Abstract
The antiestrogens tamoxifen and ICI 182,780 have been portrayed as competitive antagonists of the estrogen binding site of the α-form of the human estrogen receptor (ER). However, in functional studies, neither compound has consistently been able to block estradiol-induced transcription. In this report, three yeast genetic systems were used to investigate the effects of tamoxifen and ICI 182,780 on ER dimerization, transcriptional activation, and the interaction of the receptor with a coactivator, RIP140. Tamoxifen and ICI 182,780 were able to induce ER dimerization and ER-dependent transcription, albeit at up to 15,000-fold higher concentrations than that of estradiol. In the presence of RIP140, the transcription response maximum was increased up to 30-fold for estradiol and both antiestrogens. Whole yeast cell [3H]estradiol binding studies demonstrated that tamoxifen could displace the estradiol from the ER, whereas ICI 182,780 treatment resulted in a 4-fold increase in [3H]estradiol binding to the receptor. No antagonism of estradiol was observed with tamoxifen or ICI 182,780 in any of the yeast models employed. We have concluded that the antiestrogen activity of compounds like tamoxifen and ICI 182,780 is not caused by their ability to competitively antagonize estradiol binding to the hormone binding site, but possibly by their ability to induce ER-dependent transcription, which in mammalian systems would result in receptor down-regulation. Compounds such as tamoxifen act through the hormone binding site, whereas ICI 182,780 may cause receptor activation through an allosteric binding site.
The effect of estrogen on cells is mediated through the estrogen receptor (ER), a member of the family of ligand-dependent transcription factors (1, 2). The complexity of the responses seen with estrogen in different mammalian cells is caused in part by the existence of two receptor subtypes of the human ER, ERα and ERβ (3). Whereas the homology of the ligand binding domain of the receptor subtypes is 59% (4) and even higher for the amino acids that border the ligand binding pocket, the receptor subtypes show different transcriptional responses to antiestrogens (5, 6). The transcriptional response of the ER can be further modified by the presence of recently identified coregulatory proteins, which can enhance transcription activity (7–10). Because of the role played by estrogen in development and maintenance of some breast tumors (1), treatment with the antiestrogen tamoxifen has become a first-line therapy for controlling disease progression. However, almost all tumors that initially respond to tamoxifen therapy eventually become resistant to this antiestrogen (11, 12). This has been ascribed to the estrogen-like activity of the parent compound and its metabolites (13). As a consequence, the development of other, hopefully more potent and pure antiestrogens, such as ICI 182,780 (14), has been undertaken.
Although tamoxifen and ICI 182,780 have been portrayed as competitive antagonists of hormone binding to the ER (14, 15), their actual mechanism of action remains poorly understood. Both compounds also have been shown to produce a down-regulation of intracellular ER protein (16–18) and an impairment of both ER dimerization (19) and nucleocytoplasmic shuttling of the receptor (20). Receptor down-regulation by 17β-estradiol (E2) is well documented and postulated as a mechanism to limit the duration of hormone action in the cell (21–23). The observed receptor down-regulation by both antiestrogens and E2 (18) suggests that the induction of receptor turnover or down-regulation may require an event associated with ER activation, either receptor dimerization and/or the induction of transcription. If the antiestrogenic effects of tamoxifen and ICI 182,780 are caused by their ability to produce receptor down-regulation, they should induce either ER dimerization or activation of ER-dependent transcription, or both. In addition, if coactivators are required for ER activity, as has been postulated (7), interference by tamoxifen or ICI 182,780 with the ER-coactivator interaction may also produce an antiestrogen effect by reducing ER-dependent transcription.
To investigate whether tamoxifen or ICI 182,780 can induce or interfere with ER dimerization and transcriptional activity, we have used three yeast genetic systems in which we expressed the ERα form of the ER (3). Yeast genetic systems were used because yeast lack endogenous nuclear receptors and other receptor coregulatory proteins that are found in more complex mammalian cell lines (24), thus allowing us to isolate and study in a simple in vivo transcription system the interaction between E2 and/or antiestrogen and the ER. To investigate ligand-dependent dimerization of the ER, we used a yeast two-hybrid system (PCY2) expressing the GAL4–human ER (hER) fusion proteins (25). The ability of ER dimers to directly activate transcription was determined in the yeast RS188N, which contained the estrogen response element (ERE)–lacZ reporter system described by Lyttle et al. (26). The ability of tamoxifen or ICI 182,780 to block ER interaction with a coactivator, RIP140 (9), was tested in the yeast ERE–lacZ system. In addition, the yeast whole cell [3H]estradiol binding assay was developed in the two-hybrid yeast cell system to assess in vivo the interaction of E2, tamoxifen, and ICI 182,780 with the hormone binding site of the ER.
We demonstrate that both tamoxifen and ICI 182,780 induce ER dimerization and transcriptional activity of the ER. Tamoxifen appears to have only weak agonist activity, whereas ICI 182,780 is as efficacious as E2, although less potent. We also show in these yeast systems that antiestrogen blocks neither E2-induced ER activation nor the ER interaction with the coactivator RIP140. The binding data suggest that, whereas the actions of tamoxifen are mediated through the hormone binding site, the actions of ICI 182,780 may take place through interaction at a nonhormone binding site on the receptor.
Materials and Methods
Materials.
Tamoxifen and E2 were purchased from Sigma. ICI 182,780 was a gift from A. Wakeling (Zeneca Pharmaceuticals, Macclesfield, U.K.). The [3H]estradiol (84.1 Ci/mmol) was from DuPont/NEN.
