Abstract
Amyloid-associated diseases affect millions of people worldwide. Phenol red exhibits modest inhibition toward fibril formation of human Islet amyloid polypeptide (hIAPP) and its toxicity, which is associated with type II diabetes mellitus. However, the molecular level mechanisms of interactions remain elusive. The binding of phenol red molecules to the protofibrils of an amyloidogenic fragment (NFGAIL) of hIAPP has been investigated by molecular dynamics simulations with explicit solvent. The phenol red molecules were observed to bind primarily along either β-sheet stacking or β-strand directions. Through its three aromatic rings, the phenol red molecule preferentially interacted with the hydrophobic side chains of Phe, Leu, and Ile; and the polar sulfone and hydroxyl groups were mainly exposed in solvent. Thus, phenol red improves the solubility of the early protofibrils and represses further growth. Interestingly, there was no obvious preference toward the aromatic Phe residue in comparison to the hydrophobic Leu or Ile residues. The lack of binding along the hydrogen bond direction indicates that phenol red does not directly block the β-sheet extension. Further free energy analysis suggested that a phenol red analog may potentially improve the binding affinity.
INTRODUCTION
A number of human diseases including type II diabetes and neurodegenerative diseases such as Alzheimer's, prion, Parkinson's, and Huntington's diseases (1–4) have been associated with a common pathological process. These diseases affect a large human population worldwide with no effective treatment at present. For instance, type II diabetes alone affects more than 150 million people worldwide and the number is growing (5). In all these diseases, deposition of amyloid plaques of different proteins is found in different parts of the human body such as brain and pancreas (1–4). The observation that synthetic amyloid aggregates are toxic to the corresponding cells such as nerve cell (6) and β-islet cell (7) suggests that amyloid aggregation might be related to the diseases. Although the exact pathogenic species have not been identified, amyloid fibrils and the protofibrillar species (soluble oligomers and protofibrils) are considered the primary sources of toxicity (8–10).
There are two main classes of inhibitors against amyloidogenicity and cytotoxicity. The first class consists of short synthetic peptides from the core fragments of amyloid peptides with chemical modifications (11–15). Although the main part of the peptides may be involved in binding to amyloid target via both main-chain hydrogen bonds and specific side-chain recognition, the chemical asymmetrical modifications remove the amide hydrogen bonds and inhibit the amyloid fibril growth. The second class consists of small aromatic molecules, such as Congo red, acridine orange, rifampin, tannic acid, and ferulic acid (16–24). Several inhibition mechanisms for the second class have been proposed, including stabilization of the monomeric states to inhibit unfolding, stabilization of early small protofibrils to prevent growth of amyloid fibrils, and destabilization of amyloid fibrils.
Human islet amyloid polypeptide (hIAPP), a 37-amino-acid hormone, is the main constituent of the islet amyloid fibrils deposited in pancreas in 95% of type II diabetes mellitus (25,26). It has been established that hIAPP forms amyloid fibrils (27) in vitro which induces islet β-cell apoptosis (7). Recently, its soluble oligomer and protofibril have been suggested to be cytotoxic (28–30). The hexapeptide NFGAIL, a fragment truncated from hIAPP (residues 22–27), is one of the shortest fragments that have been shown to form amyloid fibrils similar to those formed by the full polypeptide (31) and the fibrils are cytotoxic toward the pancreatic cell line. Therefore, this hIAPP “amyloid-core” peptide has been used as a simplified model system to facilitate the discovery of key factors underlying amyloid fibril formation and the development of antiamyloid agents.
Alanine-scanning mutagenesis revealed that the Phe residue plays a critical role in the formation of the fibrils (32). This led Gazit and co-workers (14) to search for aromatic compounds to interact with Phe to inhibit the hIAPP amyloid formation and its cytotoxicity. Among the aromatic compounds, phenol red has shown moderate inhibition activity against the hIAPP fibril formation and the cytotoxicity toward pancreatic β-cells. Since phenol red has been used for many years as a diagnostic agent in kidney disease and as a weak estrogen (33), its toxicity has been well characterized. It was suggested (14) that the inhibition was due to the heteroaromatic interaction and, hence, interaction with Phe residue was thought to play crucial roles. Despite this progress, because the inhibitor-protofibril complexes are heterogeneous, high resolution structure of the complex is unavailable, hindering the effort to understand the detailed molecular level mechanisms of the interactions between the small molecule inhibitors and the early aggregation species. Furthermore, the needed high drug/hIAPP molar ratio to inhibit the hIAPP1-37 fibril formation (14) may prevent it from being a viable therapeutic agent for type II diabetes. Thus, refinement to improve its potency may be helpful.
In a series of simulation studies of the hexapeptide NFGAIL, we employed all-atom molecular dynamics with explicit solvent and particle mesh Ewald (34) method for the treatment of long-range electrostatic forces to elucidate the aggregation process from the formation of partially ordered tetramers (35) to the formation of well-ordered octamers (36) and dodecamer protofibrils (37). We now take a step further to study the interaction between phenol red and the dodecamer protofibrils and to examine modifications to improve its binding affinity.
METHOD
Systems
Three types of systems were simulated in this study, consisting of solutions of protofibril alone, of ligand alone, and of protofibril and ligand together (Table 1). In the simulations of protofibril or ligand alone, no restraints were applied. In the simulations of protofibril and ligand together, the peptide strands were restrained to the β-extended conformation by restraining the main-chain torsion angles around (Φ ≈ −110°, Ψ ≈ 135°) with a harmonic potential and a restraining force constant of 5.0 kcal/mol/rad2. Because our focus is on the ordered protofibril, this measure reduces sampling of conformational space so that the key interactions between ordered protofibril and phenol red can be studied.
TABLE 1.
