Abstract
Arabidopsis RPP5 is a member of a large class of pathogen resistance genes encoding nucleotide-binding sites and leucine-rich repeat domains. Yeast two-hybrid analysis showed that RPP5 specifically interacts with At-RSH1, an Arabidopsis RelA/SpoT homolog. In Escherichia coli, RelA and SpoT determine the level of guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp), which are the effector nucleotides of the bacterial stringent response. Functional analysis in E. coli and in Streptomyces coelicolor A3 (2) showed that At-RSH1 confers phenotypes associated with (p)ppGpp synthesis. We characterized two additional Arabidopsis RelA/SpoT homologs, At-RSH2 and At-RSH3. At-RSH genes may regulate a rapid plant (p)ppGpp-mediated response to pathogens and other stresses.
Pathogens have developed specialized infection strategies, and plants have evolved systems to rapidly detect attempted pathogen ingress (1). For example, certain pathogenic bacteria transfer effector proteins into the host cytoplasm, where they are thought to enhance virulence by subversion of the host defense machinery and other cellular functions (2). Superimposed on this basal line of host defense, plants have evolved pathogen surveillance systems comprising numerous resistance (R) proteins (3). When viral, bacterial, or fungal virulence factors are detected by the plant's surveillance system, they then become genetically defined as avirulence (Avr) products (4–8). After specific recognition of pathogen Avr products, R proteins rapidly trigger a defense response that is associated with complex cellular metabolic alterations and production of active oxygen species and nitric oxide and typically appears microscopically as host cell death at the site of pathogen ingress (9).
Arabidopsis thaliana ecotypes carrying RPP5 elicit defense responses after detection of Peronospora parasitica strains that carry the cognate Avr product (10). RPP5 is a member of a superfamily of cytoplasmic R proteins that contain nucleotide-binding (NB) sites and leucine-rich repeat (LRR) domains and is grouped further into a subclass with similarity to the effector domain of the Drosophila and human Toll and IL-1 receptors (TIR domain) (10). All NB-LRR proteins contain a central region with three conserved motifs predicted to constitute an ATP- or GTP-binding pocket and several other motifs with unknown function (3). This “NB-ARC” (NB-Apaf-1, R proteins, and CED-4) domain or “Ap-ATPase” domain is also present in several structurally related regulators of animal apoptosis, including human Apaf-1 and nematode CED-4 (11, 12), where it functions as a protein–protein interaction module (13–15). Interestingly, RPP5 family members carry diverged NB-ARC domains, possibly reflecting functional differences such as interactions with different proteins (16). To find host proteins that interact with the NB-ARC domain of RPP5, we used the yeast two-hybrid assay (17) and identified At-RSH1.
At-RSH1 is a predicted plasma membrane-anchored cytoplasmic molecule with significant homology to bacterial RelA and SpoT proteins. These enzymes determine the level of guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp), which are the effector nucleotides of the prokaryotic stringent response (18) and also play a role in antibiotic production and differentiation in Streptomyces species (19). Eukaryotic relA/spoT homologs have not been described to date. Arabidopsis contains three unlinked relA/spoT homologs (At-RSH1, 2, and 3). Here, we show that At-RSH1 confers several distinct phenotypes associated with (p)ppGpp synthesis in both Streptomyces coelicolor A3 (2) and in Escherichia coli. This functional characterization of At-RSH1 is consistent with a role for (p)ppGpp in mediating a stress-induced defense system in plants analogous to the bacterial stringent response.
Materials and Methods
Yeast Two-Hybrid Plasmids and Library.
The Matchmaker LexA Two-Hybrid system was used (CLONTECH). The plasmids pJK101 and pRFHM1 were kindly provided by R. Brent, Massachusetts General Hospital, Boston. RPP5 cDNA fragments were obtained by reverse transcription–PCR from Arabidopsis Landsberg erecta (Ler). The RPP5 baits TIR1–223, TIR-NB-ARC1–518, NB-ARC161–518, and ARC226–531 were made by fusing (EcoRI/BamHI) the cDNA fragments with the DNA-binding domain of the pLexA vector. The pLexA baits with the plant NB-LRR genes RPP1A (20), RPM1 (21), RPS4 (22), and N (23) were kindly provided by M. Botella (Sainsbury Laboratory, Norwich, U.K.), M. Grant (Wye College, Wye, U.K.), W. Gassman and B. Staskawicz (Univ. of California, Berkeley), and M. Dutton and B. Baker (Univ. of California, Berkeley), respectively.
For the two hybrid cDNA library, mRNA was isolated (Amersham Pharmacia) from healthy leaves of 4-week-old wild-type and pad4 mutant Ler plants and from leaves of these plants harvested at several time points after infection with Pseudomonas syringae pv. tomato carrying AvrRPS4 (22) or P. parasitica Noco2 (10). Directional, poly(dT)17-primed, size-selected (>0.8-kb) cDNA with EcoRI/XhoI adapters (Stratagene) was ligated in the corresponding sites of the vector pADB42 (CLONTECH) and used to transform electrocompetent E. coli DH10B cells (GIBCO/BRL). Approximately 2.5 × 106 clones were obtained, and library plasmid DNA was isolated by using Tip500 columns (Qiagen, Chatsworth, CA).
