Abstract
DNA methylation is an epigenetic modification of cytosine that is important for silencing gene transcription and transposons, gene imprinting, development, and seed viability. DNA METHYLTRANSFERASE1 (MET1) is the primary maintenance DNA methyltransferase in Arabidopsis (Arabidopsis thaliana). Reciprocal crosses between antisense MET1 transgenic and wild-type plants show that DNA hypomethylation has a parent-of-origin effect on seed size. However, due to the dominant nature of the antisense MET1 transgene, the parent with a hypomethylated genome, its gametophyte, and both the maternal and paternal genomes of the F1 seed become hypomethylated. Thus, the distinct role played by hypomethylation at each generation is not known. To address this issue, we examined F1 seed from reciprocal crosses using a loss-of-function recessive null allele, met1-6. Crosses between wild-type and homozygous met1-6 parents show that hypomethylated maternal and paternal genomes result in significantly larger and smaller F1 seeds, respectively. Our analysis of crosses between wild-type and heterozygous MET1/met1-6 parents revealed that hypomethylation in the female or male gametophytic generation was sufficient to influence F1 seed size. A recessive mutation in another gene that dramatically reduces DNA methylation, DECREASE IN DNA METHYLATION1, also causes parent-of-origin effects on F1 seed size. By contrast, recessive mutations in genes that regulate a smaller subset of DNA methylation (CHROMOMETHYLASE3 and DOMAINS REARRANGED METHYLTRANSFERASES1 and 2) had little effect on seed size. Collectively, these results show that maternal and paternal genomes play distinct roles in the regulation of seed size in Arabidopsis.
DNA methylation usually refers to a covalent addition of a methyl group to cytosine at the 5-position. DNA methylation is a heritable epigenetic process that regulates growth and development in both animals and plants (Martienssen and Colot, 2001; Reik et al., 2001; Li, 2002). In mammals, 5-methylcytosine mainly occurs at CpG dinucleotides, and plays a vital role in genome stabilization, X chromosome inactivation, silencing of transposons and endogenous retrovirus, gene expression, and imprinting (Bird and Wolffe, 1999; Bestor, 2000; Reik et al., 2001). The patterns of DNA methylation are maintained during somatic development by the DNA methyltransferase1 (Dnmt1) DNA methyltransferase (Li et al., 1992). During gametogenesis and embryogenesis, DNA methylation is lost and subsequently reestablished by the Dnmt3a and Dnmt3b DNA methyltransferases (Okano et al., 1999).
In plants, DNA methylation is involved in regulating many epigenetic phenomena (Martienssen and Colot, 2001; Bender, 2004; Gehring et al., 2004; Chan et al., 2005). These include transcriptional silencing of transposons and transgenes, defense against pathogens, regulation of imprinting, as well as the silencing of genes that control flowering time, floral organ identity, fertility, and leaf morphology (Finnegan et al., 1996; Kakutani et al., 1996; Jacobsen et al., 2000; Soppe et al., 2000; Miura et al., 2001). In Arabidopsis (Arabidopsis thaliana), CpG DNA methylation is maintained by DNA METHYLTRANSFERASE1 (MET1), a homolog of mammalian DNA methyltransferase Dnmt1 (Finnegan and Dennis, 1993; Finnegan and Kovac, 2000). In addition to CpG methylation, Arabidopsis has CpNpG and asymmetric CpNpN methylation, which are maintained by the CHROMOMETHYLASE3 (CMT3) and the de novo DNA METHYLTRANSFERASES1 and 2 (DRM1 and DRM2; Henikoff and Comai, 1998; Bartee et al., 2001; Lindroth et al., 2001; Cao and Jacobsen, 2002a, 2002b). DNA methylation is intimately associated with histone modifications, chromatin remodeling, and the accessibility of DNA to transcription factors (Martienssen and Colot, 2001; Jackson et al., 2002; Johnson et al., 2002; Soppe et al., 2002; Bender, 2004; Chan et al., 2005).
The seeds of flowering plants are derived from two fertilization events that occur in the female gametophyte. In Arabidopsis, a haploid megaspore undergoes three mitotic divisions to form an eight-nucleus, seven-cell female gametophyte containing the egg, central, synergid, and antipodal cells; the fusion of two haploid polar nuclei makes the nucleus of the central cell diploid. Fertilization of the egg cell by a sperm cell gives rise to a diploid embryo that ultimately generates the organs, tissues, and meristems of the plant. Fertilization of the central cell by a second sperm cell produces a triploid, terminally differentiated endosperm that supports embryo or seedling growth and development by producing storage proteins, lipids, and starch, and by mediating the transfer of maternal-derived nutrients to be absorbed by the embryo (Brown et al., 1999).
Arabidopsis plants with an antisense MET1 transgene or partial-loss-of-function met1 mutations caused a reduction of global DNA methylation levels, particularly at CpG dinucleotides (Finnegan et al., 1996; Kakutani et al., 1996; Ronemus et al., 1996; Kankel et al., 2003). It has also been shown that mutations in CMT3 and DRM lead to a reduction in CpNpG and asymmetric CpNpN methylation. Plants with reduced DNA methylation display pleiotropic developmental abnormalities, such as abnormal shoot and leaf development, abnormal flower organ formation, delayed flowering, reduced fertilities, and abnormal embryogenesis (Finnegan et al., 1996; Kakutani et al., 1996, 2005; Ronemus et al., 1996; Cao et al., 2003; Kankel et al., 2003; Kato et al., 2003; Saze et al., 2003; Xiao et al., 2006).