Yeast Strains.
To measure ligand-dependent ER dimerization, the yeast strain PCY2 (MATα Δgal4 Δgal80 URA3∷GAL1-lacZ lys2–801 amber his3-Δ200 trp-1Δ631 leu2 ade2–101 ochre), which is a cross between GGY1∷171 (27) and YPH499 (28), was obtained from D. Nathans and P. Chevray (Johns Hopkins University, Baltimore). The PCY2 yeast strain (GAL1) carries the GAL4 binding site upstream of a lacZ reporter. Induction of ER-dependent transcription activity was measured in yeast strain RS188N (MATα ade2–1 his3–1 leu2–112 trp1–1 ura3–1), which was a gift from T. Butt (Gene Transcription Technologies, Philadelphia; ref. 26). Yeast strains were grown in yeast extract/peptone/dextrose or supplemented synthetic dextrose media (29).
cDNA and Constructs.
Construction of the GAL4–ER fusion vectors for the yeast two-hybrid system in the GAL1 yeast was performed as described (25). The RS188N yeast/ERE–lacZ system (ER/62/ERE) was cotransformed with three vectors: (i) the lacZ reporter vector YRpE2, which contained a double copy of the ERE from the Xenopus vitellogen A gene upstream of lacZ reporter; (ii) the ER in the vector YEpE12 (gifts from T. Butt; ref. 26), which expresses the ER under the control of the yeast copper metallothionein promoter; and (iii) the empty pPC62 vector (a gift from D. Nathans and P. Chevray) for selection purposes. Transformations were performed by using the lithium acetate method with plasmid DNA (29). Yeast colonies transformed with the vectors were selected by culture on synthetic media lacking tryptophan and leucine for the GAL4–ER fusion protein/PCY2 yeast two-hybrid system and lacking tryptophan, leucine, and uracil RS188N yeast.
RIP140 cDNA in pEF-BOS vector [kindly provided by M. Parker (9)] was amplified by PCR with two primers: 5′-GCG TGC ACG CTT CTA TTG AAC ATG ACT CAT-3′ (SalI) and 5′-GGA CTA GTC CAA AAC TGG ATG GCA GGT-3′ (SpeI). The PCR-amplified fragment was cloned into pBluescript II SK+ at SalI and SpeI sites. The RIP140 coding region was released from pBluescript II SK+ by SalI and SpeI digestion and was cloned into the GAL4 expression vector pPC62. The expression vector was sequenced to confirm the correct reading frame before transforming yeast. The RS188N yeast were cotransformed with the pPC62 vector containing RIP140, the lac-Z reporter vector YRpE2, and the wild-type ER in the vector YEpE12. Colonies transformed with the vectors were selected by culture on synthetic media lacking tryptophan, leucine, and uracil.
Ligand Treatment.
Stock cultures (20 ml) of the PCY2 or RS188N yeast strains were grown in synthetic supplemented media that lacked the appropriate amino acid selection markers and contained 1% ethanol as the carbon source. Cultures were grown at 30°C until OD600 read between 1.5 and 2.5 OD600 units/ml culture. The cultures were then stored at 4°C. For ligand treatment, a volume of the stock yeast culture was added to 1 ml of the appropriate supplemented dextrose media to give an initial OD600 reading of between 0.1 and 0.15 OD600 units/ml culture. The volume of ligand or test compound added was based on this final volume (1 ml media plus volume of yeast stock culture). E2 and test compound stock solutions were made up in DMSO and diluted with DMSO such that 1 μl of test compound added to 1 ml of culture gave the desired final concentration of ligand. Control cultures received an equal volume of DMSO. The cultures were then grown at 30°C with shaking (200–240 rpm) overnight (16–18 h) after which OD600 readings and β-galactosidase (β-gal) activity were measured.
β-gal Assay.
β-gal activity was used to measure the reconstitution of the GAL4 transactivation activity via the interaction of the two GAL4–hER fusion proteins in the PCY2 yeast, and the ERE–lacZ transcription activity in the RS188N yeast. The 1.2-ml reaction volume was made with 0.1 or 0.05 ml of yeast culture, 0.7 or 0.75 ml of Z buffer (60 mM Na2HPO4/40 mM NaH2PO4/10 mM KCl/1 mM MgSO4/50 mM 2-mercaptoethanol, pH 7.0) and 0.3 ml of 0.1% SDS in Z buffer. The reaction mixture was preincubated at 30°C for 10 min. The reaction was started by the addition of 0.2 ml of 4 mg/ml o-nitrophenyl β-d-galactoside and incubated at 30°C for up to 1 h if necessary. The reaction was stopped by the addition of 0.5 ml of 1 M Na2CO3. β-gal activity (relative response unit, RRU) was determined by using A420, A550, and A600 nm readings with the following equation: RRU = 1,000 × [(A420) − (A550 × 1.75)]/[t × v × A600], where t is the time of reaction (min) and v is the volume of yeast culture used in the reaction mixture (ml). EC50 values were determined by fitting the data to a four-parameter logistic function, f(x) = [a/(1 + e ∧ b (x − c))] + d, and analyzed by sigmaplot (SPSS, Chicago). Statistical significance was determined by using an analysis of variance.
Receptor Binding Assays.
