Abstract
The Arabidopsis thaliana SOS2 and SOS3 genes are required for intracellular Na+ and K+ homeostasis and plant tolerance to high Na+ and low K+ environments. SOS3 is an EF hand type calcium-binding protein having sequence similarities with animal neuronal calcium sensors and the yeast calcineurin B. SOS2 is a serine/threonine protein kinase in the SNF1/AMPK family. We report here that SOS3 physically interacts with and activates SOS2 protein kinase. Genetically, sos2sos3 double mutant analysis indicates that SOS2 and SOS3 function in the same pathway. Biochemically, SOS2 interacts with SOS3 in the yeast two-hybrid system and in vitro binding assays. The interaction is mediated by the C-terminal regulatory domain of SOS2. SOS3 activates SOS2 protein kinase activity in a Ca2+-dependent manner. Therefore, SOS3 and SOS2 define a novel regulatory pathway important for the control of intracellular ion homeostasis and salt tolerance in plants.
The homeostasis of intracellular ion concentrations is a fundamental property of living cells. Eukaryotic cells employ primary active transport, mediated by P-type ATPases, and secondary transport, mediated by channels and cotransporters, to maintain characteristic high concentrations of K+ and low concentrations of Na+ in the cytosol (1). Intracellular K+ and Na+ homeostasis is important for the activities of many cytosolic enzymes and for maintaining membrane potential and appropriate osmoticum for cell volume regulation (1). In plants, maintenance of K+ and Na+ homeostasis is critical for salt tolerance, an important agronomic trait because salt stress significantly limits agricultural productivity in many parts of the world (2–5).
Research with Saccharomyces cerevisiae has been extremely valuable in dissecting the regulatory mechanisms of ion homeostasis and salt tolerance (6–8). The cellular mechanisms of plant salt tolerance are thought to be similar to those in yeast in which signaling through a protein phosphatase, calcineurin, modulates the expression and activities of Na+ and K+ transporters to maintain low Na+ and high K+ concentrations in the cytoplasm (6, 9, 10). The existence of calcineurin in plants has been indicated by several studies (9, 11–13). Recently, the Arabidopsis thaliana SOS3 gene product was found to share substantial sequence similarity with the regulatory subunit of yeast calcineurin (CNB) (10). Functionally, SOS3 is also similar to yeast CNB because, like the yeast cnb mutant (14–15), growth of sos3 mutant plants is hypersensitive to Na+ and Li+ inhibition (16). Despite this evidence for SOS3 participation in a calcineurin-like pathway in mediating plant salt tolerance, we report in this paper that, instead of binding and activating a protein phosphatase, SOS3 physically interacts with and activates a protein kinase encoded by the Arabidopsis SOS2 gene.
Like SOS3, the SOS2 locus is required for intracellular Na+ and K+ homeostasis (17). Mutations in SOS2 cause hypersensitivity to high Na+ and Li+ stresses, as well as to low K+ stress (17). The similar phenotypes of the sos2 and sos3 mutant plants suggest that the two genes may function in the same pathway in regulating Na+ and K+ homeostasis. As part of this study, we constructed and analyzed sos2sos3 double mutants and show that there is no additive effect when the two mutations are combined. This provides genetic evidence that SOS3 and SOS2 function in the same regulatory pathway.
To identify other components in the regulatory pathway, we used SOS3 as a bait to select interacting proteins in a yeast two-hybrid screen. The screen identified a group of related protein kinases as SOS3-interacting proteins (SIPs). Strikingly, sequence analysis revealed that the SIP kinases are in the SOS2 subfamily of protein kinases. We show that SOS2 indeed interacts with SOS3 in the yeast two-hybrid system as well as in in vitro binding assays. Furthermore, SOS2 kinase activity is activated by SOS3 in a Ca2+-dependent manner. These results illustrate that plants have evolved a novel regulatory pathway for the control of intracellular Na+ and K+ homeostasis and salt tolerance.
Materials and Methods
Plant Growth Conditions.
Conditions for plant growth in soil and in agar media were as described (17). Root growth measurement was carried out daily (16). Data presented are for growth after 1 week on the respective media.
Genetic Analysis.
For double mutant analysis, sos2-2 and sos3-1 mutants were crossed, and progeny of F1 plants were collected. A root bending assay (18) was used to select salt-hypersensitive mutants from the F2 seedlings. Double mutants were identified from the mutants by PCR genotyping. Primer pairs used to distinguish sos3-1 and its wild-type allele are 5′TCTCATGAATTTGCAGTTGC and 5′AAACTGTTTAATCTGGAGGG, resulting in 112- and 121-bp amplification products for the mutant and wild-type genes, respectively. To discern sos2-2 from its wild-type allele, primer pairs 5′AATTTGGATGATATTCGTGCAGTTTTTG and 5′TTAACATTTAAATGGAATTGACC (for wild type, resulting in a 96-bp product), and 5′CAAATTCAAGGAATCAAGAATTCGC and 5′TTAACATTTAAATGGAATTGACC (for sos2-2, yielding a 90-bp product) were used.