In addition to MET1, DECREASE IN DNA METHYLATION1 (DDM1), an ATP-dependent SWI2/SNF2 chromatin remodeling factor, is also required for normal patterns of genomic DNA methylation in Arabidopsis (Vongs et al., 1993; Jeddeloh et al., 1999). Mutations in the DDM1 gene result in a rapid loss of cytosine methylation at heterochromatic repetitive sequences and a gradual depletion of methylation at euchromatic low-copy sequences over successive generations (Kakutani et al., 1999). DDM1 has been shown to regulate gene imprinting, transposons, gene silencing, and paramutation (Vielle-Calzada et al., 1999; Hirochika et al., 2000; Singer et al., 2001).
Genetic crosses between wild-type and antisense MET1 plants revealed that DNA methylation influences F1 seed size (Adams et al., 2000; Luo et al., 2000). Pollination of antisense MET1 transgenic pistils with wild-type pollen produced large F1 seeds, and reciprocal crosses generated small F1 seeds. Thus, DNA hypomethylation has parent-of-origin effects on seed size. However, due to the dominant nature of the antisense MET1 transgene, the parent with a hypomethylated genome generates a hypomethylated gametophyte, and both the maternal and paternal genomes of the F1 embryo and seed become hypomethylated after fertilization. Thus, the distinct role played by hypomethylation at each generation is not known. To address this issue, we carried out reciprocal crosses using a complete loss-of-function allele, met1-6 (Xiao et al., 2003), to distinguish the roles of maternal versus paternal genomes in parents, gametophytes, and F1 progeny. We show that larger F1 seeds were produced when the maternal genome was hypomethylated, whereas smaller F1 seeds were generated when the male genome was hypomethylated. Reciprocal crosses between wild-type and heterozygous MET1/met1-6 plants revealed that hypomethylation in either the male or female gamete was sufficient to cause seed size variation. Moreover, reciprocal crosses between wild type and ddm1-2, which dramatically reduces genome DNA methylation, produced F1 seed that displayed parent-of-origin effects on size. By contrast, little effect on size was detected in F1 seed from reciprocal crosses between wild type and mutations with relatively smaller effects on genome DNA methylation, cmt3 and drm1 drm2. Thus, the DNA methylation status of the maternal and paternal genomes regulates seed size in Arabidopsis.
RESULTS
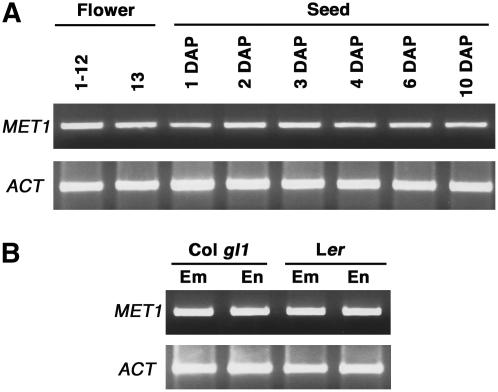
MET1 Is Expressed in the Developing Seed
To understand whether MET1 plays a role in regulating seed size, we first examined expression of MET1 in ovules and developing seeds, including embryo and endosperm. Reverse transcription (RT)-PCR analysis indicates that MET1 was expressed in ovule-containing flowers at stages 1 to 12 and stage 13, as well as in the early developing seeds at 1, 2, 3, 4, 6, and 10 d after pollination (DAP; Fig. 1A). When seed was separated into embryo and endosperm parts, we found MET1 was expressed in both embryo and endosperm of Arabidopsis ecotypes Columbia (Col)-0 and Landsberg erecta (Ler; Fig. 1B). This result indicates that MET1 is expressed in flowers prior to fertilization, as well as the developing embryo and endosperm after fertilization.
Figure 1.
Expression of the MET1 gene during plant reproduction. MET1 RNA was amplified by RT-PCR as described in “Materials and Methods.” A, RNA was isolated from flowers (stages 1–12 and 13) and developing seeds at the indicated DAP. B, Seeds from wild type (Col gl1 and Ler ecotypes) were harvested 8 DAP, dissected, and RNA was isolated from embryos (Em) and endosperm (En).
Hypomethylation of Maternal and Paternal Genomes Causes Parent-of-Origin Effects on Seed Size
To assess the effect of maternal versus paternal genome hypomethylation on seed size, we reciprocally crossed wild type with the first generation of the met1-6 homozygous plants. As shown in Figure 2, larger F1 seed was generated when the maternal parent was homozygous met1-6. By contrast, smaller F1 seed was produced when the paternal parent was homozygous for the met1-6 mutation.
Figure 2.
Parent-of-origin effects of the met1-6 mutation on seed size in Arabidopsis. For each cross, the genotype of the maternal parent is indicated first, followed by the paternal parent. WT, Wild-type parent; met1, homozygous met1-6 parent.
