrast agents as biochemical reporters. Curr. Opin. Neurobiol. 2003;13:597–602. doi: 10.1016/j.conb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Meares C.F, Chmura A.J, Orton M.S, Corneillie T.M, Whetstone P.A. Molecular tools for targeted imaging and therapy of cancer. J. Mol. Recognit. 2003;16:255–259. doi: 10.1002/jmr.634. [DOI] [PubMed] [Google Scholar]
- Michalet X, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.A, Rouze N.C, Hutchins G.D. Small animal PET imaging. In: Barone M, Borchi E, Leroy C, Rancoita P.-G, Riboni P.-L, Ruchti R, editors. Astroparticle, particle and space physics, detectors and medical physics applications. World Scientific; Singapore: 2003. pp. 381–390. [Google Scholar]
- Mitchell P. Turning the spotlight on cellular imaging. Nat. Biotechnol. 2001;19:1013–1017. doi: 10.1038/nbt1101-1013. [DOI] [PubMed] [Google Scholar]
- Moses W.M, Virador P.R.G, Derenzo S.E, Huesman R.H, Budinger T.F. Design of a high-sensitivity PET camera for human brains and small animals. IEEE Trans. Nucl. Sci. 1997;NS-44:1487–1491. [Google Scholar]
- Ntziachristos V, Yodh A.G, Schnall M, Chance B. Concurrent MRI, diffuse optical tomography of breast after indocyanine green enhancement. Proc. Natl Acad. Sci. USA. 2000;97:2767–2772. doi: 10.1073/pnas.040570597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollinger J.M, Fessler A.F. Positron emission tomography. IEEE Sig. Process Mag. 1997;14:43–55. [Google Scholar]
- Peter M, Ameer-Beg S.M. Imaging molecular interactions by multiphoton FLIM. Biol. Cell. 2004;96:231–236. doi: 10.1016/j.biolcel.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Petrovsky A, Schellenberger E, Josephson L, Weissleder R, Bogdanov A., Jr Near-infrared fluorescent imaging of tumor apoptosis. Cancer Res. 2003;63:1936–1942. [PubMed] [Google Scholar]
- Phelps M.E. Clinical brain imaging: principles and applications. F.A. Davis Company; Philadelphia: 1992. Positron emission tomography. [Google Scholar]
- Phelps M.E. Inaugural article: positron emission tomography provides molecular imaging of biological processes. Proc. Natl Acad. Sci. USA. 2000;97:9226–9233. doi: 10.1073/pnas.97.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue B.W. Focus issue: biomedical diffuse optical tomography. Opt. Express. 1999;4:230. doi: 10.1364/oe.4.000270. [DOI] [PubMed] [Google Scholar]
- Polyakov V, Sharma V, Dahlheimer J.L, Pica C.M, Luker G.D, Piwnica-Worms D. Novel Tat-peptide chelates for direct transduction of technetium-99m and rhenium into human cells for imaging and radiotherapy. Bioconjug. Chem. 2000;11:762–771. doi: 10.1021/bc000008y. [DOI] [PubMed] [Google Scholar]
- Potter K. Magnetic resonance microscopy approaches to molecular imaging: sensitivity vs. specificity. J. Cell. Biochem. Suppl. 2002;39:147–153. doi: 10.1002/jcb.10421. [DOI] [PubMed] [Google Scholar]
- Prasher D.C, Eckenrode V.K, Ward W.W, Prendergast F.G, Cormier M.J. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- Ray P, Pimenta H, Paulmurugan R, Berger F, Phelps M.E, Iyer M, Gambhir S.S. Noninvasive quantitative imaging of protein–protein interactions in living subjects. Proc. Natl Acad. Sci. USA. 2002;99:3105–3110. doi: 10.1073/pnas.052710999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberger E.A, Reynolds F, Weissleder R, Josephson L. Surface-functionalized nanoparticle library yields probes for apoptotic cells. Chembiochem. 2004;5:275–279. doi: 10.1002/cbic.200300713. [DOI] [PubMed] [Google Scholar]
- Schneider J.E, Bamforth S.D, Grieve S.M, Clarke K, Bhattacharya S, Neubauer S. High-resolution, high-throughput magnetic paragraph sign resonance imaging of mouse embryonic paragraph sign anatomy using a fast gradient-echo sequence. Magma. 2003;16:43–51. doi: 10.1007/s10334-003-0002-z. [DOI] [PubMed] [Google Scholar]
- Sharma V, Luker G.D, Piwnica-Worms D. Molecular imaging of gene expression and protein function in vivo with PET and SPECT. J. Magn. Reson. Imaging. 2002;16:336–351. doi: 10.1002/jmri.10182. [DOI] [PubMed] [Google Scholar]