Summary of simulated systems
| ID | Content | No. of water molecules | Box* size (Å) | Concentration (mM) | No. of simulations | Length of each |
|---|---|---|---|---|---|---|
| 1 | Protofibril1+PR† | 7165 | 67.4 | 7.0 | 2 | 20 ns |
| 2 | Protofibril2+PR | 7165 | 67.4 | 7.0 | 2 | 20 ns |
| 3 | Protofibril3+PR | 7165 | 67.4 | 7.0 | 2 | 20 ns |
| 4 | Protofibril 1 | 2965 | 50.8 | 16.5 | 1 | 10 ns |
| 5 | Protofibril 2 | 2965 | 50.8 | 16.5 | 1 | 10 ns |
| 6 | Protofibril 3 | 2965 | 50.8 | 16.5 | 1 | 10 ns |
| 7 | PR, acidic | 788 | 31.9 | 42.0 | 1 | 10 ns |
| 8 | PR, basic | 788 | 31.9 | 42.0 | 1 | 10 ns |
Triclinic box equivalent to the true truncated octahedral box.
PR, phenol red.
Na+ is added to neutralize the charge of basic form (Fig. 4).
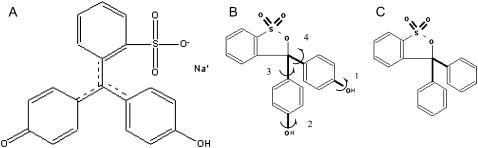
Phenol red comprises three aromatic rings and has both acidic and basic forms (see Fig 4, A and B) with pKa of 7.4. Thus, the acidic form is the main species (∼90%) in the experiment (14) of Gazit and co-workers (pH 6.5). In our simulations, we chose to focus on the acidic species. The interaction between the basic form and protofibril was postevaluated in the calculation of the binding free energy by substituting the acidic form with the basic form.
FIGURE 4.
Phenol red in basic form (A) or in acidic form (B) and its analogue (C). Four rotatable bonds of basic form are labeled.
In the simulations of phenol red and protofibril, each system consists of four phenol red molecules in acidic form, a protofibril, and water molecules. The four phenol red molecules were initially placed 15 Å away from a preformed 12-peptide protofibril (37) at four corners of the plane perpendicular to the β-sheet plane. They were then immersed into a triclinic box of 7165 water molecules, equivalent to a truncated-octahedral box with dimensions of a = b = c = 67.35 Å, α = β = γ = 109.47°. The periodic water box was constructed such that the solute was at least 10 Å away from the box surface and the minimum distance between solute and the image was 20 Å. The effective concentrations of phenol red and the protofibril were ∼28 mM and ∼7 mM, respectively. In comparison, the concentrations of phenol red and peptides in the experiment of Gazit and co-workers were, respectively, 40 μM and 4 μM14. High concentration (∼1000 fold higher) and high temperature (320 K) were used in the simulations to bypass the slow diffusion step and to speed up the binding process. In addition, the presence of four phenol red molecules in each simulation system allows sampling of multiple phenol red conformations in a single trajectory and enhances sampling in comparison to systems with a single phenol red molecule.
The Duan et al. all-atom point-charge force field (38) (Amber ff03) was chosen to represent the peptide; and the N- and C-termini were blocked respectively by acetyl (ACE) and amine groups (NHE). The partial charges of the phenol red molecule were obtained by fitting its gas-phase electrostatic potential calculated at HF/6-31G* level of quantum mechanical theory using RESP (39) in both acidic and basic forms (Fig. 4, A and B) subjected to geometry optimization at the level of HF/6-31G*. The other parameters of phenol red molecule were taken from the Amber GAFF (40) parameter set. The solvent was explicitly represented by the TIP3P (41) water model. To validate their force field parameters, a simulation (10 ns) was conducted on each form at 320 K. Their bond length and bond angles in crystal form (42) were well maintained in the molecular dynamics simulations (data not shown), and the torsion angles of the rotatable bonds were well sampled as indicated by the sampling of a complete 360° (Fig. S2, Supplementary Material).
Molecular dynamics simulation
The Amber simulation package was used in both molecular dynamics simulations and data processing (43). The protofibril-ligand-water system was subjected to periodic boundary conditions via both minimum image and discrete Fourier transform as part of the particle mesh Ewald method (34). After the initial energy minimization, two simulation runs for each system were performed starting from the same coordinates but with different initial random velocities by choosing different random number seeds. The initial velocities were generated according to the Boltzmann's distribution at 500 K. The simulations started from a 10.0-ps run at 500 K to randomize the orientations and positions of the four phenol red molecules. A short 1.0-ns molecular dynamics at 320 K in the NPT ensemble (constant number of atoms in the box, constant pressure and temperature) was performed to adjust system size and density and to equilibrate the solvent. The simulations were continued at 320 K for 19 ns in the NVT ensemble (constant number of atoms in the box, constant volume, and constant temperature). The particle mesh Ewald method (34) was used to treat the long-range electrostatic interactions. SHAKE (44) was applied to constrain all bonds connecting hydrogen atoms, and a time step of 2.0 fs was used. To reduce the computation, nonbonded forces (van der Waals and electrostatic forces) were calculated using a standard two-stage RESPA approach (45) where the forces within a 10-Å radius were updated every step and those beyond 10 Å were updated every two steps. Temperature was controlled at 320 K by using Berendsen's algorithm (46) with a coupling constant of 2.0 ps. The center of mass translation and rotation were removed every 500 steps. Studies have shown this removes the “block of ice” problem (47,48). The trajectories were saved at 2.0-ps intervals for further analysis. The same protocol was used in the simulation of the ligand and protofibril alone system, except that the heating step at 500 K was skipped.
Binding free energies
The binding free energies of phenol red and an analog were evaluated on the representative structures of the populated complex clusters from the protofibril-ligand simulations using the MM-GBSA (molecular mechanics-generalized Born/surface area) approach (49). The solvation free energy was represented by a combination of the GBSA method using the GBSA module in the Amber package. The phenol red analog (3H-2,1-benzoxathiole, 3,3-diphenyl-, 1,1-dioxide) is generated by substituting the phenol rings of phenol red molecule with benzene rings. The force field parameters of the analog were obtained by following the same protocol as that of phenol red. The complex free energies were evaluated after 2000 steps of energy minimization.