We quantitatively measured F1 seed size from reciprocal crosses by sifting them through sieves with different size openings, weighing seeds, and measuring seed length and width. As shown in Table I, when wild-type pistils were pollinated with pollen from a homozygous met1-6 plant, F1 seed size was significantly reduced with 68% and 28% of the seed retained on sieve numbers 60 (250-μm opening) and 70 (212-μm opening), respectively. For the wild-type control cross, 64% and 35% F1 seed was retained on sieve numbers 50 (300-μm opening) and 60, respectively. Pollination of met1-6 pistils with wild-type pollen produced the largest F1 seed, in which 10% seed was retained on sieve number 45 (355-μm opening), 84% was retained on sieve number 50, and only 6% was retained on sieve number 60 (Table I).
Table I.
Parent-of-origin effects of the met1-6 mutation on seed size
| Genetic Cross
|
Percentage of Seeds Retained on Indicated Sievea
|
Weight | |||||
|---|---|---|---|---|---|---|---|
| Maternal Parent | Paternal Parent | No. 45 355 μm | No. 50 300 μm | No. 60 250 μm | No. 70 212 μm | No. 80 180 μm | |
| mgb | |||||||
| Wild type | met1-6 | 0 | 0 | 68 ± 4 | 28 ± 6 | 4 ± 2 | 1.6 ± 0.1 |
| Wild type | Wild type | 0 | 64 ± 8 | 35 ± 8 | 0 | 0 | 2.8 ± 0.1 |
| met1-6 | Wild type | 10 ± 2 | 84 ± 5 | 6 ± 4 | 0 | 0 | 3.9 ± 0.2 |
| Wild type | MET1/met1-6 | 0 | 37 ± 6 | 47 ± 5 | 12 ± 7 | 4 ± 2 | 2.2 ± 0.3 |
| MET1/met1-6 | Wild type | 0 | 72 ± 7 | 27 ± 7 | 0 | 0 | 3.0 ± 0.3 |
Two hundred seeds were passed through sieves. Standard deviation for three replicates is shown.
Two hundred seeds were weighed. Weight per 100 seeds is shown. Standard deviation for three replicates is shown.
The F1 seed size of the different crosses was reflected in their seed mass. Average weight of 100 F1 seeds of the wild-type control cross was 2.8 mg. Pollination of wild-type pistils with met1-6 pollen produced F1 seeds with an average weight of 1.6 mg per 100 seeds, whereas the reciprocal cross generated 3.9 mg per 100 F1 seeds.
We also measured F1 seed length and width of the above three crosses. As shown in Table II, the wild-type F1 seed from control crosses were on average 497-μm long and 302-μm wide. Pollination of wild-type pistils with met1-6 pollen produced F1 seeds that were on average 404-μm long and 245-μm wide, approximately 19% smaller than F1 wild-type seeds. By contrast, F1 seed from the reciprocal cross were approximately 27% longer and 23% wider. Taken together, the above results indicate that hypomethylation of the maternal genome results in larger and heavier seeds, whereas hypomethylation of the paternal genome results in smaller, lighter seed.
Table II.
Parent-of-origin effects of the met1-6 mutation on F1 seed length and width
| Genetic Cross
|
Lengtha | Changeb | Widtha | Changeb | |
|---|---|---|---|---|---|
| Maternal Parent | Paternal Parent | ||||
| μm | μm | ||||
| Wild type | met1-6 | 404 ± 20 | −19% | 245 ± 22 | −19% |
| Wild type | Wild type | 497 ± 27 | 302 ± 19 | ||
| met1-6 | Wild type | 631 ± 35 | +27% | 372 ± 22 | +23% |
Average length and width of 50 F1 seeds is shown.
Change is relative to wild-type F1 seed.
Hypomethylation of Maternal and Paternal Genomes Causes Parent-of-Origin Effects on Endosperm Structure and Development
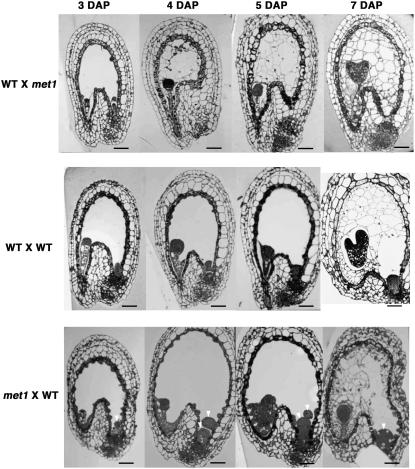
We next examined F1 seed to see if parental hypomethylation affects structure and the timing of seed development. We sectioned F1 seed at 3, 4, 5, and 7 DAP (Fig. 3). We found that development of small F1 seed with a hypomethylated paternal genome was accelerated compared to wild-type controls. For example, in F1 seeds with a paternal hypomethylated genome, endosperm started to cellularize at 4 DAP and almost finished cellularization at 5 DAP, whereas wild-type endosperm had not started to cellularize at 5 DAP. Smaller F1 seed also displayed a smaller endosperm volume than wild-type seeds. By contrast, larger F1 seed with a hypomethylated maternal genome displayed delayed development and had a larger endosperm volume when compared to wild-type controls. In particular, these F1 seed had enlarged chalazal endosperm and nodule at the chalazal end of the seed (Fig. 3). These results show that maternal and paternal genome hypomethylation results in distinct patterns of endosperm development within F1 seed.
Figure 3.
Structure differences of F1 seeds from reciprocal crosses between wild-type and met1-6 mutant parents. Thin sections of seeds were isolated, fixed, and stained as described in “Materials and Methods.” For each cross, the genotype of the maternal parent is indicated first, followed by the paternal parent. WT, Wild-type parent; met1, homozygous met1-6 parent. Arrowheads indicate large chalazal endosperm and nodule at the chalazal end. Scale bars represent 50 μm.