Yeast Two-Hybrid Screen and Interaction Assay.
The SOS3 coding region was amplified by PCR with primers containing restriction sites and was cloned in frame between the NcoI and SalI sites of pAS2, to form pAS-SOS3 as the bait to screen an Arabidopsis expression library in pACT1. The λACT cDNA expression library (19) was converted to a pACT plasmid library by infecting Escherichia coli BNN132 cells (20). pAS-SOS3 was transformed into yeast Y190, which was then analyzed by Western blotting using α-HA antibody (Babco, Berkeley, CA) for detection of SOS3 fusion protein expression. Transformation and screening were according to Bai and Elledge (21). Approximately 1.6 × 106 transformants were plated and selected on SC agar medium, which lacks leucine, tryptophan, and histine and contains 25 mM 3-amino-1,2,4-triazole (triple selection medium). Large colonies were streaked and subjected to a 5-bromo-4-chloro-3-indolyl β-d-galactoside filter assay. Seventy-eight clones showing blue color were chosen for further analysis by growing them in SC medium minus leucine, which favors the loss of the bait plasmid, and isolating the pACT-SIP plasmids for sequence determination. To confirm the interaction between putative SIP clones and SOS3, purified pACT-SIP plasmids were retransformed into the yeast Y190 harboring pAS-SOS3 and were subjected to growth on the triple selection medium and β-galactosidase assay. For control interactions with the identified prey proteins, empty vector pAS2, pAS-RB [maize retinoblastoma protein (22), a gift from B. Larkins, University of Arizona, Tucson, AZ] and pAS-p53 (21) (a gift from S. J. Elledge, Baylor College of Medicine, Houston, TX) were used as baits.
The N-terminal (residues 1–260) and C-terminal (residues 257–446) parts of SOS2 were amplified by PCR with primers containing restriction sites and were cloned into BamHI-EcoRI of pACT2 to test interaction with pAS-SOS3.
Protein Expression.
GST-SOS2, GST-SOS2(K40N), and GST-SOS2(G197E) were produced as described by Liu et al. (23). To produce bacterially expressed GST-SOS3, the coding region of SOS3 cDNA was amplified by PCR with primers harboring restriction sites, cloned in frame into BamHI-EcoRI of pGEX-2TK (Amersham Pharmacia) and transformed into E. coli BL21 DE3 cells (Amersham Pharmacia). E. coli cultures were induced with 0.5 mM IPTG, and recombinant proteins were affinity-purified from bacterial lysates with glutathione-Sepharose beads (Amersham Pharmacia).
Kinase Assay.
GST-fusion proteins were incubated in kinase buffer (20 mM Tris⋅HCl, pH 8.0/5 mM MgCl2/1 mM CaCl2/1 mM DTT). The kinase reaction was started by adding [γ-32P] ATP and was transferred to 30°C for 30 min. The reaction was stopped by adding 4× SDS-sample buffer and was analyzed by SDS/PAGE and autoradiography. Oligopeptide phosphorylation was assayed in a final reaction volume of 25 μl in the kinase buffer with 200 μM oligopeptides and 1 μg of GST-SOS2 and with or without 1 μg of GST-SOS3. Peptides p1 (LRRASLG) and p2 (VRKRTLRRL) were from Sigma. Peptide p3 (ALARAASAAALARRR) was custom synthesized by Research Genetics (Huntsville, AL). Incubation was for 30 min at 30°C. Fusion proteins (GST-SOS2 and GST-SOS3) bound to glutathione-Sepharose were pelleted by low speed centrifugation. Fifteen microliters of supernatant were used for detection of peptide phosphorylation according to Davies et al. (24). For Ca2+-free conditions, CaCl2 was omitted, and 10 mM EGTA was included in the kinase buffer.
In Vitro Binding Assays.
For pull-down assays, radiolabeled SOS3 protein was produced from pET14b-SOS3 (NcoI-BamHI) by using an in vitro transcription and translation assay kit with 3H-leucine as the sole source of leucine (TNT Coupled Reticulocyte Lysate System, Promega), following the manufacturer's instructions. Equal amounts of 3H-leucine-SOS3 were incubated under constant rocking for 1 h at 4°C in 150 μl of binding buffer (20 mM Tris⋅HCl, pH 7.5/150 mM NaCl/1 mM CaCl2/0.1% Nonidet P-40) with 2 μg each of GST-SOS2, GST-RB [GST fusion of the maize retinoblastoma protein (22)], or GST bound to glutathione-Sepharose beads. GST-RB and GST were used as controls in the binding assays. The Sepharose beads were pelleted and washed extensively with the binding buffer and were analyzed by SDS/PAGE and fluorography.