RESULTS
Three representative protofibrils were selected in this study from a pool of about eight ordered aggregates produced in an earlier study (37). They are hereafter referred to as “protofibrils 1–3” as illustrated in Fig. S1 (Supplementary Material). To test their stability, simulations (10 ns each) without any restraints were conducted at 320 K. At the end of the simulations, minor changes of the protofibril were observed, as expected under thermal fluctuation. For example, some peptides located at the ends of the β-sheets changed to coil; two ending peptides of the middle layer in protofibril 3 moved to the upper layer (supplemental Fig. S1). However, the three-layer architecture of the protofibrils was well maintained and the majority of peptides were still in β-sheet conformation. Because we intend to study the interaction between the phenol red and the ordered protofibrils, we chose to restrain the peptides to the extended conformation in protofibril + ligand simulations. However, the interpeptide distances were not restrained. Nevertheless, the restraints prevented phenol red from inducing conformational changes of peptides.
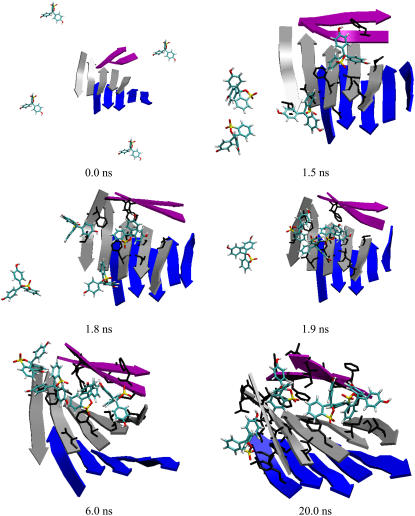
For each of the three protofibril oligomers, starting from a 12-peptide protofibril in three-layer β-sheet configuration, four phenol red molecules were placed 15 Å away from the protofibril (e.g., see Fig. 3) that were separated by water molecules, allowing simultaneous sampling of multiple phenol red conformations in a single simulation trajectory. Two independent simulation runs (20 ns each) were conducted for each of the three systems. Thus, the six simulations allowed us to sample 24 phenol red conformations simultaneously. We examined the conformations of the phenol red molecules in both the free and the bound forms in the complex simulations and compared to those in the ligand simulations. The fact that all four rotatable dihedral angles completely sampled the entire 360° suggests a reasonable sampling (supplemental Fig. S2). After binding to protofibril, the phenol red became slightly more rigid, indicated by somewhat restricted sampling of its torsion angles 3 and 4 (supplemental Fig. S2). The close resemblance of the torsion angle distributions to those in the free ligand simulation is indicative of a good sampling in the bound state.
FIGURE 3.
Snapshots from a trajectory of protofibril 3. The three layers of the protofibril are in cartoon. The phenol red were initially placed 15 and 10 Å away from the protofibril and water-box surface, respectively. For clarity, water molecules are not shown.
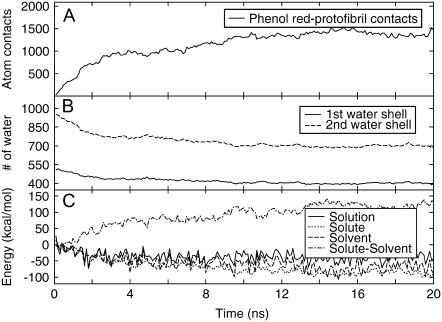
In the presence of phenol red, the overall structures of the three protofibril oligomers were well maintained in the simulations with restraints. For protofibrils 1 and 2, the main-chain heavy atom RMSD of the protofibrils relative to their initial structures remained 2–4 Å in the simulations (data not shown). An exception was the top layer of protofibril 3 that was partially peeled off from the protofibril, resulting in a larger RMSD (discussed later). At the end of 20.0 ns, all phenol red molecules bound to the protofibrils. The systems were close to steady state, as indicated by the relatively steady system energy (Fig. 1 C) and number of atom contacts between four phenol red molecules and protofibril (Fig. 1 A).
FIGURE 1.
Properties of the protofibril-inhibitor-water system averaged over all simulations. (Top) Atom contacts between the protofibril and four phenol red molecules (within 5 Å cutoff). (Middle) Number of water molecules in the first (0.0–3.4 Å) and second (3.4–5.0 Å) solvation shells of the solute (the protofibril and the phenol red molecules). (Bottom) The potential energy changes of the solution, solute, solvent, and the interaction energy change of solute-solvent upon the binding of the phenol red molecules to the protofibril.
Overall, the total energy of the system (Fig. 1 C) dropped to the steady-state level within 2.0 ns of simulation, whereas the individual components continued to develop thereafter. These changes are primarily characterized by the rise of solute-solvent interaction energy coupled with reduction in both solvent and solute energies, a hallmark of the desolvation process. In this process, as solute molecules form closer contacts, water molecules were released from the solute surface to the bulk of solvent and the number of water molecules in the first two solvation shells are reduced (Fig. 1 B). In fact, the protofibril-inhibitor complex lost ∼20% of the water molecules in the first two solvation shells during the process; the number of water molecules in the first solvation shell dropped to ∼400 from initially ∼500, and those in the second shell dropped to ∼725 from initially ∼925. The effect is even more dramatic if one considers that most of the reduction of the solvation shells was mainly due to desolvation of four phenol red molecules since the peptides started from the oligomeric complexes and only surface peptides were marginally solvated. Evidentially, this suggests strongly favorable entropic and enthalpic contributions upon releasing water molecules into the bulk solvent.
The enthalpic contribution of the solvent in this binding process was further evaluated by decomposing the potential energy of the solution into the potential energies of solute (protofibril plus phenol red) and solvent (waters) as well as the solute-solvent interaction energy (Fig. 1 C). Despite the large fluctuation, the total energy decreased by about ∼9.6 kcal/mol when averaged over the first and last 4.0 ns. The solute energy decreased by ∼53.2 kcal/mol, indicating favorable interactions (electrostatic and van der Waal interactions) between phenol red molecules and the protofibril. In the meantime, the interaction energies between the solute and water increased by 80.4 kcal/mol due to desolvation of the polar groups of both phenol red molecules and the protofibril upon the binding. Since the phenol red and the protofibril were well solvated in water before they bound to each other and their polar groups formed good hydrogen bonds with water, desolvation effectively removed some of these favorable interactions and resulted in large unfavorable water-solute interaction energy. This unfavorable energy term (80.4 kcal/mol) was compensated partially by the favorable energy term of phenol red-protofibril interactions (−53.2 kcal/mol). With the favorable water-water interactions (−36.7 kcal/mol), the total potential energy of the system becomes weakly favorable toward the binding (−9.6 kcal/mol). The large favorable water-water energy (−36.7 kcal/mol) demonstrates the substantial enthalpic contribution from releasing water molecules to the bulk solvent in the binding process because of the increased water-water hydrogen bonds formed by the released water. Other contributions include the favorable entropy increase by releasing water from the hydrophobic surface and unfavorable entropy loss due to binding of the solute molecules.