DNA Methylation Status of Gametes Affects Seed Size
To investigate whether hypomethylation in male or female gametes is sufficient to affect seed size, we reciprocally crossed wild type with the heterozygous MET1/ met1-6 plants. Wild-type pistils pollinated with pollen from a heterozygous MET1/ met1-6 plant produced a subset of smaller F1 seeds compared to wild-type controls (Table I). That is, nearly half of the F1 seed (47%) was retained on sieve number 60 (250 μm), and there was also 12% and 4% seed retained on sieve numbers 70 (212 μm) and 80 (180 μm), respectively. By contrast, when MET1/ met1-6 pistils were pollinated with wild-type pollen, we detected a subtle increase in seed size. Most F1 seeds (72%) were retained on sieve number 50 (300 μm), and no seed was retained on sieve number 70 or number 80.
To examine the correlation between changes in seed size and gamete hypomethylation, we determined the genotype of F1 seeds produced by reciprocal crosses between wild-type and heterozygous MET1/ met1-6 plants. Wild-type pistils were pollinated with pollen from a MET1/ met1-6 plant, F1 seed were visually divided into two size categories (medium-sized and small-sized seeds), and the genotype of seeds was determined as described in “Materials and Methods.” We found that 92% of the medium-sized seeds inherited the wild-type MET1 allele from the paternal parent. The probability that this deviation from 1:1 segregation of paternal-derived MET1 and met1-6 alleles within the medium-sized population occurred by chance is extremely low (paternal MET1:met1-6, 106:9, χ2 = 82, P ≪ 0.005). Moreover, 84% of the small-sized F1 seeds inherited the mutant met1-6 allele from the paternal parent (paternal MET1:met1-6, 14:73, χ2 = 40, P ≪ 0.005). These results suggest that hypomethylation in the male gametophyte is sufficient to reduce F1 seed size. We also determined the genotype of F1 seed from the reciprocal cross, MET1/ met1-6 pistils pollinated with wild-type pollen. We found that 78% of the medium-sized F1 seed inherited the paternal wild-type MET1 allele. The probability that this deviation from 1:1 segregation of maternal-derived MET1 and met1-6 alleles within the medium-sized population occurred by chance is again extremely low (maternal MET1:met1-6, 78:22, χ2 = 36, P ≪ 0.005). Moreover, 75% of the large F1 seed inherited the mutant met1-6 allele (maternal MET1:met1-6, 25:75, χ2 = 21, P ≪ 0.005). Thus, hypomethylation in the gametophyte generation is sufficient to affect F1 seed size.
Hypomethylation of the Maternal or Paternal Genome by the ddm1-2 Mutation Affects Seed Size
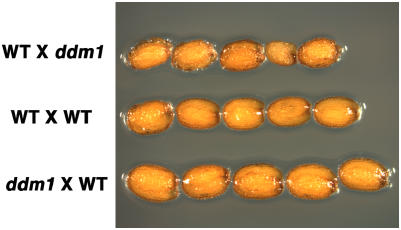
Does met1-6 mutant hypomethylation uniquely influence seed size? To address this question, we looked at the effect of mutations in the DDM1 gene on seed size. DDM1, an ATP-dependent SWI2/SNF2 chromatin remodeling protein, also is required for genomic DNA methylation. Mutations in DDM1 result in a rapid loss of cytosine methylation at repetitive sequences and a gradual depletion of methylation at low-copy sequences (Jeddeloh et al., 1999). We reciprocally crossed the recessive ddm1-2 mutant with wild-type plants and analyzed F1 seed size by measuring their width and length. As shown in Figure 4 and Table III, when pistils were pollinated with pollen from a homozygous ddm1-2 mutant male plant, a modest decrease in F1 seed size was detected. By contrast, F1 seeds from the reciprocal cross were significantly larger than the wild-type controls (Fig. 4; Table III). This result is consistent with the increased size of F1 seeds that bear mutations in DDM1 as well as in the MEDEA (MEA) Polycomb group gene (Vielle-Calzada et al., 1999). Thus, a hypomethylated maternal or paternal genome caused by either met1-6 or ddm1-2 mutation is sufficient to influence F1 seed size.
Figure 4.
Parent-of-origin effects of the ddm1-2 mutation on seed size in Arabidopsis. For each cross, the genotype of the maternal parent is indicated first, followed by the paternal parent. WT, Wild-type parent; ddm1, homozygous ddm1-2 parent.
Table III.
Parent-of-origin effects of the ddm1-2 mutation on F1 seed length and width
| Genetic Cross
|
Lengtha | Changeb | Widtha | Changeb | |
|---|---|---|---|---|---|
| Maternal Parent | Paternal Parent | ||||
| μm | μm | ||||
| Wild type | ddm1-2 | 462 ± 32 | −6% | 271 ± 24 | −9% |
| Wild type | Wild type | 491 ± 27 | 299 ± 16 | ||
| ddm1-2 | Wild type | 607 ± 38 | +24% | 346 ± 21 | +16% |
Average length and width of 50 F1 seeds is shown.
Change is relative to wild-type F1 seed.