- Sharpe J. Optical projection tomography as a new tool for studying embryo anatomy. J. Anat. 2003;202:175–181. doi: 10.1046/j.1469-7580.2003.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe J, Ahlgren U, Perry P, Hill B, Ross A, Hecksher-Sorensen J, Baldock R, Davidson D. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- Siegel A.M, Marota J.J.A, Boas D.A. Design and evaluation of a continuous-wave diffuse optical tomography system. Opt. Express. 1999;4:287–298. doi: 10.1364/oe.4.000287. [DOI] [PubMed] [Google Scholar]
- Sipkins D.A, Cheresh D.A, Kazemi M.R, Nevin L.M, Bednarski M.D, Li K.C. Detection of tumor angiogenesis in vivo by alphaVbeta3-targeted magnetic resonance imaging. Nat. Med. 1998;4:623–626. doi: 10.1038/nm0598-623. [DOI] [PubMed] [Google Scholar]
- Smith A.M, Gao X, Nie S. Quantum-dot nanocrystals for in-vivo molecular and cellular imaging. Photochem. Photobiol. 2004;80:377–385. doi: 10.1562/0031-8655(2004)080<0377:QDNFIV>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Tang Y, Shah K, Messerli S.M, Snyder E, Breakefield X, Weissleder R. In vivo tracking of neural progenitor cell migration to glioblastomas. Hum. Gene Ther. 2003;14:1247–1254. doi: 10.1089/104303403767740786. [DOI] [PubMed] [Google Scholar]
- Venugopalan V. 2004,. Optical society of America BIOMED topical meeting tutorial on tissue optics. pp. 1–32. [Google Scholar]
- Walter G, Barton E.R, Sweeney H.L. Noninvasive measurement of gene expression in skeletal muscle. Proc. Natl Acad. Sci. USA. 2000;97:5151–5155. doi: 10.1073/pnas.97.10.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward K.M, Aletras A.H, Balaban R.S. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST) J. Magn. Reson. 2000;143:79–87. doi: 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- Weissleder R. A clearer vision for in vivo imaging. Nat. Biotechnol. 2001;19:316–317. doi: 10.1038/86684. [DOI] [PubMed] [Google Scholar]
- Weissleder R, Mahmood U. Molecular imaging. Radiology. 2001;219:316–333. doi: 10.1148/radiology.219.2.r01ma19316. [DOI] [PubMed] [Google Scholar]
- Weissleder R, Elizondo G, Wittenberg J, Rabito C.A, Bengele H.H, Josephson L. Ultrasmall superparamagnetic iron oxide: characterization of a new class of contrast agents for MR imaging. Radiology. 1990;175:489–493. doi: 10.1148/radiology.175.2.2326474. [DOI] [PubMed] [Google Scholar]
- Weissleder R, Tung C.H, Mahmood U, Bogdanov A., Jr In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat. Biotechnol. 1999;17:375–378. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- Whitaker M. Fluorescent tags of protein function in living cells. Bioessays. 2000;22:180–187. doi: 10.1002/(SICI)1521-1878(200002)22:2<180::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Wickline S.A, Lanza G.M. Molecular imaging, targeted therapeutics, and nanoscience. J. Cell. Biochem. Suppl. 2002;39:90–97. doi: 10.1002/jcb.10422. [DOI] [PubMed] [Google Scholar]
- Winter P.M, et al. Molecular imaging of angiogenesis in nascent Vx-2 rabbit tumors using a novel alpha(nu)beta3-targeted nanoparticle and 1.5 tesla magnetic resonance imaging. Cancer Res. 2003;63:5838–5843. [PubMed] [Google Scholar]
- Wolff S.D, Balaban R.S. Magnetization transfer contrast (MTC) and tissue water proton relaxation in vivo. Magn. Reson. Med. 1989;10:135–144. doi: 10.1002/mrm.1910100113. [DOI] [PubMed] [Google Scholar]
- Wu J.C, Sundaresan G, Iyer M, Gambhir S.S. Noninvasive optical imaging of firefly luciferase reporter gene expression in skeletal muscles of living mice. Mol. Ther. 2001;4:297–306. doi: 10.1006/mthe.2001.0460. [DOI] [PubMed] [Google Scholar]
- Wu J.C, Inubushi M, Sundaresan G, Schelbert H.R, Gambhir S.S. Optical imaging of cardiac reporter gene expression in living rats. Circulation. 2002;105:1631–1634. doi: 10.1161/01.cir.0000014984.95520.ad. [DOI] [PubMed] [Google Scholar]
- Yu Y.A, Szalay A.A, Wang G, Oberg K. Visualization of molecular and cellular events with green fluorescent proteins in developing embryos: a review. Luminescence. 2003;18:1–18. doi: 10.1002/bio.701. [DOI] [PubMed] [Google Scholar]
- Zaccolo M. Use of chimeric fluorescent proteins and fluorescence resonance energy transfer to monitor cellular responses. Circ. Res. 2004;94:866–873. doi: 10.1161/01.RES.0000123825.83803.CD. [DOI] [PubMed] [Google Scholar]
- Zhang S, Merritt M, Woessner D.E, Lenkinski R.E, Sherry A.D. PARACEST agents: modulating MRI contrast via water proton exchange. Acc. Chem. Res. 2003;36:783–790. doi: 10.1021/ar020228m. [DOI] [PubMed] [Google Scholar]