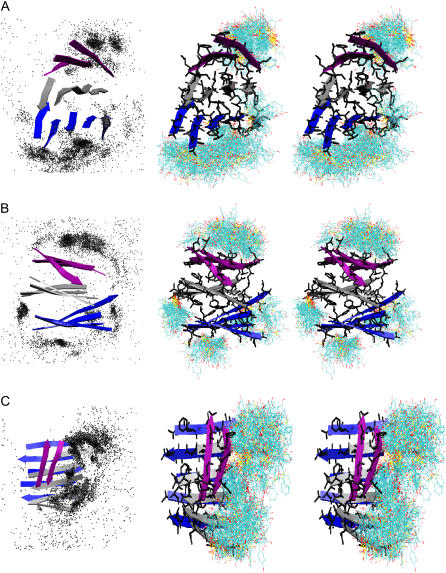
To illustrate the interactions between phenol red and the protofibril oligomers, we superimpose the phenol red molecule images from the simulation snapshots by aligning the protofibril structures. As shown in Fig. 2, phenol red molecules were bound to the protofibrils and formed tight clusters, indicating stable binding. Notably, most phenol red molecules were bound to the top or bottom faces of the oligomers and rarely bound to the peptide main-chain hydrogen bond face. For protofibril 1 (Fig. 2 A), which is organized as three-layer ordered β-sheets with each layer a four-strand β-sheet, the prevalent binding modes were at the top and bottom with only sporadic binding at the main-chain hydrogen bond direction. In protofibril 2 (Fig. 2 B) with a similar architecture, there was notable binding at the peptide terminal faces along the β-strand direction in addition to the binding at the top and bottom faces. Again, there was virtually no binding at the β-sheet extension direction along backbone hydrogen bonds. Therefore, the prevalent mode of binding was at the β-sheet stacking direction perpendicular to the β-sheet planes. Incidentally, these protofibril oligomers have comparable exposed surfaces in all three directions (i.e., β-sheet stacking direction perpendicular to the β-sheet planes, β-sheet extension direction along backbone hydrogen bonds, and β-strand direction). Hence the binding preference observed in the simulations was an indication of the underlying energetic bias toward those directions. The lack of interaction with the main-chain hydrogen bonds is attributable to the physicochemical property of phenol red molecules for its lack of ability to form stable hydrogen bonds with the peptides in solution.
FIGURE 2.
Phenol red molecules observed around the NFGAIL protofibrils. The protofibrils are in cartoon. (Left) Each dot represents a phenol red molecule conformation. (Right) Stereoscopic plots of the populated clusters. Phenol red molecules observed in the simulations are represented by lines.
Clustering analysis is used to illuminate the detailed interactions between phenol red molecules and the protofibril. In this analysis, the protofibril was first aligned and the phenol red molecules were clustered using the distance between their geometry centers with a cutoff of 5 Å. The representative structures of the top 10 clusters from the combined two simulation runs of a protofibril are shown in Fig. S3 (Supplementary Material). In addition to the interesting features noted before, the polar OH and SO3 groups of phenol red molecules tend to stay in contact with water, indicating they were still solvated by water molecules upon binding. They also formed hydrogen bonds with the side chain of Asn and NH2 of C-termini. In contrast, the apolar aromatic rings of phenol red molecules tend to cluster around the apolar side chains. For example, the three rings of phenol red molecule formed a hydrophobic cluster with the side chains of Leu, Phe, and Ala in most clusters (e.g., A1, B1, and B3, and many others). Phenol red molecule was also seen to bind between the top two layers of the oligomer and formed the hydrophobic contacts with the side chains of Phe and Ile in cluster C1. In all these cases, the sulfone group pointed away from the peptides.
As summarized in Table 2, the phenol red molecules clustered around the stacking faces of the protofibrils, showing the tendency to bind on the β-sheet surface and, to a less extent, to the ends of the strands. In fact, the aggregated population of the clusters where phenol red molecules bound to the protofibrils from the β-sheet stacking and β-strand directions was close to ∼60% in the simulations of protofibril 1 and 2 (∼64% and ∼69%, respectively). In comparison, almost no phenol red molecule bound to the protofibrils from the β-sheet extension (hydrogen bond) direction. This may stem from the geometry of a phenol red molecule which formed ∼200 atom contacts with the side chains and only ∼50 with the main chain (data not shown). Since the three polar groups (two OH groups on the phenol rings and the SO3 group) of phenol red molecule points to three orthogonal directions, it is difficult to simultaneously form three hydrogen bonds with the peptide main chain. In other words, due to lack of favorable interactions with peptide main chains it is less favorable for phenol red molecules to bind to the protofibrils from the backbone hydrogen bond direction. In contrast, the size and shape of phenol red molecules are compatible with the amino acid side chains (e.g., Phe, Leu, and Ile) and favorable interactions with side chains via aromatic and/or hydrophobic contacts lead to the binding to the protofibril preferentially from β-sheet stacking and, to a lesser degree, β-strand directions.
TABLE 2.
Binding of phenol red to protofibrils in different directions (details are in supplemental Fig. S3)
| β-Sheet stacking direction (top, down) | β-Strand direction (back, front) | β-Sheet extension direction (left, right) | Others* | |
|---|---|---|---|---|
| Protofibril 1 | 58% | 8% | 0% | 34% |
| Protofibril 2 | 52% | 17% | 0% | 31% |
Sum of all small clusters (population <2%).