Plants have CpNpG and asymmetric CpNpN DNA methylation that are mainly maintained by CMT3 and DRM2. Does non-CpG DNA methylation play a role in determining seed size? To answer this, we reciprocally crossed recessive mutants cmt3-7 and drm1 drm2 with their corresponding wild-type plants, and then compared F1 seed size with wild-type control F1 seed. We did not observe a significant change in F1 seed size from these crosses (data not shown). This suggests loss of CpNpG and CpNpN methylation has little effect on seed size.
DISCUSSION
DNA methylation is essential for regulating development in plants (Martienssen and Colot, 2001; Bender, 2004; Gehring et al., 2004; Chan et al., 2005). DNA methylation silences transposons, regulates gene imprinting, and influences flowering time, floral organ identity, fertility, embryogenesis, and leaf morphology (Finnegan et al., 1996; Kakutani et al., 1996; Jacobsen et al., 2000; Soppe et al., 2000; Miura et al., 2001). Many of these phenomena resulted from gene silencing by DNA methylation and gene activation by DNA hypomethylation. Here, we show that maternal or paternal genome hypomethylation has a parent-of-origin effect on seed size in Arabidopsis (Table I; Fig. 2). Maternal genome hypomethylation causes seeds to be larger, whereas paternal genome hypomethylation generates smaller seeds.
Role of Continuing Genome Hypomethylation in F1 Seed
F1 seed from crosses between wild type and plants bearing an antisense MET1 transgene displayed parent-of-origin effects on seed size (Adams et al., 2000; Luo et al., 2000). In these experiments, due to the dominant nature of the antisense MET1 transgene, the parent with a hypomethylated genome generates a hypomethylated gametophyte, and both the maternal and paternal genomes of the F1 embryo and seed become hypomethylated after fertilization. This is because reduction of MET1 expression in the seed by the antisense MET1 transgene would be expected to reduce the level of methylation of both maternal and paternal genomes. Thus, the distinct role played by hypomethylation at each generation is not known. We have carried out reciprocal crosses between wild-type plants and plants homozygous for a recessive met1-6 allele. In these experiments, the F1 zygote has a hypomethylated set of maternal-derived or paternal-derived chromosomes. This state is preserved because the F1 embryo and endosperm have a wild-type MET1 allele. Wild-type MET1 protein maintains the DNA methylation on chromosomes inherited from the wild-type parent. However, methylation of chromosomes from the met1-6 parent, in particular, CpG methylation, is not restored by de novo DNA methyltransferases (Cao and Jacobsen, 2002b; Chan et al., 2006). Thus, the continuous hypomethylation of both maternal and paternal chromosomes in the F1 seed is not a prerequisite for parent-of-origin effects on seed size.
Role of Genome Hypomethylation in the Preceding Sporophytic Generation and Its Gametophyte
During embryogenesis and gametogenesis in mammals, patterns of DNA methylation at imprinted loci are erased and then reestablished (Reik et al., 2001; Li, 2002). Dnmt1 and de novo methyltransferases Dnmt3a and Dnmt3b are important for maintenance of DNA methylation in oocytes and spermatogenesis (Chen and Li, 2006). By contrast, in flowering plants there is no evidence for erasure and reestablishment of DNA methylation during gametogenesis or embryogenesis (Cao and Jacobsen, 2002b). Rather, DNA methylation is maintained in the gametophyte and sporophyte generations by DNA methyltransferases, primarily by MET1, and also by CMT3 and DRM2 (Kankel et al., 2003; Saze et al., 2003; Xiao et al., 2003; Kinoshita et al., 2004).
In Arabidopsis, there are two or three mitotic cell divisions during male or female gametogenesis, respectively. In met1-6 mutant gametophytes, each cell division results in an approximate halving of the genome DNA methylation (Saze et al., 2003). Due to the small number of cell divisions, genome hypomethylation in the met1-6 male and female gametophytes is incomplete and varies from gamete to gamete (Saze et al., 2003). After fertilization, the wild-type MET1 allele in the heterozygous F1 seed produces MET1 protein that maintains DNA methylation inherited from gametes. Reciprocal crosses with wild-type and heterozygous MET1/ met1-6 plants indicated that this hypomethylation within the gametophyte is sufficient to cause changes in F1 seed size. Thus, genome hypomethylation during the parental sporophyte generation is not required to generate parent-of-origin effects on seed size. Genome hypomethylation in the female or male gametophyte, which is preserved in the F1 seed, is sufficient.
Modulation of Seed Size Requires Genome Hypomethylation at CpG Sites
Both MET1 and DDM1 are required for maintaining genome methylation patterns in Arabidopsis (Kakutani et al., 1999; Lindroth et al., 2001; Xiao et al., 2003). MET1 is a DNA methyltransferase, whereas DDM1 is an ATP-dependent SWI2/SNF2 chromatin remodeling protein and the mechanism by which DDM1 regulates DNA methylation is likely indirect (Verbsky and Richards, 2001). Here, we show that mutations in either MET1 or DDM1 affect seed size. By contrast, mutations in CMT3 and DRM1/DRM2, which primarily reduce CpNpG and CpNpN methylation and have a modest effect on overall genome methylation (Cao and Jacobsen, 2002a), have little effect on seed size. One possibility is that significant hypomethylation of all three classes of plant DNA methylation is a prerequisite for alterations in seed size. This may reflect the redundant suppression of gene expression by the three classes of DNA methylation. Alternatively, CpG DNA methylation may be particularly important for regulation of genes that influence seed size. Indeed, CpG DNA methylation is important in the regulation of genes imprinted in the endosperm, MEA, FWA, and FIS2 (Kinoshita et al., 2004; Gehring et al., 2006; Jullien et al., 2006b).