The interactions of the various test compounds and E2 with the ER hormone binding site were determined in a yeast whole cell binding assay in the PCY2 yeast by using [3H]estradiol. The yeast cultures used for the binding assays were prepared from the stock cultures maintained at 4°C by starting fresh cultures in the appropriate volume of supplemented dextrose media (minus tryptophan and leucine) at an OD600 reading of 0.1–0.15 OD600 unit/ml cultures. The cultures were grown to an OD600 reading of 0.2–0.4 OD600 unit/ml cultures and used immediately. This density was found to give optimum specific binding.
Saturation analysis of [3H]estradiol binding was performed by incubating 80 μl of the yeast culture with various concentrations of [3H]estradiol (0.1–10 nM final concentration added in a 20-μl volume) for 3 h at room temperature. The total assay volume was 200 μl and was made up with yeast culture media. Nonspecific binding at each concentration of ligand was determined by parallel incubations run in the presence of 5 μM unlabeled E2. At the indicated time, the cells were harvested by filtration onto GFB membranes. The specific binding was analyzed by the method of Scatchard (30) to obtain linear regressions on a plot of bound/free vs. bound. The apparent dissociation constant (Kd) was calculated from the slope of the regression line and the number of binding sites per 1 million cells calculated from the x-axis intercept.
Competition binding assays were performed in the PCY2 yeast by using the same protocol as above, except that cells were incubated in the presence of 1 nM [3H]estradiol and various concentrations of test compounds made up in DMSO at a concentration of 1 μl of compound/1 ml of incubation volume to give the correct final concentration. The total assay volume was 200 μl. Control cells for total binding were incubated under identical conditions with an equivalent volume of DMSO. IC50 values were determined by fitting the data to the four-parameter logistic function shown above. Ki values were calculated by using the method of Cheng and Prusoff (31).
Results
Ligand-Induced Dimerization of the GAL4–hER Fusion Proteins.
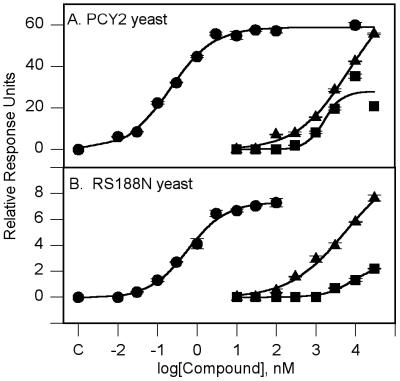
Both tamoxifen and ICI 182,780 induced dimerization of the GAL4–hER fusion proteins (Fig. 1A). No β-gal activity was seen in the absence of added ligand, a result which indicated that dimerization was indeed a ligand-dependent event. Tamoxifen and ICI 182,780 had EC50 values of 1.53 ± 0.83 μM and 2.35 ± 0.23 μM, respectively. E2 had an EC50 value of 0.16 ± 0.02 nM (Table 1). ICI 182,780 gave activation levels equal to E2 maximal induction, but tamoxifen appeared to only partially activate the system, being able to induce β-gal activity to only 50% of the E2 maximum. In preliminary experiments run in the PCY2 yeast, neither tamoxifen nor ICI 182,780 had any effect on the E2 dose-response curve (data not shown). This was in contrast to our report of this yeast system (25). However, the β-gal assay used in this report has been modified from our previous report by the elimination of chloroform from the assay buffer. This resulted in a dramatic increase in sensitivity to E2. The EC50 value reported here for E2 is 1,000-fold lower than reported in our previous paper. The potency of E2 in this system raised the concern that the PCY2 yeast may contain spare receptors (32) that would increase the efficacy of E2 as an agonist. This could explain the inability of either tamoxifen or ICI 182,780 to block the effects of E2. The ability of the antiestrogens to block the action of E2 was therefore investigated in the RS188N yeast, where receptor expression levels could be controlled.
Figure 1.
Dose response for E2, tamoxifen, and ICI 182,780. (A) PCY2 yeast. (B) RS188N yeast. The figure is representative of three to four independent experiments. The points shown are means ± SEM. The curves areas follows: E2 (●), ICI 182,780 (▴), and tamoxifen (■). Point C is the response in the absence of added E2.
Table 1.
Summary of the effects of E2, tamoxifen, and ICI 182,780 on the hER in yeast genetic systems
| Treatment | ER/62/ERE yeast
|
||||||
|---|---|---|---|---|---|---|---|
| Two-hybrid yeast
|
No CuSO4
|
+ 100 μM CuSO4
|
|||||
| EC50, nM | β-gal max, RRU | Ki, nM | EC50, nM | β-gal max, RRU | EC50, nM | β-gal max, RRU | |
| E2 | 0.16 (0.02) | 43.8 (7.5) | 3.47 (0.12) | 1.23 (0.23) | 9.03 (1.30) | 1.18 (0.18) | 178.2 (13.7) |
| Tamoxifen | 1,536 (830) | 23.0 (2.6) | 2,610 (190) | >30,000 | ND | >30,000 | ND |
| ICI 182,780 | 2,355 (233) | 44.6 (6.6) | 5,230 (1,240) | 806 (320) | 7.92 (1.23) | 3,400 (250) | 222.4 (5.8) |
Values are means (SEM) from three or four separate experiments. ND, not determined.
Induction of ERE–lacZ Transcription in the RS188N Yeast.