Yeast Two-Hybrid Assays.
All RPP5 baits repressed the pJK101 lacZ reporter in yeast EGY48, indicating that the LexA-RPP5 fusion proteins are expressed and transferred to the nucleus (17). The RPP5 bait constructs, empty vectors, and several unrelated baits and preys (e.g., pLexLam, pRFHM1, pLex53, pB42-T) were tested in all combinations, but no activation of the reporter genes was observed, and hence, no indications were obtained for nonspecific interactions with RPP5 baits. Transformation of EGY48 (p8op-lacZ) carrying the RPP5 bait TIR-NB-ARC1–518 with the Arabidopsis cDNA library resulted in ≈3.5 million primary transformants. After amplification, ≈38 million colony-forming units (cfu) were analyzed for activation of the LEU2 and lacZ reporters. The NB-ARC161–518 bait was used to screen ≈4.5 million EGY48 (p8op-lacZ) library transformants, and ≈45 million cfu were analyzed for activation of the LEU2 and lacZ reporters. Library plasmids were rescued in E. coli strain XL-1 Blue MRF′ (Stratagene). Yeast transformations and liquid β-galactosidase assays were done as described in ref. 17 and CLONTECH's Yeast Protocols.
Nucleic Acid Analysis.
Recombinant plasmids were made according to standard procedures (17). DNA sequence reactions (Perkin–Elmer) were run on a 377 DNA sequencer (Applied Biosystems). The cDNA sequences were extended by using the 5′ rapid amplification of cDNA ends system (GIBCO/BRL). Other DNA and RNA manipulations were done essentially as described previously (10). DNA sequences and predicted gene products were aligned by using the clustalw algorithm (24), and phylogenetic analysis was done with the neighbor-joining method (25), with 1,000 bootstrap replicates.
Bacterial Expression and Complementation Analysis.
For expression in E. coli, the 1.6-kb 5′ region (NdeI/XbaI) of the At-RSH1 cDNA was cloned in pT7–7 under the control of the heat-inducible T7 RNA polymerase promoter system (26). The E. coli wild-type strain CF1648 and its derived mutants, CF1652 (relA∷kan) and CF1693 (relA∷kan, spoT∷cam) (27), were kindly provided by M. Cashel, National Institutes of Health, Bethesda, MD.
For expression in S. coelicolor, the full-length At-RSH1 cDNA (NdeI/NcoI) and the 1.6-kb 5′ portion (NdeI/XbaI) were cloned in the thiostrepton-inducible expression vector pIJ8600 (28). Plasmids were transferred to S. coelicolor strain M600 as described previously (29). Cloning details are available upon request.
Results
The NB-ARC Domain of RPP5 Interacts with At-RSH1 in Yeast.
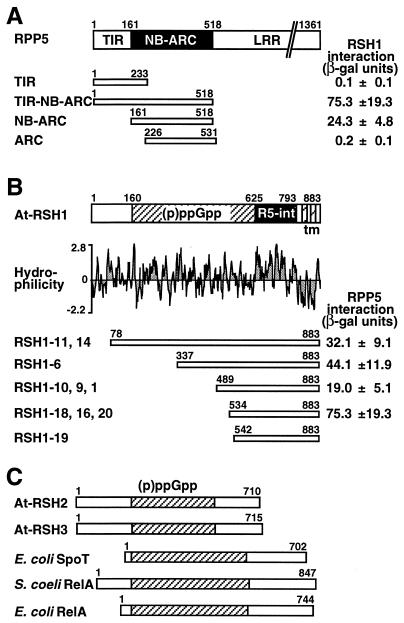
We used the yeast two-hybrid assay to identify RPP5-interacting protein(s). With the TIR-NB-ARC bait (Fig. 1A), 19 colonies were identified that conferred both leucine prototrophy and β-galactosidase activity. The 3′ ends of 10 clones were identical, and the corresponding gene was designated A. thaliana RelA/SpoT homolog 1 (At-RSH1). The different lengths of the inserts indicated the isolation of five independent At-RSH1 clones (Fig. 1B). In a second screen, the NB-ARC bait (Fig. 1A) identified 15 clones. DNA sequence analysis showed that 12 cDNA inserts were identical to the RSH1–18/16/20 group identified in the previous screen (Fig. 1B).
Figure 1.