Mechanisms for the Control of Seed Size by DNA Methylation
How does hypomethylation of maternal and paternal genomes influence seed size? Genomic imprinting in the endosperm has been shown to regulate seed size and development in plants (Haig and Westoby, 1991; Kohler et al., 2005; Baroux et al., 2006; Gehring et al., 2006; Jullien et al., 2006a, 2006b). One theory to explain the evolution of genomic imprinting is the parental conflict theory (Haig and Westoby, 1991). If a mother has offspring by more than one father, the theory predicts that alleles of some genes active in the offspring in allocating resources from maternal tissue to offspring will have a different pattern of expression depending on the parent of origin of those alleles. For endosperm activators, the maternal allele is likely silenced and paternal allele expressed so that mother has resources for each individual of her offspring. By contrast, for the endosperm repressors, the maternal allele is expressed and the paternal allele is silenced. DNA methylation usually represses gene transcription and loss of DNA methylation might activate silenced genes or alleles. In the cross of the met1-6 female with the wild-type male plant, considering the nature of the loss-of-function recessive met1-6 allele, only the maternal genome derived from the met1-6 mutant plant becomes hypomethylated, and the paternal genome derived from wild-type plant is not epigenetically changed by hypomethylated maternal genome during or after fertilization. Thus, the maternal alleles of endosperm activators, which are normally silenced, are expressed and lead to larger F1 seeds (Fig. 2; Table II). In the cross of a wild-type female with the met1-6 male parent, the maternal-derived, wild-type genome is not modified and the paternal-derived genome is hypomethylated. Thus, normally silenced paternal alleles of endosperm repressors are expressed, which leads to repressed endosperm development and smaller seeds (Fig. 2; Table II).
Another hypothesis to explain endosperm imprinting and seed development is dosage effect (Haig and Westoby, 1991; Scott et al., 1998; Dilkes and Comai, 2004). In this case, seed development is hypothesized to depend on a ratio of maternal and paternal genomes in endosperm. The ratio is normally two maternal genomes to one paternal genome. If the 2:1 ratio is disturbed, expression from maternal and paternal genomes may be imbalanced, and a breakdown of endosperm or even failure of seed development might occur (Dilkes and Comai, 2004). The cross between a diploid female parent and tetraploid male parent produces larger F1 seeds, whereas the reciprocal cross generates smaller F1 seeds (Scott et al., 1998). Paternal excess promotes growth of seed, whereas maternal excess inhibits seed development (Haig and Westoby, 1991; Adams et al., 2000). In reciprocal crosses of the met1-6 mutant with wild-type plants, inheritance of the hypomethylated paternal met1-6 allele resembles maternal genome excess, resulting in smaller seed (Figs. 2 and 3). By contrast, inheritance of the hypomethylated maternal met1-6 allele resembles paternal genome excess, thus resulting in larger seed.
DNA methylation likely regulates genes and pathways affecting seed size. The target genes and pathways are not known. However, the study of Polycomb group proteins may provide clues. Polycomb group proteins are epigenetic regulators that form complexes that alter chromatin structure and silence gene expression (Lund and van Lohuizen, 2004). A direct connection between gene silencing by DNA methyltransferases and Polycomb group proteins has recently been shown (Vire et al., 2006). In Arabidopsis, mutations in the FIE, FIS2, and MEA Polycomb Group genes cause parent-of-origin effects on seed viability (Hsieh et al., 2003), as well as endosperm cell proliferation (Kiyosue et al., 1999), a key component in the regulation of seed size (Fig. 3). Also, genetic interactions have been detected between MET1 and FIE (Vinkenoog et al., 2000), FIS2 (Jullien et al., 2006b), and DME, a regulator of MEA gene imprinting (Xiao et al., 2003). Thus, it is possible that there is a relationship between DNA methylation and Polycomb group proteins in the parent-of-origin regulation of seed size.
MATERIALS AND METHODS
Plant Materials and Growth Condition
Arabidopsis (Arabidopsis thaliana) plants were grown in greenhouses under continuous light at 23°C. The heterozygous MET1/met1-6 plants (Col gl1 ecotype) were obtained in the original genetic screen for suppressors of dme-mediated seed abortion (Lindroth et al., 2001; Xiao et al., 2003) and maintained in the heterozygous state by backcrossing with wild-type Col gl1. The homozygous met1-6/met1-6 plants we used were selected from the first generation progeny produced by self-pollinated heterozygous MET1/met1-6 plants. Wild-type plants (Col gl1) were used in crosses with met1-6 (Col gl1) plants. Wild-type Col-0 plants were used in crosses with ddm1-2 (Col gl1; Jeddeloh et al., 1999). Genotyping plants for the met1-6 was performed as described (Lindroth et al., 2001; Xiao et al., 2003).