RS188N yeast was used to determine whether the ability to induce dimerization of the ER observed in the two-hybrid yeast did in fact result in ER dimers that were able to activate ERE–lacZ transcription. The ER was expressed in this yeast system as a fusion protein with ubiquitin under the expression control of a copper-inducible metallothionein promoter. The attachment of ubiquitin to the amino terminus of proteins has been reported to improve both the quality and quantity of the protein when expressed in yeast and Escherichia coli (26). In yeast, the ubiquitin is removed from the fusion protein after expression, releasing the ER. This promoter gave a dose-dependent response to copper (0.01–100 μM) in the amount of β-gal activity produced by 100 nM E2 (data not shown). There were very low levels of β-gal induction in the absence of CuSO4, indicating a low level of “leakage” from the metallothionein promoter. β-gal activity in the absence of added copper was approximately 5% of that seen at 100 μM CuSO4, yet reproducible dose-response curves were possible (Fig. 1B).
Tamoxifen and ICI 182,780 were able to induce ERE–lacZ transcriptional activity (Fig. 1B). The EC50 values for E2 and ICI 182,780 were 1.23 ± 0.23 nM and 0.806 ± 0.32 μM, respectively (Table 1). As in the PCY2 yeast, ICI 182,780 treatment resulted in a maximum β-gal activity equal to that produced by E2. Tamoxifen, at its highest concentration of 30 μM, although able to induce β-gal activity under both growth conditions, gave activity of only 10–20% of the E2 maximum; consequently, an EC50 value could not be calculated. In the presence of 100 μM CuSO4, E2 and ICI 182,780 had EC50 values of 1.18 ± 0.18 nM and 3.40 ± 0.25 μM, respectively.
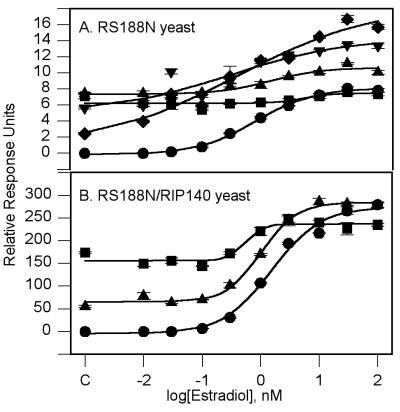
The ability of either tamoxifen or ICI 182,780 to block the E2 dose response was investigated in RS188N yeast grown in the absence of CuSO4. Tamoxifen, at 10 μM, had no effect on the E2 EC50 value or its maximum response (Table 1). The E2 dose-response curve was also run in the presence of 0.3, 1.0, 3.0, and 10.0 μM ICI 182,780 (Fig. 2A). Increasing concentrations of ICI 182,780 did increase starting levels of β-gal activity as expected, because ICI 182,780 could activate the ER on its own. However, coincubation with ICI 182,780 did not significantly change the EC50 value for E2 (Table 2). Interestingly, ICI 182,780 caused a significant increase in the maximum β-gal response. This increase followed an inverse dose-response relationship. At 0.3, 1.0, and 3 μM ICI 182,780, the levels of β-gal activity seen with E2 concentrations above 10 nM were significantly higher than those seen with E2 alone. These data may indicate that ICI 182,780 is capable of further activation of the ER through a nonhormone binding site.
Figure 2.
Effect of antiestrogens on E2 dose response. (A) RS188N yeast. The curves are as follows: E2 (●); + ICI 182,780: 0.3 μM (♦), 1.0 μM (▾), 3.0 μM (▴), and 10 μM (■). (B) RS188N/RIP140 yeast. The curves are as follows: E2 (●); + 10.0 μM ICI 182,780 (▴) or + 10.0 μM tamoxifen (■). Point C is the response in the absence of added E2. The values shown are means ± SEM from a single experiment.
Table 2.
Effect of anti-estrogens on E2 dose response in ER/62/ERE yeast
| E2 | + 10 μM TMX | + ICI
|
|||
|---|---|---|---|---|---|
| 10.0 μM | 3.0 μM | 1.0 μM | 0.3 μM | ||
| EC50 (nM) | 1.15 | ND | 1.76 | 0.51 | 0.52 |
| (0.12) | (0.31) | (0.20) | (0.11) | ||
| β-gal max (RRU) | 7.86 | 9.76 | 10.52* | 13.34* | 16.50* |
| (0.43) | (2.90) | (0.04) | (1.04) | (2.00) | |
Values are means (SEM) from three or four separate experiments. Each concentration of the dose response curves was run in triplicate. The ER/62/ERE yeast used in the E2 dose response studies with added antiestrogen were grown in the absence of CuSO4. TMX, tamoxifen; ND, not determined; *, P < 0.5 vs. E2 only.
[3H]Estradiol Binding Studies in Whole PCY2 Yeast Cells.