For gel blot overlay assays, the SOS3 ORF was cloned into the pGEX-2TK vector to allow direct labeling of recombinant GST-SOS3 with 32P because it contains the recognition sequence for protein kinase A between the GST domain and the multiple cloning sites. The recombinant GST-SOS3 was purified by affinity chromatography on glutathione-Sepharose resin and was labeled by using protein kinase A and γ-32P-ATP following published procedures (25). The GST was then cleaved from GST-SOS3 by using thrombin. GST-SOS2, GST-SOS2(K40N), GST-SOS2(G197E), and control proteins were fractionated by SDS/PAGE and were electroblotted onto nitrocellulose membrane. The blot was incubated with 32P-labeled SOS3 in TBST [20 mM Tris⋅HCl, pH 7.8/0.5 M NaCl/0.15% (vol/vol) Tween 20], was washed with TBST, and was exposed to x-ray film.
Results
Genetic Interaction Between sos2 and sos3.
To determine genetically whether SOS2 and SOS3 function in the same pathway, we constructed sos2sos3 double mutants by crossing the deletion alleles sos2-2/sos2-2 and sos3-1/sos3-1 and selecting sos mutants from the F2 generation. Because SOS2 and SOS3 are closely linked on chromosome V, we devised a PCR-based screen for the relatively rare occurrence of double mutants. Two lines of sos2sos3 double mutants were identified from screening 86 sos mutants from the F2 population by the presence of sos2-2- and sos3-1-specific PCR products and the lack of PCR products corresponding to the respective wild-type alleles. The double mutants were confirmed by test crosses with the single mutants.
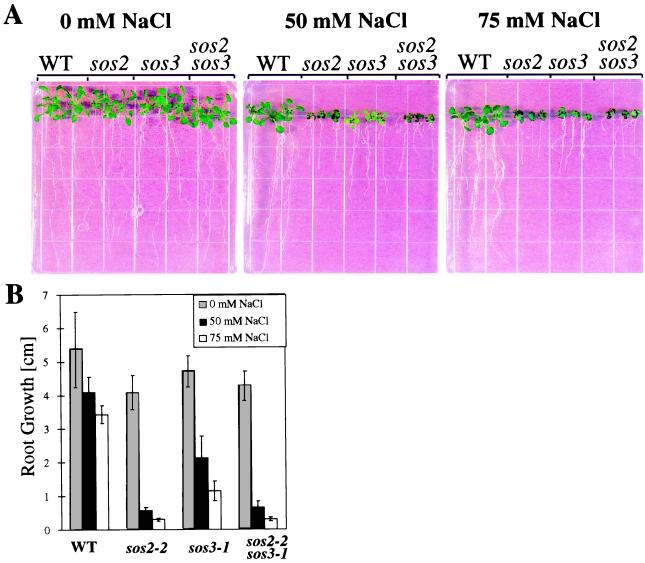
We tested the growth of the single mutants and sos2sos3 double mutants on media containing 0, 50, or 75 mM NaCl. Consistent with previous results (17), both sos2 and sos3 mutants show hypersensitivity to NaCl stress, although sos2 displays greater sensitivity than sos3 (Fig. 1). The double mutants resembled sos2-2 plants in appearance (e.g., dark curly leaves under 50 mM NaCl) and in shoot and root growth (Fig. 1A). Root growth measurements show that the double mutant was more severely inhibited than sos3-1 but was indistinguishable from the sos2-2 mutant under all conditions (Fig. 1B). The lack of additive effects of the mutations is consistent with SOS2 and SOS3 functioning in the same pathway.
Figure 1.
Genetic interaction between sos2 and sos3. (A) Seedling phenotypes of wild-type (WT) plants and sos2-2, sos3-1, and sos2sos3 double mutants treated with various levels of NaCl. (B) Root growth measurement. Five-day-old seedlings were transferred from normal nutrient medium to media supplemented with different concentrations of NaCl, and root elongation after 7 days is presented. Error bars represent the standard deviation (n = 12). The pictures were taken 10 days after the NaCl treatments.
Two-Hybrid Screen for SOS3-Interacting Proteins Identifies a Family of Protein Kinases That Are Similar to SOS2.