The polar SO3 and OH groups of phenol red were mostly solvated in water and formed hydrogen bonds with the peptides mainly to the NH2 of C-termini and the side chain of Asn. As shown in Table S1 before binding to the protofibrils when averaged from the first 0.1 ns, the SO3 and the two OH groups formed, respectively, ∼3.4 and ∼4.7 hydrogen bonds with water molecules that were reduced to, respectively, ∼2.4 and ∼3.6 when averaged from the last 5 ns. Only ∼1 hydrogen bond was lost for each of the SO3 and two OH groups. Therefore, most of these polar groups were still solvated by water molecules. In contrast, the three aromatic rings lost ∼143 (∼40%) atom-atom contacts with water molecules.
On the other hand, the three aromatic rings of the phenol red molecule formed ∼102 atom-atom contacts with the protofibril, as shown in Table S1, and interacted mostly with the aromatic and hydrophobic residues and formed aromatic and hydrophobic clusters with the side chains of Phe, Leu, Ile, and, to a lesser degree, Ala. In more detail, the rings formed ∼20, ∼24, and ∼23 atom-atom contacts with Phe, Ile, and Leu, respectively, whereas they formed ∼9, ∼10, and ∼4 atom-atom contacts with Ala, Asn, and Gly, respectively. The comparable level of contacts with three hydrophobic residues, Phe and Leu or Ile, implies that phenol red has a similar preference toward them. In contrast, the difference between hydrophobic and hydrophilic contacts indicated that the three aromatic rings of phenol red interacted mostly with the aromatic and hydrophobic side chains of Phe, Leu, and Ile. Therefore, besides aromatic interactions, hydrophobic interactions also played an important role, which is complementary to the explanation by Gazit and co-workers (14). Thus, phenol red protects the protofibril oligomers from further aggregation by interacting with the aromatic and hydrophobic side chains through its phenol and benzene rings and with water molecules through its polar sulfone and hydroxyl groups.
Although protofibril 3 is also organized as a three-layer β-sheet oligomer, its upper layer consists of only two peptides (Fig. 2 C), rendering it a relatively weaker layer. In the simulations of protofibril 3, an interesting binding mode was observed in which the phenol red molecules competed with the two peptide strands on the top layer for binding to the middle layer of the oligomer. A result of such competition was the insertion of phenol red molecules into the area between the top and middle layers, giving the appearance of partial removal of the top layer from the protofibril (supplemental Fig. S3 C1). Snapshots from the trajectory are shown in Fig. 3. In this trajectory, the two β-strands of the upper layer of the protofibril opened up at ∼2.0 ns and two nearby phenol red molecules moved into the gap and prevented the layer from closing back. At 6.0 ns, the last phenol red molecule bound to the left edge of the middle layer. These phenol red molecules stayed at the site throughout the rest of the simulation. In contrast, in an early study (37) without phenol red, although the upper layer exhibited fluctuation, it stayed as an integral part of the protofibril in the last 60 ns till the completion of the 100.0-ns trajectory. Thus, the fact that the phenol red molecules were able to move into the gap within 2.0 ns indicates that the phenol red may play a role in assisting the partial removal of the upper layer by exploiting weakness of the structure. The fact that insertion of the phenol red molecules occurred only in one of the six simulations indicates that such events are rare within the simulation timescales.
DISCUSSION
The elevated peptide and ligand concentrations in the simulations in comparison to experimental concentrations contributed to the significant reduction in the computational cost. At the experimental concentrations (14), we would have to increase the system size by ∼1000 times with a corresponding increase in computer time. The concentration also has direct influence on the kinetic rate of reaction. With the decrease in concentration by 1000 times, we would expect an increase in timescales also by 1000 times for the binding to occur. Thus, the combined increase in computer time would be ∼106 times and the simulations would be intractable. Therefore, we intentionally selected higher concentration to allow simulations to be completed in a reasonable time. However, because of the increase in concentrations, the timescales and stability of binding have been changed. Thus, our study serves to qualitatively characterize the process.
We evaluated the interaction between the protofibrils and phenol red in basic form that constitutes ∼10% in the solution of pH 6.5. In these calculations, the acidic form phenol red in the simulated trajectories was substituted by the basic form and the binding free energy was calculated using the MM-GBSA method (49). In comparison, the basic form has a weaker binding free energy (−11.3 kcal/mol) than (−13.2 kcal/mol) acidic form (Table 3). The difference came from the unfavorable electrostatic part (∼2 kcal/mol) of the binding free energy, indicating negative charge of the basic form does not enhance the binding to the hydrophobic peptide. We like to note that although the MM-GBSA calculations may overestimate the absolute binding free energy due to the missing terms (e.g., conformational entropy change of the solute upon binding) and underestimate the desolvation free energy, they usually give a reasonable qualitative estimate on the relative binding free energy when two similar ligands are compared (50).
TABLE 3.
Binding free energy of phenol red and its analog
| Unit (kcal/mol) | ΔEGAS | ΔEGB | ΔESUR | ΔETOT |
|---|---|---|---|---|
| Phenol red in basic form* | −23.8 ± 9.0 | 15.2 ± 7.9 | −2.7 ± 0.5 | −11.3 ± 3.3 |
| Phenol red in acidic form† | −24.9 ± 7 .0 | 14.4 ± 5.2 | −2.7 ± 0.6 | −13.2 ± 3.5 |
| Phenol red‡ | −24.7 | 14.5 | −2.7 | −13.0 |
| Phenol red analog* | −20.7 ± 5.4 | 9.4 ± 3.3 | −2.8 ± 0.5 | −14.1 ± 3.7 |
| Change (ΔΔE) | 4.2 | −5 | 0.1 | −0.9 |
ΔEGAS, gas phase potential energy.
ΔEGB, GB reaction field energy.
ΔESUR, surface area energy.
ΔETOT = ΔEGAS + ΔEGB + ΔESUR potential energy in water.
The representative structures of the populated clusters are shown in supplemental Fig. S3.
The complex structures were obtained by substituting the acidic form in supplemental Fig. S3.
∼10% of basic form and ∼90% of acidic form at pH = 6.5.