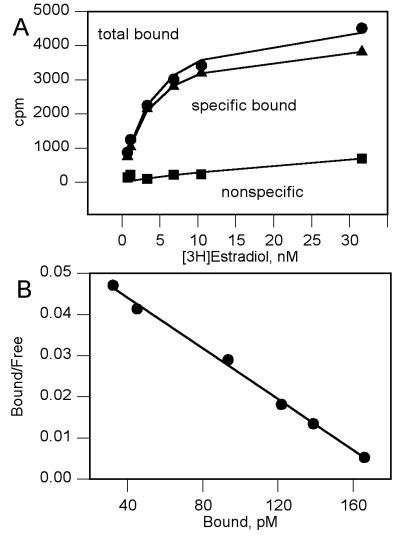
Whole cell competition binding studies with [3H]estradiol were performed to investigate whether the activation of ER-dependent transcription by the antiestrogens was through interaction with the hormone binding site on the receptor. The PCY2 yeast was used for the competition studies because preliminary experiments showed better levels of specific binding when compared with the RS188N yeast (data not shown). There was no specific [3H]estradiol binding in the two-hybrid yeast in the absence of the human estrogen receptor (Table 3). Saturation analysis with [3H]estradiol binding in the PCY2 yeast revealed a single class of binding sites with a Kd and Bmax value of 3.00 ± 0.24 nM and 9.39 ± 3.5 fmol receptor per million cells, respectively (Fig. 3). This Kd value was similar to reported Ki values from in vitro binding studies by using the mouse (14, 15) and hER (5, 33). This indicated that E2 had no problem penetrating the yeast cell wall.
Table 3.
[3H]Estradiol binding (cpm) in the two-hybrid yeast in the presence and absence of the hER
| [3H]Estradiol, nM | Absence of ER
|
Presence of ER
|
||||
|---|---|---|---|---|---|---|
| Total bound | Nonspecific bound | Specific bound | Total bound | Nonspecific bound | Specific bound | |
| 0.067 | 59 | 72 | 175 | 49 | 126 | |
| 1.0 | 68 | 115 | 289 | 57 | 232 | |
| 3.3 | 154 | 134 | 20 | 627 | 168 | 459 |
| 6.7 | 154 | 160 | 937 | 174 | 763 | |
| 10.0 | 183 | 245 | 28 | 1,105 | 224 | 881 |
| 30.0 | 470 | 645 | 1,679 | 666 | 1,013 | |
Values are the means of six to eight determinations at each concentration of [3H]estradiol. The yeast without the ER contained the empty 86 and 62 vectors. Nonspecific binding was determined in the presence of 1 μM unlabeled E2.
Figure 3.
Analysis of [3H]estradiol binding in the PCY2 yeast. (A) Saturation binding. (B) Scatchard analysis. The Kd value was 3.00 ± 0.24 nM and Bmax was 9.39 ± 3.5 fmol per million cells. Both figures are representative of three separate experiments with each concentration of [3H]estradiol run in quadruplicate.
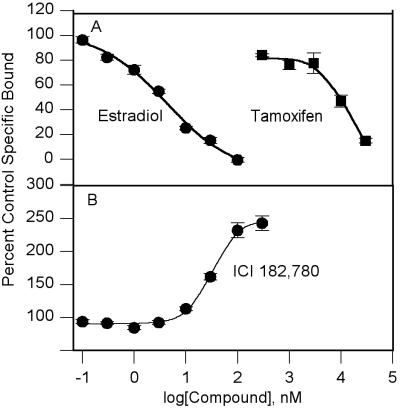
Competition binding studies were run in the presence of 1 nM [3H]estradiol. E2 had a Ki value of 3.47 ± 0.12 nM (Fig. 4A and Table 1). Tamoxifen had a Ki value of 2.61 ± 0.19 μM. The Ki value for inhibition of [3H]estradiol binding by tamoxifen and its EC50 value for inducing ER dimerization in the two-hybrid yeast were similar, at 2.61 ± 0.19 μM and 1.50 ± 0.80 μM, respectively.
Figure 4.
Whole cell competition binding studies for [3H]estradiol in the PCY2 yeast. (A) E2 and tamoxifen. The curves are as follows: E2 (●); tamoxifen (■). The Ki values are 3.47 ± 0.12 nM for E2 and 2.61 ± 0.19 μM for tamoxifen. (B) ICI 182,780. The ED50 for increasing [3H]estradiol binding was 52.3 ± 12.4 μM. The curves are representative of three to four independent experiments, with compound concentrations run in quadruplicate. The points shown are means ± SEM.
ICI 182,780, however, did not inhibit [3H]estradiol binding at concentrations up to 10 μM. At concentrations above 10 μM, ICI 182,780 was found to increase [3H]estradiol binding in a dose-dependent manner (Fig. 4B) up to 4-fold above control values. ICI 182,780 had an ED50 value of 52.3 ± 12.4 μM for this stimulation effect.
Effect of Tamoxifen and ICI 182,780 on the Interaction Between ER and the Coactivator RIP140.
To test whether the antiestrogenic effects of tamoxifen or ICI 182,780 could be caused by their ability to block the interaction of the ER with a coactivator, the RS188N yeast were cotransformed with the ER, the ERE–lac-Z plasmid and the RIP140 cDNA in the pPC62 vector (RS188N/RIP140). When grown in the absence of CuSO4, the E2 maximum response in the RS188N/RIP140 yeast was greater than 20-fold the maximum response to E2 in the RS188N yeast in the absence of RIP140, 278 ± 24 RRU and 9.49 ± 1.06 RRU, respectively (Fig. 2 A and B). The EC50 value for E2 in the RS188N/RIP140 yeast was 1.49 ± 0.28 nM, which was similar to the EC50 value in the RS188N yeast.
The effect of tamoxifen or ICI 182,780 (at 10 μM) on induction of β-gal activity was increased by a similar magnitude in the RS188N/RIP140 yeast. ERE–lacZ transcriptional activity at 10 μM tamoxifen or ICI 182,780 in the RS188N vs. RS188N/RIP140 yeast strains was 1.33 ± 0.11 RRU vs. 54.9 ± 2.5 RRU and 7.84 ± 0.6 RRU vs. 173.5 ± 2.3 RRU, respectively. Neither tamoxifen nor ICI 182,780 had an effect on the E2 EC50 value or the maximum level of β-gal induction (Fig. 2B).