The entire SOS3 ORF was fused in frame to the C terminus of the DNA-binding domain of GAL4 to create a bait in the plasmid pAS2 and was transformed into the yeast strain Y190 containing two reporter genes, His3 and LacZ (21). The bait plasmid pAS-SOS3 by itself did not activate transcription of the two reporter genes. For the prey library, we used the λ-ACT cDNA library prepared from mRNA isolated from young Arabidopsis seedlings (19). The prey plasmids were excised and transformed into the yeast strain harboring pAS-SOS3. Of ≈1.6 × 106 transformants plated, 78 clones were confirmed positive for His3 and LacZ expression. Prey plasmids were isolated from the clones, and sequence analysis revealed that 39 encode proteins with significant sequence similarities to serine/threonine protein kinases. Based on sequence alignments, the 39 SOS3-interacting (SIP) kinase clones define a family of seven proteins (i.e., SIP1 to SIP7) that are closely related to each other. At least one pACT-SIP plasmid representing each of the seven SIP kinases was reintroduced into the yeast strain Y190 to test for autoactivation of the reporter genes. They were also introduced into the yeast Y190 containing the control bait pAS-p53, pAS-RB, or the empty bait vector pAS2 to examine the specificity of the interactions. None of them was found to activate the reporter genes when expressed alone or in combination with pAS2 or the control baits.
Complete amino acid sequences were obtained for SIP1, SIP2, SIP3, and SIP4 by searching the GenBank database. SIP2 corresponds to a serine/threonine protein kinase-like sequence (26) previously identified through homology cloning. SIP1, SIP3, and SIP4 correspond to genomic sequences (GenBank accession numbers AB019228, AL022198.1, and AC002338.1, respectively) released by the Arabidopsis Genome Initiative.
Remarkably, when SOS2 was cloned recently (23), we found that the deduced amino acid sequence of SOS2 is very similar to the SIP kinases. The alignment of the SOS2 amino acid sequence with four of the SIP kinases is shown in Fig. 2. Like SOS2, the putative kinase catalytic domain of the SIPs resides in the N-terminal portion of the proteins and is most similar to the yeast SNF1 and mammalian AMPK kinases (27, 28) (alignments not shown). Similarities between SOS2 and the SIP kinases are found in not only the N-terminal catalytic domain but also in the C-terminal regulatory domain.
Figure 2.
Amino acid alignment of SOS2 with SIP1, SIP2, SIP3, and SIP4. Amino acids identical in SOS2 and at least another two proteins are highlighted in black and conservative substitutions in gray.
SOS2 Interacts with SOS3 in the Yeast Two-Hybrid System.
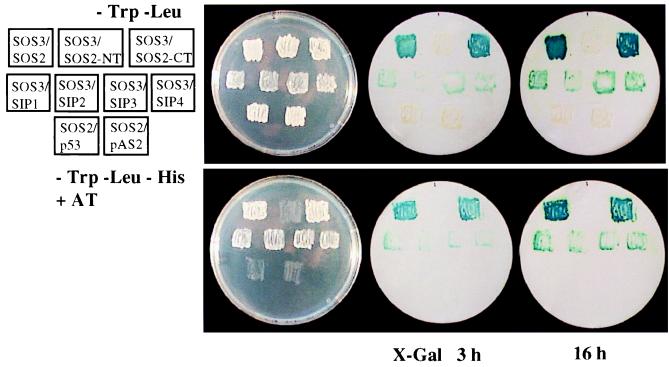
Although SOS2 was not among the SIP kinases identified by the yeast two-hybrid screen, its high sequence similarity with the SIP kinases indicates that it may interact with SOS3. To test this hypothesis, SOS2 was cloned in frame in the pACT2 vector and was introduced into the yeast strain Y190 containing the bait pAS-SOS3, or pAS2 vector or pAS-p53 as controls. Indeed, SOS2 interacted with SOS3 in the yeast two-hybrid system, and its interaction appeared stronger than that of the other SIP kinases, as suggested by the faster and stronger color appearance in β-galactosidase assays (Fig. 3). Expression of pACT-SOS2 alone (not shown) or in combination with pAS2 bait vector or with the control bait pAS-p53 did not activate the reporter gene expression (Fig. 3). Thus, SOS2 specifically interacts with SOS3 in the yeast two-hybrid system. Because the SOS2 gene is expressed at a very low level, it would not be unexpected that SOS2 message would be underrepresented or even absent in the Arabidopsis λ-ACT cDNA expression library used for yeast two-hybrid screening. Indeed, PCR amplification using SOS2-specific primers on plasmid DNA of this library yielded no product whereas using SIP-clone specific primers resulted in good amplification (not shown).
Figure 3.
Interaction between SOS3 and SOS2, SOS2 N-terminal (SOS2-NT) and C-terminal (SOS2-CT) domains, and SIP1, SIP2, SIP3, or SIP4 in the yeast two-hybrid assay. Yeast strains containing pAS-SOS3 and pACT-SOS2, pACT-SOS2-NT, pACT-SOS2-CT, pACT-SIP1-1, pACT-SIP2-1, pACT-SIP3-1, pACT-SIP4-1, respectively, were assayed for LacZ expression. pAS-p53 and pAS2 were used as negative controls in combination with pACT-SOS2. (Upper Right) Yeast grown on SC medium minus tryptophan and leucine to select for both the bait and prey proteins. (Lower Right) SC medium minus tryptophan, leucine, histidine, and plus 25 mM 3-amino-1,2,4-triazole (AT) to allow the growth of only positively interacting clones.