Sectioning and Microscopy
Thin-section studies of seeds were carried out using methods as described (Brown et al., 1999). Briefly, seeds were fixed in 4% glutaraldehyde in 0.1 m phosphate buffer, pH 6.9, postfixed in osmium ferricyanide, dehydrated through a graded acetone series, and infiltrated with Spurr's resin (EMS). Seeds were sectioned sagittally in the plane of the micropyle and stalk with a LKB historange microtome equipped with glass knives. Then 2.0- to 5.0-μm sections were mounted on glass slides (Brown and Lemmon, 1995) and stained with Polychrome stain (Fox, 1997). Thin sections were observed and photographed under light microscopy. The approximate volume of endosperm was estimated by observations of the area of endosperm in photographs of sectioned seeds.
MET1 Expression Analysis
RT-PCR analysis was performed as described (Kinoshita et al., 1999). Total RNA was isolated from flowers at flower stages 1 to 12 and stage 13 and seeds at 1, 2, 3, 4, 6, and 10 DAP, and the ecotype used was Col gl1. For MET1 expression in embryo and endosperm, seeds of Col-0 and Ler at 8 DAP were used. Seed was dissected into embryo and endosperm parts as described (Kinoshita et al., 1999). Primers used in the experiment were as follows: MET1: DD-F4, 5′-TTAAACGATCCTGACAGCGG-3′ and DD-R1, 5′-GGATTCAGATTCAGTTCCTTC-3′; ACT: ACT.con.F, 5′-GATTTGGCATCACACTTTCTACAATG-3′ and ACT.con.R, 5′-GATTTGGCATCACACTTTCTACAATG-3′. Primer pairs spanned intron sequences so that amplification of RNA could be distinguished from amplification of any contaminating DNA.
Sieving, Weighing, and Measuring Seed Size
Mature F1 dry seeds in batches of 200 were weighed using an analytical balance and sieved through a series of fine wire sieves (nos. 40, 45, 50, 60, 70, and 80 with openings of 425, 355, 300, 250, 212, and 180 μm, respectively; USA Standard Testing Sieve; Fisher Scientific). Seeds retained by each sieve were counted. Three replicates were done for all crosses. For measuring seed length and width, photos of seed population of each cross were taken under a microscope (model SZX-ILLB100; Olympus), then seed length and width were measured using NIH Image 1.63. Fifty seeds from each cross were measured for seed size.
Determining the Genotype of F1 Seed
After pollinating wild-type pistils with pollen from MET1/ met1-6 plants, we separated F1 seeds into two categories: medium- and small-sized seeds. F1 seeds were placed on Murashige and Skoog medium plates, treated for 3 d at 4°C, and then incubated in a growth chamber for 10 d at 22°C. Young seedlings were transferred to soil in pots and grown in the greenhouse at 22°C. A rosette leaf from each 4-week-old plant was harvested. Genomic DNA was isolated, and determining the genotype of MET1 and met1-6 alleles was performed as described previously (Lindroth et al., 2001; Xiao et al., 2003). For the cross between the heterozygous MET1/met1-6 female and wild-type male parent, the same procedure was used except that we separated F1 seeds into medium- and large-sized categories.
Acknowledgments
We thank Jon Penterman, Tzung-Fu Hsieh, and Daphna Michaeli for critically reading this manuscript. We thank Eric Richards for providing ddm1-2 mutant seeds.
This work was supported by the National Institutes of Health (grant no. GM069415 to R.L.F.) and the U.S. Department of Agriculture (grant no. 2005–02355 to R.L.F.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Robert L. Fischer (rfischer@berkeley.edu).
References
- Adams S, Vinkenoog R, Spielman M, Dickinson HG, Scott RJ (2000) Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 127: 2493–2502 [DOI] [PubMed] [Google Scholar]
- Baroux C, Gagliardini V, Page DR, Grossniklaus U (2006) Dynamic regulatory interactions of Polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis. Genes Dev 20: 1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee L, Malagnac F, Bender J (2001) Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev 15: 1753–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J (2004) DNA methylation and epigenetics. Annu Rev Plant Biol 55: 41–68 [DOI] [PubMed] [Google Scholar]
- Bestor TH (2000) The DNA methyltransferases of mammals. Hum Mol Genet 14: 2395–2402 [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP (1999) Methylation-induced repression—belts, braces, and chromatin. Cell 99: 451–454 [DOI] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE (1995) Methods in plant immunolight microscopy. Methods Cell Biol 49: 85–107 [DOI] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Nguyen H, Olsen O-A (1999) Development of endosperm in Arabidopsis thaliana. Sex Plant Reprod 12: 32–42 [Google Scholar]
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE (2003) Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 13: 2212–2217 [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE (2002. a) Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA 99: 16491–16498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE (2002. b) Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 12: 1138–1144 [DOI] [PubMed] [Google Scholar]
- Chan SW-L, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6: 351–360 [DOI] [PubMed] [Google Scholar]
- Chan SW-L, Henderson IR, Zhang X, Shah G, Chien JS-C, Jacobsen SE (2006) RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in Arabidopsis. PLoS Genet 2: 791–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Li E (2006) Establishment and maintenance of DNA methylation patterns in mammals. Curr Top Microbiol Immunol 301: 179–201 [DOI] [PubMed] [Google Scholar]
- Dilkes BP, Comai L (2004) A differential dosage hypothesis for parental effects in seed development. Plant Cell 16: 3174–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES (1993) Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res 21: 2383–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Kovac KA (2000) Plant DNA Methyltransferases. Plant Mol Biol 43: 189–201 [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES (1996) Reduced DNA methylation in Arabidopsis results in abnormal plant development. Proc Natl Acad Sci USA 93: 8449–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox LM (1997) Microscopy 101. Polychrome stain for epoxy sections. Microscopy Today 97: 21 [Google Scholar]