Discussion
In the present study, we used three yeast genetic systems to look at the interaction of two antiestrogens, tamoxifen and ICI 182,780, with the human ERα to investigate whether the compounds functioned solely as competitive antagonists at the hormone binding site, or whether their antiestrogen activity was caused by another mechanism.
Tamoxifen and ICI 182,780 were able to induce ER dimerization and ER-dependent transcription as demonstrated in the PCY2 and RS188N yeast systems, respectively, although this did require concentrations of the antiestrogens that were up to 15,000 times higher than for E2. Other laboratories have reported that tamoxifen, 4-hydroxytamoxifen, ICI 182,780, or the closely related compound, ICI 164,384, could induce ER-dependent transcription (5, 26, 33, 34). The PCY2 yeast data indicate that this activation of transcription occurs because of the ability of the compounds to induce the formation of ER dimers. Consequently, the antiestrogen activity of ICI 182,780 or tamoxifen is not caused by inhibition of ER dimerization as was proposed by Fawell et al. (19).
The large difference in potency between E2 and the antiestrogens may be caused by a difficulty in penetrating the yeast cell wall by the antiestrogens. From the binding studies in the PCY2 yeast system, the Kd and Ki values for E2 were similar to the reported values from in vitro binding assays (5, 14, 15, 33), indicating that E2 did not have trouble crossing the yeast cell wall. However, the Ki value for tamoxifen binding to the ER in yeast was up to 8-fold higher than reported values for binding to mouse or hERα in cell-free systems (5, 35). This result indicates that tamoxifen did have trouble penetrating the yeast and may partially account for its high EC50 value and weak potency.
We were unable to conclude whether ICI 182,780 had difficulty crossing the yeast cell wall. In contrast to tamoxifen, ICI 182,780 did not inhibit [3H]estradiol binding to the ER in the PCY2 yeast in which it was able to induce ER dimerization. The observed increase in [3H]estradiol bound to the ER could indicate that there is an alternate binding site on the ER, through which ICI 182,780 could activate the ER and influence E2 binding. The γ-aminobutyric acid-benzodiazepine interaction in the γ-aminobutyric acid A receptor complex is a well known example of positive allosteric interactions between receptor ligands and receptor activation (36). The ability of ICI 182,780 to increase the E2-induced β-gal maximum response in this study may be an indication of a positive allosteric interaction between the E2 and ICI 182,780 binding sites. Binding to the allosteric site would still only result in ER dimerization and induction of transcription. The fact that this increased activation occurred in an inverse dose response requires further investigation. Zysk et al. (33) also have reported a statistically significant increase in [3H]estradiol binding in yeast in the presence of ICI 182,780. This observation supports previous reports indicating that there may indeed be an alternate activation site on the ER (37, 38).
In this study, neither tamoxifen nor ICI 182,780 were able to antagonize the effect of E2. As mentioned, this is in contrast to our prior report with the PCY2 yeast (25) and reports in mammalian cell systems with ERα (5). Our inability to demonstrate antagonism of E2 activity may be because of changes in the assay methodology or experimental limitations in using yeast genetic systems.
The elimination of chloroform from the assay stopping solution did result in a decrease in E2 EC50 values from approximately 1 μM to 0.16 nM. However, this 20-fold difference between E2 EC50 and Kd or Ki values obtained from [3H]estradiol binding studies in the PCY2 yeast may be indicative of the presence of spare ER receptors in that yeast system. The overproduction of ER would result in a situation where only a small fraction of the available receptors need to bind the agonist to fully saturate the coupled response system. The ability of spare receptors (32) to enhance agonist potency has been well documented in the study of cell surface G-protein-coupled receptors. Conversely, the ability of an antagonist to block agonist activation of the response in such a system becomes extremely difficult.
With the RS188N yeast system grown in the absence of CuSO4, we had a yeast with approximately 15% of the maximum E2-induced β-gal activity of the PCY2 yeast, yet we were still unable to block the effect of E2 with tamoxifen or ICI 182,780. The reduced ability of tamoxifen to penetrate the yeast cell wall may account for the inability of this compound to antagonize E2 action. It does not explain the results seen with ICI 182,780. In agreement with our results, Lyttle et al. (26) and Kohno et al. (34) have also shown that the related antiestrogen ICI 164,384 did not antagonize E2 effects in yeast.
The discovery of coactivator proteins such as RIP140 (7–10) that bind to and enhance the activity of the ER introduces the possibility that interference with the coactivator-receptor binding surfaces by antiestrogens may block E2 action. The coactivator RIP140 has been reported to enhance ER function slightly in mammalian systems. It is postulated that its presence may be mandatory for ER activity (9). In the RS188N/RIP140 yeast, there was a dramatic increase in E2 responsiveness; the β-gal maximum value was increased greater than 20-fold. The EC50 for E2 was unchanged, indicating that RIP140 has no effect on E2 affinity for the receptor. The ERE–lacZ transcriptional response of tamoxifen and ICI 182,780 were also enhanced in this yeast system (Fig. 2B; points in the absence of E2) compared with their response RS188N yeast. As in the RS188N, neither tamoxifen nor ICI 182,780 at 10 μM blocked E2 activity in the RS188N/RIP140 yeast, indicating that neither compound interfered with the coactivator–receptor interaction.