SOS3 Interacts with the C-Terminal Regulatory Domain of SOS2.
All SIP kinase clones encoded only truncated proteins, and the positions of N-terminal truncations for SIP1, SIP2, SIP3, and SIP4 clones are shown in Table 1. It appears that protein kinase activity is not required for interaction with SOS3 because many of the interacting clones have their putative catalytic domain truncated (Table 1). To determine whether SOS3 interacts with only the regulatory domain of SOS2, the N-terminal kinase catalytic domain (residues 1–260) and the C -terminal regulatory domain (257–446) were each cloned in frame into the prey vector pACT2, and their interaction with SOS3 was determined by using the yeast two-hybrid assay. Expression of the preys in yeast cells were confirmed by using antibodies against the HA-tag, which is part of the prey fusion proteins (21). Fig. 3 shows that SOS3 interacted with the C-terminal but not the N-terminal domain of SOS2.
Table 1.
Positions of N-terminal truncations of SIP1, SIP2, SIP3, and SIP4 kinase clones
| Clone no. | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| SIP1 | Δ276 | Δ283 | Δ288* | |||
| SIP2 | Δ4 | Δ237 | Δ251† | |||
| SIP3 | Δ223 | Δ300‡ | ||||
| SIP4 | Δ86 | Δ186§ | Δ219 | Δ221 | Δ293 | Δ300 |
Given are the number of amino acids missing from the N-terminal part of the protein.
*Clones represented three times.
†Clones represented three times.
‡Clones represented two times.
§Clones represented four times.
SOS2 Interacts with SOS3 in Vitro.
To corroborate the interaction between SOS2 and SOS3 observed in the yeast two-hybrid system, we tested whether the two proteins bind to each other in vitro. In a pull-down assay, [3H]leucine-labeled SOS3 protein was first synthesized by in vitro transcription and translation, and then was incubated with GST-SOS2, GST-RB, or GST that were bound to glutathione-Sepharose beads. The beads were pelleted and washed extensively, and the bound proteins were resolved by SDS/PAGE and were detected by fluorography. SOS3 was found to bind to GST-SOS2 but not to the control GST or GST-RB (Fig. 4A).
Figure 4.
SOS2 interacts with SOS3 in vitro. (A) Interaction between SOS3 and SOS2 in a pull-down assay. [3H]leucine-labeled SOS3 was incubated with glutathione-Sepharose immobilized GST, GST fusion proteins of SOS2, or RB. Proteins bound to the Sepharose beads were pelleted, washed thoroughly, electrophoresed, and detected by fluorography. (B) Interaction between SOS3 and SOS2 in a gel blot overlay assay. GST, partially purified GST-RB, GST-SOS2, GST-SOS2(K40N), GST-SOS2(G197E), and protein size markers (M) were separated by SDS/PAGE. Two identical gels were run; one was stained with Coomassie blue (Left), and the other was electroblotted and probed with [32P]labeled SOS3 (Right).
In addition, we conducted a gel blot overlay assay in which 32P-labeled SOS3 was used to probe a Western blot containing GST-SOS2 and, as controls, GST, GST-RB, and protein size markers (Fig. 4B). Again, SOS3 was found to bind to GST-SOS2 but not to GST, GST-RB, or the molecular markers (Fig. 4B). To determine whether protein kinase activity is required for SOS2 interaction with SOS3, recombinant proteins of the two inactive SOS2 mutants, GST-SOS2(K40N) and GST-SOS2(G197E) (23), were tested in the overlay assay. SOS3 was able to bind to the inactive SOS2 mutants (Fig. 4B), demonstrating that protein kinase activity is not required for SOS2-SOS3 interaction. The binding between SOS3 and SOS2 in vitro does not seem to require Ca2+ because, when the pull-down assay was conducted in the absence of Ca2+ and in the presence of 5 mM EGTA, identical results were obtained (results not shown).
SOS3 Activates SOS2 Kinase Activity.
We tested various proteins and peptides as substrates for SOS2 in kinase assays. SOS2 did not phosphorylate histone H1 or casein (results not shown). However, several synthetic peptides based on the recognition sequences of protein kinase C or SNF1/AMPK are strongly phosphorylated by SOS2 (Fig. 5A). SOS2 can phosphorylate either a serine or threonine residue because both the serine-containing (p1: LRRASLG and p3: ALARAASAAALARRR) and threonine-containing (p2: VRKRTLRRL) peptides are recognized as substrates (Fig. 5A). Phosphorylation of the peptides by SOS2 depends on the presence of SOS3. Without SOS3, little phosphorylation of the peptides was detected (Fig. 5A).