In this study, we focused on the interaction between phenol red molecules and early protofibrils. The majority of the trajectories indicated that the phenol red molecules bind to the protofibrils and may prevent formation of larger protofibrils by blocking the hydrophobic surface, disrupting the surface pattern, and increasing the solubility of small protofibrils. The simulations suggested that the phenol red molecules interact strongly with the side chains of Phe, Leu, and Ile through its aromatic phenol and benzene rings. The polar SO3 and OH groups are mainly exposed to the solvent and form hydrogen bonds. The result is a highly soluble complex of protofibril and phenol red molecules. Furthermore, this effect may be applicable to the nucleation phase of the early oligomer formation in which peptide monomers undergo conformation conversion and nucleation. The binding of phenol red molecules to peptide monomers could stabilize the monomeric peptide and delay/reduce the nucleation. This is consistent with the observation that chaperones (51) and surfactants (52–54) also demonstrate inhibition activity against amyloid β aggregation.
We shall note that the proposed interaction mechanism of phenol red deviates somewhat from the initial motivation of Gazit and co-workers (14), who targeted the aromatic Phe side chains as the primary binding sites. However, in addition to the aromaticity, Phe side chains are also very hydrophobic. Thus, our results are complementary to the experimental findings. Here, our simulation suggests that phenol red also targets to the hydrophobic nature of Phe. In addition, it is interesting that phenol red did not bind along the hydrogen bond direction, indicating interaction at the β-sheet extension side is not the primary mechanism.
Further enhancement in binding affinity may be contemplated by improving solubility of the aggregates and by enhancing the interactions with the side chains. Along this line, we substituted the phenol rings of the phenol red with the benzene rings to enhance their interactions with the peptide side chains (Fig. 4 C). Removal of the polar hydroxyl groups is expected to directly enhance the hydrophobicity of the two aromatic rings and to indirectly make it easier for the sulfone group to solvate in water. As a result, this new compound may have a higher binding affinity to the amyloid protofibrils. Indeed, the binding free energy of this new compound to the protofibrils was enhanced by −0.9 kcal/mol, as estimated by the GBSA free energy calculation (Table 3) with less favorable (−5.0 kcal/mol) solvation. This compound is commercially available with Chemical Abstract registry number 15448-98-3. Its solubility in water is reported by AsInEx (5 Gabrichevskogo St., Bldg. 8, Moscow, 125367) as 113.0764 mg/L, which is slightly lower than that of phenol red (140.1104 mg/L). Taken together, this new compound may have higher potency to inhibit the amyloid aggregation, fibril formation, and cytotoxicity.
CONCLUSION
We have conducted a set of simulations to study the interaction between phenol red and the protofibrils of an amyloidogenic peptide NFGAIL. The favorable binding direction of phenol red molecules to the protofibrils was along either the β-sheet stacking direction or the β-strand direction, whereas binding along the β-sheet extension direction was sporadic. Although multiple binding sites existed, phenol red molecules mainly interacted with the aromatic and hydrophobic side chains of Phe, Leu, Ile, and, to a lesser extent, Ala through its aromatic rings. Although the sulfone and hydroxyl groups of phenol red sometimes formed hydrogen bonds with the amide group of peptide termini Asn and the blocking unit Nhe, most of the time these polar groups were exposed to solvent and the effect is an improved solubility of the aggregate complex. The lack of binding along the hydrogen-bonding direction suggests that phenol red does not block β-sheet extension directly. Alternatively, phenol red may act as a detergent to prevent the formation of larger protofibrils by the blocking hydrophobic surface and improving the solubility of small peptide aggregates. Based on the interaction mechanism revealed in the simulations, an analog is suggested by substituting the phenol rings with benzene rings that may enhance binding affinity by ∼−0.9 kcal/mol as calculated using the GBSA method.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
Usage of graphics packages, including PyMol, VMD, and RasMol, is gratefully acknowledged.
This work was supported by research grants from the National Institutes of Health (GM64458 and GM67168 to Y.D.).
References
- 1.Thirumalai, D., D. K. Klimov, and R. I. Dima. 2003. Emerging ideas on the molecular basis of protein and peptide aggregation. Curr. Opin. Struct. Biol. 13:146–159. [DOI] [PubMed] [Google Scholar]
- 2.Rochet, J. C., and P. T. Lansbury. 2000. Amyloid fibrillogenesis: themes and variations. Curr. Opin. Struct. Biol. 10:60–68. [DOI] [PubMed] [Google Scholar]
- 3.Dobson, C. M. 1999. Protein misfolding, evolution and disease. Tr. in Biochem. Sci. 24:329–332. [DOI] [PubMed] [Google Scholar]
- 4.Kelly, J. W. 1998. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr. Opin. Struct. Biol. 8:101–106. [DOI] [PubMed] [Google Scholar]
- 5.Zimmet, P., K. Alberti, and J. Shaw. 2001. Global and societal implications of the diabetes epidemic. Nature. 414:782–787. [DOI] [PubMed] [Google Scholar]