The turnover of hormone-bound receptor has been proposed as a mechanism to limit the hormone signal in the target tissue (21–23) and several studies with ICI 164,384 have shown a rapid loss of ER protein (both mouse and human) in as little as 1 h after treatment in mammalian systems (16–18). One could conclude that the induction of ER down-regulation requires ER dimerization and activation of transcription. Consequently, the antiestrogen activity of tamoxifen and ICI 182,780 could be caused by their ability to induce ER dimerization and ER-dependent transcription. In mammalian systems, this would activate an endogenous receptor metabolic pathway. Yeast, however, do not appear to possess this metabolic pathway (34). There was no loss of ER protein in yeast expressing the mouse ER after exposure to E2 or ICI 164,384. This would account for our inability in this study, as well as all other studies in yeast, to demonstrate antagonism of E2-induced transcription by an antiestrogen.
If induction of ER down-regulation is the mechanism underlying antiestrogen activity, the potency of antiestrogens to induce ER dimerization in yeast should correlate with their ability to induce receptor turnover in mammalian systems and their potential as antiestrogens in vivo. Our rank order of potency for ER dimerization in the PCY2 yeast was E2 > ICI 182,780 ≫ tamoxifen. This is similar to the rank order of efficacy of related compounds reported by Borras et al. (18) in their study of induction of ER turnover, E2 > ICI 164,384 ≫ 4-hydroxy-tamoxifen. If coactivator proteins, such as RIP140, are also present in the E2 target tissue, then the antiestrogen activity of all compounds could be magnified, even that of a very weak agonist such as tamoxifen.
It is undoubtedly possible that other mechanisms specific to mammalian cells underlie the antiestrogen activity of compounds like tamoxifen and ICI 182,780, such as differences in compound-receptor binding caused by different cytoplasmic components, cytoplasm-to-nuclear transport systems, and cell line differences. The use of the yeast genetic systems in our study and other studies allows us to look directly at ligand–receptor interaction in the absence of many confounding influences found in mammalian systems. The common element in all the studies in yeast is the ability of the antiestrogens to induce ER-dependent transcription because, as we show, of their ability to induce ER dimerization. Consequently, we purpose that the antiestrogen activity of tamoxifen and ICI 182,780, and other similar compounds, may be mediated by their ability to induce ER down-regulation through the formation of transcriptionally active ER dimers. Whereas tamoxifen does interact with the ER hormone binding site, indications are that ICI 182,780 may bind to an allosteric site on the receptor. These data also serve to underscore that currently there are no classical “antagonists” to E2 binding available, i.e., compounds that block E2 binding but do not themselves induce receptor activation.
Acknowledgments
We thank Drs. P. Chevray and D. Nathans (Johns Hopkins University, Baltimore) for the yeast two-hybrid system; Dr. T. Butt (Gene Transcription Technologies, Philadelphia) for the yeast strains, the yeast ERE reporter, and the inducible ER expression vectors; and B. Katzenellenbogen (University of Illinois, Urbana–Champaign) for the ER plasmid. We also thank Drs. E. Jensen (Karolinska Institute, Huddinge, Sweden), and W. Pike and S. Barton (University of Cincinnati College of Medicine, Cincinnati) for critically reading the manuscript and providing helpful suggestions. This work was supported by grants from the National Institutes of Health (CA72039) and the American Cancer Society (96–075).
Abbreviations
- ER
estrogen receptorα
- ERE
estrogen response element
- E2
17β-estradiol
- β-gal
β-galactosidase
- RRU
relative response unit
- hER
human ER
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040558197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040558197
References
- 1.Limieux P, Fuqua S. J Steroid Biochem Mol Biol. 1996;56:87–91. doi: 10.1016/0960-0760(95)00269-3. [DOI] [PubMed] [Google Scholar]
- 2.Luisi B F, Schwabe J W R, Freedman L P. Vitam Horm. 1994;49:1–47. doi: 10.1016/s0083-6729(08)61145-0. [DOI] [PubMed] [Google Scholar]
- 3.Kuiper G G J M, Enmark E, Pelto-Huikko M, Nilsson F, Gustafsson J-A. Proc Natl Acad Sci USA. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enmark E, Pelto-Huikko M, Grandien K, Lagercrantz S, Lagercrantz J, Nordenskjold M, Gustafsson J-A. J Clin Endocrinol Metab. 1997;82:4258–4265. doi: 10.1210/jcem.82.12.4470. [DOI] [PubMed] [Google Scholar]