Figure 5.
SOS2 kinase activity is activated by SOS3 in a Ca2+-dependent manner. (A) SOS2 phosphorylation of peptide substrates is activated by SOS3. Oligopeptides p1, p2, and p3 were incubated in the presence of [γ-32P] ATP and Ca2+ in a kinase buffer with GST-SOS2, and with or without GST-SOS3. γ-32P-incorporation was measured by scintillation counting. (B) SOS3 activation of SOS2 is Ca2+-dependent. Oligopeptide p3 was phosphorylated by GST-SOS2 in the presence of GST-SOS3, and with or without free Ca2+. Error bars represent the standard deviation (n = 3).
SOS3 Activation of SOS2 Kinase Depends on Calcium.
The predicted amino acid sequence of SOS3 suggested that it is a calcium-binding protein (10). 45Ca2+-overlay assays confirmed that SOS3 is capable of binding Ca2+ (J.-K.Z., unpublished results). We tested whether SOS3 activation of SOS2 kinase depends on Ca2+. Ca2+-free conditions in the kinase assay were achieved by the presence of a large excess of EGTA (10 mM). Without free Ca2+, phosphorylation of p3 was substantially reduced (Fig. 5B), suggesting that Ca2+ is required for SOS3 activation of SOS2 kinase.
Discussion
We employed two different approaches to identify proteins that function in the SOS3 pathway for intracellular ion homeostasis. In one approach, SOS3 was used as a bait in the yeast two-hybrid system to isolate interacting proteins (i.e., SIPs). In an alternative approach, we isolated the SOS2 gene through positional cloning (23). The two approaches converged when we found that SOS2 encodes a protein kinase highly similar to a group of SOS3-interacting proteins identified in the two-hybrid screen. SOS2 interacts with SOS3 in vitro as well as in the yeast two-hybrid assay. SOS2 was not identified in the original yeast two-hybrid screening because it did not appear to be represented in the prey library, which is likely attributable to its very low level of expression (23). The C-terminal regulatory domain of SOS2 is responsible for interaction with SOS3, and the interaction does not depend on SOS2 kinase activity or Ca2+. In the presence of Ca2+, SOS3 activates SOS2 kinase activity. Double mutant analysis indicates that the sos2 and sos3 mutations are not additive. Together, these results strongly suggest that SOS2 kinase acts as a downstream component in the SOS3 pathway leading to K+ and Na+ homeostasis and plant salt tolerance.
Many plant ion transporters have recently been cloned and characterized. Knowledge of the regulatory mechanisms of transporter abundance and activities in response to environmental, hormonal, and developmental signals is essential for understanding plant growth and development. SOS3 is the first regulatory protein known to control intracellular K+ and Na+ homeostasis and salt tolerance in plants. It is a myristoylated (J.-K.Z., unpublished data) calcium-binding protein with sequence similarities to CNB and animal neuronal calcium sensors. Because of the sequence and functional similarities of SOS3 with yeast CNB, a calcineurin-like pathway had been anticipated to mediate plant salt tolerance (6, 10). However, as yet, no protein phosphatase has been found to interact with SOS3. Instead, SOS3 was found to interact with and activate a protein kinase encoded by SOS2. The strong and stable binding between SOS2 and SOS3 suggests the interesting possibility that the two proteins form a novel protein kinase complex. Such a complex would be reminiscent of multisubunit protein kinases such as calmodulin-dependent kinases that consist of calmodulin as a regulatory subunit plus a kinase catalytic subunit, although very little sequence similarity exists between components of the two types of kinase complexes. The SOS3/SOS2 kinase complex is also different from calmodulin-dependent kinases in that the binding between calmodulin and the kinase is calcium-dependent. The phenotypes of the sos mutants suggest that the SOS3/SOS2 kinase complex functions specifically in the regulation of intracellular Na+ and K+ homeostasis and salt tolerance.
It is unlikely that SOS3 also activates a calcineurin A (CNA)-like protein phosphatase to mediate plant salt tolerance. Despite pharmacological evidence supporting the existence of calcineurin in plants (11, 12), no genes similar to CNA have been identified from plants, yet. Our results imply that plant salt tolerance may be mediated not by a calcineurin-like protein phosphatase but, rather, by the SOS3/SOS2 kinase complex. Nevertheless, the possible existence and involvement of calcineurin-like phosphatases in other plant processes cannot be ruled out. Recently, another CNB-like protein from Arabidopsis was characterized (13). Its induction by cold, drought, and wounding stresses suggested that it does not function like yeast CNB or Arabidopsis SOS3 in regulating intracellular Na+ and K+ homeostasis, although it was able to mediate Ca2+ signaling through the rat calcineurin A phosphatase when both were expressed in yeast. Previous pharmacological studies (11, 12) indicated the existence of calcineurin in guard cell regulation but did not address a role in salt tolerance. Pardo et al. (9) observed improved salt tolerance caused by ectopic expression of a constitutively active yeast calcineurin in tobacco plants. Because the mode of action of the yeast protein phosphatase in plant cells is not known, it may function by a mechanism other than ion homeostasis.