- 6.Hashimoto, M., E. Rockenstein, L. Crews, and E. Masliah. 2003. Role of protein aggregation in mitochondrial dysfunction and neurodegeneration in Alzheimer's and Parkinson's diseases. Neuromol. Med. 4:21–35. [DOI] [PubMed] [Google Scholar]
- 7.Lorenzo, A., B. Razzaboni, G. C. Weir, and B. A. Yankner. 1994. Pancreatic islet cell toxicity of amylin associated with type-2 diabetes mellitus. Nature. 368:756–760. [DOI] [PubMed] [Google Scholar]
- 8.Hardy, J., and D. J. Selkoe. 2002. Medicine—the amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 297:353–356. [DOI] [PubMed] [Google Scholar]
- 9.Bucciantini, M., E. Giannoni, F. Chiti, F. Baroni, L. Formigli, J. S. Zurdo, N. Taddei, G. Ramponi, C. M. Dobson, and M. Stefani. 2002. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 416:507–511. [DOI] [PubMed] [Google Scholar]
- 10.Bucciantini, M., G. Calloni, F. Chiti, L. Formigli, D. Nosi, C. M. Dobson, and M. Stefani. 2004. Prefibrillar amyloid protein aggregates share common features of cytotoxicity. J. Biol. Chem. 279:31374–31382. [DOI] [PubMed] [Google Scholar]
- 11.Kapurniotu, A., A. Schmauder, and K. Tenidis. 2002. Structure-based design and study of non-amyloidogenic, double N-methylated IAPP amyloid core sequences as inhibitors of IAPP amyloid formation and cytotoxicity. J. Mol. Biol. 315:339–350. [DOI] [PubMed] [Google Scholar]
- 12.Tatarek-Nossol, M., L. M. Yan, A. Schmauder, K. Tenidis, G. Westermark, and A. Kapurniotu. 2005. Inhibition of NAPP amyloid-fibril formation and apoptotic cell death by a designed hIAPP amyloid-core-containing hexapeptide. Chem. Biol. 12:797–809. [DOI] [PubMed] [Google Scholar]
- 13.Gilead, S., and E. Gazit. 2004. Inhibition of amyloid fibril formation by peptide analogues modified with alpha-aminoisobutyric acid. Angew. Chem. Internat. Ed. 43:4041–4044. [DOI] [PubMed] [Google Scholar]
- 14.Porat, Y., Y. Mazor, S. Efrat, and E. Gazit. 2004. Inhibition of islet amyloid polypeptide fibril formation: a potential role for heteroaromatic interactions. Biochemistry. 43:14454–14462. [DOI] [PubMed] [Google Scholar]
- 15.Scrocchi, L. A., Y. Chen, S. Waschuk, F. Wang, S. Cheung, A. A. Darabie, J. McLaurin, and P. E. Fraser. 2002. Design of peptide-based inhibitors of human islet amyloid polypeptide fibrillogenesis. J. Mol. Biol. 318:697–706. [DOI] [PubMed] [Google Scholar]
- 16.Aitken, J. F., K. M. Loomes, B. Konarkowska, and G. J. S. Cooper. 2003. Suppression by polycyclic compounds of the conversion of human amylin into insoluble amyloid. Biochem. J. 374:779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Felice, F. G., M. N. N. Vieira, L. M. Saraiva, J. D. Figueroa-Villar, J. Garcia-Abreu, R. Liu, L. Chang, W. L. Klein, and S. T. Ferreira. 2004. Targeting the neurotoxic species in Alzheimer's disease: inhibitors of Abeta oligomerization. FASEB J. 18:1366–1372. [DOI] [PubMed] [Google Scholar]
- 18.Lin, S. J., Y. J. Shiao, C. W. Chi, and L. M. Yang. 2004. Abeta aggregation inhibitors. Part 1: Synthesis and biological activity of phenylazo benzenesulfonamides. Bioorg. Med. Chem. Lett. 14:1173–1176. [DOI] [PubMed] [Google Scholar]
- 19.Lorenzo, A., and B. A. Yankner. 1994. Beta-amyloid neurotoxicity requires fibril formation and is inhibited by Congo red. PNAS, USA. 91:12243–12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ono, K., K. Hasegawa, H. Naiki, and M. Yamada. 2004. Anti-amyloidogenic activity of tannic acid and its activity to destabilize Alzheimer's beta-amyloid fibrils in vitro. Biochim. Biophys. Acta-Mol. Bas. Dis. 1690:193–202. [DOI] [PubMed] [Google Scholar]
- 21.Ono, K., Y. Yoshiike, A. Takashima, K. Hasegawa, H. Naiki, and M. Yamada. 2004. Vitamin A exhibits potent antiamyloidogenic and fibril-destabilizing effects in vitro. Exp. Neurol. 189:380–392. [DOI] [PubMed] [Google Scholar]
- 22.Ono, K., K. Hasegawa, H. Naiki, and M. Yamada. 2004. Curcumin has potent anti-amyloidogenic effects for Alzheimer's beta-amyloid fibrils in vitro. J. Neurosci. Res. 75:742–750. [DOI] [PubMed] [Google Scholar]
- 23.Ono, K., M. Hirohata, and M. Yamada. 2005. Ferulic acid destabilizes preformed beta-amyloid fibrils in vitro. Biochem. Biophys. Res. Com. 336:444–449. [DOI] [PubMed] [Google Scholar]
- 24.Tomiyama, T., H. Kaneko, K. Kataoka, S. Asano, and N. Endo. 1997. Rifampicin inhibits the toxicity of pre-aggregated amyloid peptides by binding to peptide fibrils and preventing amyloid-cell interaction. Biochem. J. 322:859–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westermark, P., C. Wernstedt, T. D. O'Brien, D. W. Hayden, and K. H. Johnson. 1987. Islet amyloid in type 2 human diabetes mellitus and adult diabetic cats contains a novel putative polypeptide hormone. Am. J. Pathol. 127:414–417. [PMC free article] [PubMed] [Google Scholar]
- 26.Hoppener, J. W. M., B. Ahren, and C. J. M. Lips. 2000. Islet amyloid and type 2 diabetes mellitus. N. Engl. J. Med. 343:411–419. [DOI] [PubMed] [Google Scholar]
- 27.Makin, O. S., and L. C. Serpell. 2004. Structural characterisation of islet amyloid polypeptide fibrils. J. Mol. Biol. 335:1279–1288. [DOI] [PubMed] [Google Scholar]
- 28.Porat, Y., S. Kolusheva, R. Jelinek, and E. Gazit. 2003. The human islet amyloid polypeptide forms transient membrane-active prefibrillar assemblies. Biochemistry. 42:10971–10977. [DOI] [PubMed] [Google Scholar]
- 29.Anguiano, M., R. J. Nowak, and P. T. Lansbury. 2002. Protofibrillar islet amyloid polypeptide permeabilizes synthetic vesicles by a pore-like mechanism that may be relevant to type II diabetes. Biochemistry. 41:11338–11343. [DOI] [PubMed] [Google Scholar]