- 5.Barkhem T, Carlsson B, Nilsson Y, Enmark E, Gustafsson J-A, Nilsson S. Mol Pharmacol. 1998;54:105–112. doi: 10.1124/mol.54.1.105. [DOI] [PubMed] [Google Scholar]
- 6.Paech K, Webb P, Kuiper G G J M, Nilsson S, Gustafsson A-K, Kushner P J, Scanlon T S. Science. 1997;277:1508–1510. doi: 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- 7.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 8.Cavailles V, Dauvois S, Daniellian P S, Parker M G. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavailles V, Dauvois S, L'Horset F, Lopez G, Hoare S, Kushner P J, Parker M G. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onate S A, Tsai S Y, Tsai M-J, O'Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 11.Osborne C K, Coronado E, Allred D C, Wiebe V, DeGregario M. J Natl Cancer Inst. 1991;83:1477–1482. doi: 10.1093/jnci/83.20.1477. [DOI] [PubMed] [Google Scholar]
- 12.Osborne C K, Wiebe V J, McGuire W L, Ciocca D R, DeGregorio M W. J Clin Oncol. 1992;10:304–310. doi: 10.1200/JCO.1992.10.2.304. [DOI] [PubMed] [Google Scholar]
- 13.Howell A, Dodwell D, Laidlaw I, Anderson H, Anderson E. In: Endocrine Therapy of Breast Cancer IV. Goldhirsch A, editor. Berlin: Springer; 1990. pp. 49–58. [Google Scholar]
- 14.Wakeling A E, Dukes M, Bowler J. Cancer Res. 1991;51:3867–3873. [PubMed] [Google Scholar]
- 15.Weatherill P J, Wilson A M P, Nicholson R I, Davies P, Wakeling A E. J Steroid Biochem. 1988;30:263–266. doi: 10.1016/0022-4731(88)90103-3. [DOI] [PubMed] [Google Scholar]
- 16.Gibson M K, Nemmers L A, Beckman W C, Jr, Davis V L, Curtis S W, Korach K S. Endocrinology. 1991;129:2000–2010. doi: 10.1210/endo-129-4-2000. [DOI] [PubMed] [Google Scholar]
- 17.Dauvois S, Danielian P S, White R, Parker M G. Proc Natl Acad Sci USA. 1992;89:4037–4041. doi: 10.1073/pnas.89.9.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borras M, Laios I, El Khissiin A, Seo H-S, Lempereur F, Legros N, Leclercq G. J Steroid Biochem Mol Biol. 1996;57:203–213. doi: 10.1016/0960-0760(95)00272-3. [DOI] [PubMed] [Google Scholar]
- 19.Fawell S E, White R, Hoare S, Sydenham M, Page M, Parker M G. Proc Natl Acad Sci USA. 1990;87:6883–6887. doi: 10.1073/pnas.87.17.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dauvois S, White R, Parker M G. J Cell Sci. 1993;106:1377–1388. doi: 10.1242/jcs.106.4.1377. [DOI] [PubMed] [Google Scholar]
- 21.Read L D, Snider C E, Miller G L, Katzenellenbogen B S. Mol Endocrinol. 1988;2:263–271. doi: 10.1210/mend-2-3-263. [DOI] [PubMed] [Google Scholar]
- 22.Bellingham D L, Dar M, Cidlowski J A. Mol Endocrinol. 1992;6:2090–2102. doi: 10.1210/mend.6.12.1491690. [DOI] [PubMed] [Google Scholar]
- 23.Wolf D A, Herzingert T, Hermeking H, Blachke G, Horz W. Mol Endocrinol. 1993;7:924–936. doi: 10.1210/mend.7.7.8413317. [DOI] [PubMed] [Google Scholar]
- 24.Walfish P G, Yoganathan T, Yang Y-F, Hong H, Butt T R, Stallcup M R. Proc Natl Acad Sci USA. 1997;94:3697–3702. doi: 10.1073/pnas.94.8.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Peters G A, Zeng X, Tang M, Ip W, Khan S. J Biol Chem. 1995;270:23322–23329. doi: 10.1074/jbc.270.40.23322. [DOI] [PubMed] [Google Scholar]
- 26.Lyttle R C, Damian-Matsumura P, Juul H, Butt T R. J Steroid Biochem Mol Biol. 1992;42:677–685. doi: 10.1016/0960-0760(92)90108-u. [DOI] [PubMed] [Google Scholar]
- 27.Gill G, Ptashne M. Cell. 1987;51:121–126. doi: 10.1016/0092-8674(87)90016-x. [DOI] [PubMed] [Google Scholar]
- 28.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arbuckle N D, Dauvois S, Parker M G. Nucleic Acids Res. 1992;20:3839–3844. doi: 10.1093/nar/20.15.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scatchard G. Ann NY Acad Sci. 1949;51:660–672. [Google Scholar]
- 31.Cheng Y C, Prusoff W H. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 32.Limbird L E. Cell Surface Receptors: A Short Course on Theory and Methods. Boston: Martius Nijhoff; 1986. [Google Scholar]
- 33.Zysk J R, Johnson B, Ozenberger B A, Bingham B, Gorski J. Endocrinology. 1995;136:1323–1326. doi: 10.1210/endo.136.3.7867588. [DOI] [PubMed] [Google Scholar]
- 34.Kohno H, Gandini O, Curtis S W, Korach K S. Steroids. 1994;59:572–578. doi: 10.1016/0039-128x(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 35.Wakeling A E, Bowler J. J Endocrinol. 1987;112:R7–R10. doi: 10.1677/joe.0.112r007. [DOI] [PubMed] [Google Scholar]
- 36.Braestrup C, Nielsen M, Honore T, Jensen L H, Petersen E N. Neuropharmacology. 1983;22:1451–1457. doi: 10.1016/0028-3908(83)90113-2. [DOI] [PubMed] [Google Scholar]
- 37.Hedden A, Muller V, Jensen J V. Ann NY Acad Sci. 1995;261:109–120. doi: 10.1111/j.1749-6632.1995.tb31373.x. [DOI] [PubMed] [Google Scholar]
- 38.Jensen E V. Ann NY Acad Sci. 1996;284:1–17. doi: 10.1111/j.1749-6632.1996.tb16223.x. [DOI] [PubMed] [Google Scholar]