Although SOS3 also shares sequence similarity with animal neuronal calcium sensors (10), they too appear to function differently. Neuronal calcium sensor 1 can replace calmodulin in the activation of calcineurin (29). Another member of the neuronal calcium sensor superfamily, recoverin, functions by inhibiting rhodopsin kinase (30, 31).
The physical interaction between SOS3 and SOS2 is consistent with genetic evidence that suggests that the SOS genes function in the same pathway (Fig. 1). We propose that, in response to a cytosolic Ca2+ signal generated by Na+ stress (32), SOS3 activates SOS2 kinase, which then phosphorylates downstream effectors of salt tolerance (Fig. 6). SOS3 activation, rather than inhibition, of SOS2 kinase (Fig. 5) is consistent with the observation that both SOS3 and SOS2 are positive regulators of salt tolerance (10, 17) and with the indication from analyses of the sos2-5 mutant allele that an active SOS2 kinase is required for its function as a positive regulator (23). The physiological substrates of the SOS2 kinase are not yet known but may include certain Na+ and K+ transporters (4, 33–36) and transcriptional factor(s) that mediate the expression of these transporters under salt stress.
Figure 6.
Proposed regulatory pathway for intracellular Na+ and K+ homeostasis and Na+ tolerance in plants.
The physiological significance of the interaction between SOS3 and other SIP kinases observed in the yeast two-hybrid system is still unclear. The interaction between SOS3 and the other SIP kinases is much weaker compared with the interaction with SOS2 (Fig. 2). In addition, possible differences in the temporal and spatial expression between SOS3 and the other SIP kinases may preclude their interaction in planta. The natural interaction partners of some of the other SIP kinases might be SOS3-like calcium-binding proteins (13). We propose that SOS3-like calcium-binding proteins each interact with certain members of the SIP family of protein kinases to form specific kinase complexes that may perform a broad range of functions in mediating Ca2+ signaling and the regulation of ion transport or other processes in various plant cells. For example, SOS3 and SOS2-like proteins (i.e., SIPs) expressed in guard cells may interact to mediate Ca2+ signaling in stomatal regulation.
Acknowledgments
We sincerely thank R. T. Leonard, R. A. Bressan, H. Bohnert, and J. Lasswell for critical reading of the manuscript; B. A. Larkins for the gift of GST-RB1 and pAS-RB1; and B. Dilkes, R. Dante, and P. Hughes for helpful discussions. This work was supported by National Institutes of Health Grant R01GM59138 and U.S. Department of Agriculture/National Research Initiative Grant 9801270.
Abbreviation
- SIP
SOS3-interacting protein
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040577697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040577697
References
- 1.Stein W D. Channels, Carriers and Pumps: An Introduction to Membrane Transport. New York: Academic; 1990. [Google Scholar]
- 2.Apse M P, Aharon G S, Snedden W A, Blumwald E. Science. 1999;285:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- 3.Niu X, Bressan R A, Hasegawa P M, Pardo P M. Plant Physiol. 1995;109:735–742. doi: 10.1104/pp.109.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubio F, Gassmann W, Schroeder J I. Science. 1995;270:1660–1663. doi: 10.1126/science.270.5242.1660. [DOI] [PubMed] [Google Scholar]
- 5.Epstein E, Norlyn J D, Rush D W, Kingsbury R W, Kelley D B, Cunningham G A, Wrona A F. Science. 1980;210:399–404. doi: 10.1126/science.210.4468.399. [DOI] [PubMed] [Google Scholar]
- 6.Bressan R A, Pardo J M, Hasegawa P M. Trends Plant Sci. 1998;3:411–412. doi: 10.1016/s1360-1385(00)01692-7. [DOI] [PubMed] [Google Scholar]
- 7.Serrano R, Gaxiola R. Crit Rev Plant Sci. 1994;13:121–138. [Google Scholar]