- 30.Janson, J., R. H. Ashley, D. Harrison, S. McIntyre, and P. C. Butler. 1999. The mechanism of islet amyloid polypeptide toxicity is membrane disruption by intermediate-sized toxic amyloid particles. Diabetes. 48:491–498. [DOI] [PubMed] [Google Scholar]
- 31.Tenidis, K., M. Waldner, J. Bernhagen, W. Fischle, M. Bergmann, M. Weber, M. L. Merkle, W. Voelter, H. Brunner, and A. Kapurniotu. 2000. Identification of a penta- and hexapeptide of islet amyloid polypeptide (IAPP) with amyloidogenic and cytotoxic properties. J. Mol. Biol. 295:1055–1071. [DOI] [PubMed] [Google Scholar]
- 32.Azriel, R., and E. Gazit. 2001. Analysis of the minimal amyloid-forming fragment of the islet amyloid polypeptide—an experimental support for the key role of the phenylalanine residue in amyloid formation. J. Biol. Chem. 276:34156–34161. [DOI] [PubMed] [Google Scholar]
- 33.Berthois, Y., J. A. Katzenellenbogen, and B. S. Katzenellenbogen. 1986. Phenol red in tissue-culture media is a weak estrogen—implications concerning the study of estrogen-responsive cells in culture. PNAS, USA. 83:2496–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Essmann, U., L. Perera, M. L. Berkowitz, T. A. Darden, H. Lee, and L. G. Pedersen. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103:8577–8593. [Google Scholar]
- 35.Wu, C., H. Lei, and Y. Duan. 2004. Formation of partially ordered oligomers of amyloidogenic hexapeptide (NFGAIL) in aqueous solution observed in molecular dynamics simulations. Biophys. J. 87:3000–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, C., H. Lei, and Y. Duan. 2005. The role of Phe in the formation of well-ordered oligomers of amyloidogenic hexapeptide (NFGAIL) observed in molecular dynamics simulations with explicit solvent. Biophys. J. 88:2897–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, C., H. Lei, and Y. Duan. 2005. Elongation of ordered peptide aggregate of an amyloidogenic hexapeptide (NFGAIL) observed in molecular dynamics simulations with explicit solvent. J. Am. Chem. Soc. 127:13530–13537. [DOI] [PubMed] [Google Scholar]
- 38.Duan, Y., S. Chowdhury, G. Xiong, C. Wu, W. Zhang, T. Lee, P. Cieplak, J. Caldwell, R. Luo, J. Wang, and P. A. Kollman. 2003. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase QM calculations. J. Comp. Chem. 24:1999–2012. [DOI] [PubMed] [Google Scholar]
- 39.Bayly, C. I., P. Cieplak, W. D. Cornell, and P. A. Kollman. 1993. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges—the RESP model. J. Phys. Chem. 97:10269–10280. [Google Scholar]
- 40.Wang, J. M., R. M. Wolf, J. W. Caldwell, P. A. Kollman, and D. A. Case. 2004. Development and testing of a general Amber force field. J. Comp. Chem. 25:1157–1174. [DOI] [PubMed] [Google Scholar]
- 41.Jorgensen, W. L., J. Chandrasekhar, J. D. Madura, R. W. Impey, and M. L. Klein. 1983. Comparisons of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935. [Google Scholar]
- 42.Yamaguchi, K., Z. Tamura, and M. Maeda. 1997. Molecular structure of the zwitterionic form of phenolsulfonphthalein. Anal. Sci. 13:521–522. [Google Scholar]
- 43.Case, D. A., T. E. Cheatham, T. Darden, H. Gohlke, R. Luo, K. M. Merz, A. Onufriev, C. Simmerling, B. Wang, and R. J. Woods. 2005. The Amber biomolecular simulation programs. J. Comput. Chem. 26:1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryckaert, J.-P., G. Ciccotti, and H. J. C. Berendsen. 1977. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comp. Phys. 23:327–341. [Google Scholar]
- 45.Procacci, P., and B. J. Berne. 1994. Multiple time-scale methods for constant-pressure molecular-dynamics simulations of molecular-systems. Mol. Phys. 83:255–272. [Google Scholar]
- 46.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, A. DiNola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Comp. Phys. 81:3684–3690. [Google Scholar]
- 47.Chiu, S. W., M. Clark, S. Subramaniam, and E. Jakobsson. 2000. Collective motion artifacts arising in long-duration molecular dynamics simulations. J. Comp. Chem. 21:121–131. [Google Scholar]
- 48.Harvey, S. C., R. K. Z. Tan, and T. E. Cheatham. 1998. The flying ice cube: velocity rescaling in molecular dynamics leads to violation of energy equipartition. J. Comp. Chem. 19:726–740. [Google Scholar]
- 49.Kollman, P. A., I. Massova, C. Reyes, B. Kuhn, S. Huo, L. Chong, M. Lee, T. Lee, Y. Duan, W. Wang, O. Donini, P. Cieplak, et al. 2000. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 33:889–897. [DOI] [PubMed] [Google Scholar]
- 50.Chong, L. T., Y. Duan, L. Wang, I. Massova, and P. A. Kollman. 1999. Molecular dynamics and free-energy calculations applied to affinity maturation in antibody 48G7. Proc. Natl. Acad. Sci. USA. 96:14330–14335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gestwicki, J. E., G. R. Crabtree, and I. A. Graef. 2004. Harnessing chaperones to generate small-molecule inhibitors of amyloid beta aggregation. Science. 306:865–869. [DOI] [PubMed] [Google Scholar]
- 52.Wang, S. S. S., Y. T. Chen, and S. W. Chou. 2005. Inhibition of amyloid fibril formation of beta-amyloid peptides via the amphiphilic surfactants. Biochim. Biophys. Acta-Mol. Bas. Dis. 1741:307–313. [DOI] [PubMed] [Google Scholar]
- 53.Lomakin, A., D. S. Chung, G. B. Benedek, D. A. Kirschner, and D. B. Teplow. 1996. On the nucleation and growth of amyloid beta-protein fibrils: detection of nuclei and quantitation of rate constants. PNAS, USA. 93:1125–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sabate, R., and J. Estelrich. 2005. Stimulatory and inhibitory effects of alkyl bromide surfactants on beta-amyloid fibrillogenesis. Langmuir. 21:6944–6949. [DOI] [PubMed] [Google Scholar]