- Gehring M, Choi Y, Fischer RL (2004) Imprinting and seed development. Plant Cell (Suppl) 16: S203–S213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL (2006) DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D, Westoby M (1991) Genomic imprinting in endosperm: its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Philos Trans R Soc Lond B Biol Sci 333: 1–14 [Google Scholar]
- Henikoff S, Comai L (1998) A DNA methyltransferase homolog with a chromodomain exists in multiple polymorphic forms in Arabidopsis. Genetics 149: 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H, Okamoto H, Kakutani T (2000) Silencing of retrotransposons in arabidopsis and reactivation by the ddm1 mutation. Plant Cell 12: 357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Hakim O, Ohad N, Fischer RL (2003) From flour to flower: how Polycomb group proteins influence multiple aspects of plant development. Trends Plant Sci 8: 439–445 [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Sakai H, Finnegan EJ, Cao X, Meyerowitz EM (2000) Ectopic hypermethylation of flower-specific genes in Arabidopsis. Curr Biol 10: 179–186 [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Stokes TL, Richards EJ (1999) Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet 22: 94–97 [DOI] [PubMed] [Google Scholar]
- Johnson LM, Cao X, Jacobsen SE (2002) Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr Biol 12: 1360–1367 [DOI] [PubMed] [Google Scholar]
- Jullien PE, Katz A, Oliva M, Ohad N, Berger F (2006. a) Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Curr Biol 16: 486–492 [DOI] [PubMed] [Google Scholar]
- Jullien PE, Kinoshita T, Ohad N, Berger F (2006. b) Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18: 1360–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh JA, Flowers SK, Munakata K, Richards EJ (1996) Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci USA 93: 12406–12411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T, Kato M, Kinoshita T, Miura A (2005) Control of development and transposon movement by DNA methylation in Arabidopsis thaliana. Cold Spring Harb Symp Quant Biol LXIX: 139–143 [DOI] [PubMed] [Google Scholar]
- Kakutani T, Munakata K, Richards EJ, Hirochika H (1999) Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics 151: 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ (2003) Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163: 1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Miura A, Bender J, Jacobsen SE, Kakutani T (2003) Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr Biol 13: 421–426 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T (2004) One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303: 521–523 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Yadegari R, Harada JJ, Goldberg RB, Fischer RL (1999) Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm. Plant Cell 11: 1945–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T, Ohad N, Yadegari R, Hannon M, Dinneny J, Wells D, Katz A, Margossian L, Harada J, Goldberg RB, et al (1999) Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc Natl Acad Sci USA 96: 4186–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C, Page DR, Gagliardini V, Grossniklaus U (2005) The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat Genet 37: 28–30 [DOI] [PubMed] [Google Scholar]
- Li E (2002) Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet 3: 662–673 [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE (2001) Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Lund AH, van Lohuizen M (2004) Polycomb complexes and silencing mechanisms. Curr Opin Cell Biol 16: 239–246 [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A (2000) Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA 97: 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen RA, Colot V (2001) DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science 293: 1070–1074 [DOI] [PubMed] [Google Scholar]
- Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T (2001) Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411: 212–214 [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 247: 247–257 [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J (2001) Epigenetic reprogramming in mammalian development. Science 293: 1089–1093 [DOI] [PubMed] [Google Scholar]
- Ronemus MJ, Galbiati M, Ticknor C, Chen J, Dellaporta SL (1996) Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273: 654–657 [DOI] [PubMed] [Google Scholar]
- Saze H, Scheid OM, Paszkowski J (2003) Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet 34: 65–69 [DOI] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Bailey J, Dickinson HG (1998) Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125: 3329–3341 [DOI] [PubMed] [Google Scholar]
- Singer T, Yordan C, Martienssen RA (2001) Robertson's Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev 15: 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe WJJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJM (2000) The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell 6: 791–802 [DOI] [PubMed] [Google Scholar]
- Soppe WJJ, Jasencakova Z, Andreas H, Tetsuji K, Armin M, Huang MS, Jacobsen SE, Ingo S, Fransz PF (2002) DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J 21: 6549–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbsky ML, Richards EJ (2001) Chromatin remodeling in plants. Curr Opin Plant Biol 4: 494–500 [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada J-P, Thomas J, Spillane C, Coluccio A, Hoeppner MA, Grossniklaus U (1999) Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev 13: 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkenoog R, Spielman M, Adams S, Fischer RL, Dickinson HG, Scott RJ (2000) Hypomethylation promotes autonomous endosperm development and rescues postfertilization lethality in fie mutants. Plant Cell 12: 2271–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Eynde AV, Bernard D, Vanderwinden J-M, et al (2006) The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439: 871–874 [DOI] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ (1993) Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928 [DOI] [PubMed] [Google Scholar]
- Xiao W, Custard KD, Brown RC, Lemmon BE, Harada JJ, Goldberg RB, Fischer RL (2006) DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell 18: 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Gehring M, Choi Y, Margossian L, Pu H, Harada JJ, Goldberg RB, Pennell RI, Fischer RL (2003) Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev Cell 5: 891–901 [DOI] [PubMed] [Google Scholar]