- 8.Mulet J M, Leube M P, Kron S J, Rios G, Fink G R, Serrano R. Mol Cell Biol. 1999;19:3328–3337. doi: 10.1128/mcb.19.5.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pardo J M, Reddy M P, Yang S, Maggio A, Huh G-H, Matsumoto T, Coca M A, Paino-D'Urzo M, Koiwa H, Uan D-J, et al. Proc Natl Acad Sci USA. 1998;95:9681–9686. doi: 10.1073/pnas.95.16.9681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Zhu J-K. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- 11.Luan S, Li W, Rusnak F, Assmann S M, Schreiber S L. Proc Natl Acad Sci USA. 1993;90:2202–2206. doi: 10.1073/pnas.90.6.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen G J, Sanders D. Plant Cell. 1995;7:1473–1483. doi: 10.1105/tpc.7.9.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kudla J, Xu Q, Harter K, Gruissem W, Luan S. Proc Natl Acad Sci USA. 1999;96:4718–4723. doi: 10.1073/pnas.96.8.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakamura T Y, Liu Y, Hirata D, Namba H, Harada S, Hirokawa T, Miyakawa T. EMBO J. 1993;12:4063–4071. doi: 10.1002/j.1460-2075.1993.tb06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendoza I, Rubio F, Rodriguez-Navarro A, Pardo J M. J Biol Chem. 1994;269:8792–8796. [PubMed] [Google Scholar]
- 16.Liu J, Zhu J-K. Proc Natl Acad Sci USA. 1997;94:14960–14964. doi: 10.1073/pnas.94.26.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J-K, Liu J, Xiong L. Plant Cell. 1998;10:1181–1192. doi: 10.1105/tpc.10.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu S-J, Ding L, Zhu J-K. Plant Cell. 1996;8:617–627. doi: 10.1105/tpc.8.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J, Harter K, Theologis A. Proc Natl Acad Sci USA. 1997;94:11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 21.Bai C, Elledge S J. Methods Enzymol. 1996;273:331–347. doi: 10.1016/s0076-6879(96)73029-x. [DOI] [PubMed] [Google Scholar]
- 22.Grafi G, Burnett R J, Helentjaris T, Larkins B A, DeCaprio J A, Sellers W R, Kaelin W G., Jr Proc Natl Acad Sci USA. 1996;93:8962–8967. doi: 10.1073/pnas.93.17.8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, J., Ishitani, M., Halfter, U., Kim, C.-S. & Zhu, J.-K. (March 21, 2000) Proc. Natl. Acad. Sci. USA, 10.1073/pnas.060034197. http://www.pnas.org/cgi/doi/10.1073/pnas.060034197
- 24.Davies S P, Carling D, Hardie D G. Eur J Biochem. 1989;186:123–128. doi: 10.1111/j.1432-1033.1989.tb15185.x. [DOI] [PubMed] [Google Scholar]
- 25.Kaelin W G, Jr, Krek W, Sellers W R, DeCaprio J A, Ajchenbaum F, Fuchs C S, Chittenden T, Li Y, Farnham P J, Blanar M A. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 26.Mizoguchi T, Hayashida N, Yamaguchi-Shinizaki K, Kamada H, Shinosaki K. Plant Physiol. 1994;106:1229–1230. doi: 10.1104/pp.106.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Celenza J L, Carlson M. Science. 1986;233:1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- 28.Mitchelhill K I, Stapleton D, Gao G, House C, Michell B, Katsis F, Witters L A, Kemp B E. J Biol Chem. 1994;269:2361–2364. [PubMed] [Google Scholar]
- 29.Schaad N K, De Castro E, Nef S, Hegi S, Hinrichsen R, Martone M E, Ellisman M H, Sikkink R, Rusnak F, Sygush J, Nef P. Proc Natl Acad Sci USA. 1996;93:9253–9258. doi: 10.1073/pnas.93.17.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klenchin V A, Calvert P D, Bownds M D. J Biol Chem. 1995;270:16147–16152. doi: 10.1074/jbc.270.27.16147. [DOI] [PubMed] [Google Scholar]
- 31.Chen C K, Inglese J, Lefkowitz R J, Hurley J B. J Biol Chem. 1995;270:18060–18066. doi: 10.1074/jbc.270.30.18060. [DOI] [PubMed] [Google Scholar]
- 32.Lynch J, Polito V S, Lauchli A. Plant Physiol. 1989;90:1271–1274. doi: 10.1104/pp.90.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson J A, Huprikar S S, Kochian L V, Lucas W J, Gaber R F. Proc Natl Acad Sci USA. 1992;89:3736–3740. doi: 10.1073/pnas.89.9.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirsch R E, Lewis B D, Spadling E P, Sussman M R. Science. 1998;280:918–921. doi: 10.1126/science.280.5365.918. [DOI] [PubMed] [Google Scholar]
- 35.Kim E J, Kwak J M, Uozumi N, Schroeder J I. Plant Cell. 1998;10:51–62. doi: 10.1105/tpc.10.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaxiola R A, Rao R, Sherman A, Grisafi P, Alper S L, Fink G R. Proc Natl Acad Sci USA. 1999;96:1480–1485. doi: 10.1073/pnas.96.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]