Overview of RPP5/At-RSH1 interactions and (p)ppGpp synthetase domain-containing proteins. (A) Domain structure of RPP5 and interaction of RPP5 bait constructs with At-RSH1 (RSH1–18) in the yeast two-hybrid assay. β-Galactosidase ± SD in Miller units are averages from two replicates with three transformants. (B) Domain structure of At-RSH1, hydrophilicity analysis, and the five groups with At-RSH1 clones. R5-int, RPP5 interaction domain; TM, two transmembrane segments. (C) Domain structure of At-RSH2 and At-RSH3 with the (p)ppGpp synthetase domains aligned relative to that of At-RSH1, E. coli RelA (GenBank J04039) and SpoT (M24503), and S. coelicolor RelA (X87267).
Retransformation of yeast cells with the TIR-NB-ARC or the NB-ARC baits with four different At-RSH1 library plasmids showed that the LEU2 and lacZ reporters were consistently activated only in the presence of galactose, indicating that GAL4 promoter-driven expression of the At-RSH1 cDNAs is required. In cells with either the TIR-NB-ARC bait or the NB-ARC bait, the four RSH1 library plasmids differentially activated the LEU2 and lacZ reporters in a similar manner (Fig. 1B). Furthermore, the TIR bait and the ARC bait did not interact with any RSH1 clone (Fig. 1A), indicating that the NB-ARC domain of RPP5 is required and sufficient for interaction with At-RSH1.
We then examined whether At-RSH1 interacts with other NB-LRR proteins. We made an identical bait construct with the expressed RPP5 homolog from the Columbia ecotype (Col-0), RPP5-ColF (87% identity, 92% similarity), of unknown function (16). In addition, we obtained four other comparable NB-ARC domain-containing baits derived from the Arabidopsis RPP1A (20), RPM1 (21), and RPS4 genes (22) and the tobacco N gene (23). Based on the pJK101 repression assay (17), all fusion proteins expressed from these NB-LRR baits were transferred to the nucleus. However, none of these baits activated the reporter genes upon coexpression with any of the RSH1 clones, indicating that At-RSH1 specifically interacts with RPP5 bait constructs.
At-RSH1 Gene Structure and Predicted Functional Domains.
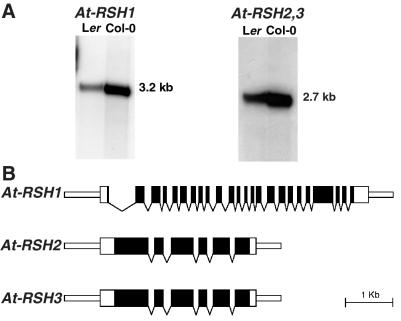
DNA blot analysis (2× SSC, 50°C) showed that At-RSH1 is a single-copy gene in several Arabidopsis ecotypes. The Col-0 At-RSH1 genome sequence (GenBank accession no. AF075597) showed that At-RSH1 resides at 14 cM from the top of chromosome 4. Hybridization of At-RSH1 to poly(A)-enriched leaf RNA (0.2× SSC, 65°C) revealed a single transcript of 3.2 kb (Fig. 2A). No induction of the At-RSH1 transcript was observed in 10 mg of total RNA isolated from leaves harvested at several time points after infection with P. syringae DC3000, P. syringae DC3000 carrying AvrRPS4 (22), forceps wounding, or treatment with 0.5 mM salicylic acid or 1 mM methyl jasmonate. A full length At-RSH1 cDNA (3,137 bp) was obtained by 5′ rapid amplification of cDNA ends and encodes a 2,649-bp ORF. Alignment of the At-RSH1 gene (5,729 bp) with the corresponding cDNA revealed 23 introns (Fig. 2B). The 3′ end of At-RSH1 corresponds to two expressed sequence tags (ESTs) (GenBank accession nos. Z34756 and Z34769).
Figure 2.
At-RSH1, 2, and 3 transcript analysis and gene structures. (A) Poly(A) RNA gel blot with leaf RNA isolated from the Arabidopsis Landsberg erecta (Ler) and Columbia (Col-0) ecotypes, hybridized to radiolabeled cDNA sequences corresponding to At-RSH1 and At-RSH2; transcript sizes indicated in kilobases. (B) Intron/exon structures of At-RSH1, 2, and 3. Wide rectangles indicate transcribed regions, and solid rectangles constitute coding sequence. Narrow rectangles indicate 5′ and 3′ sequences, and intron positions are marked by arrowheads.
At-RSH1 codes for an 883-aa residue protein of 98.6 kDa. Secondary structure and hydropathy analysis (Fig. 1B) suggested two C-terminal transmembrane segments at residues 811–827 and 848–864. Topology predictions indicated that At-RSH1 is cytoplasmically localized but anchored at the plasma membrane. The C-terminal portion of At-RSH1 is sufficient for interaction with RPP5 (Fig. 1B); it contains a hydrophilic solvent-exposed region of ≈160 aa (residues 634–793) that may function as the RPP5-interacting domain (Fig. 1B). Database searches using tblastn (30) failed to reveal any sequences with significant homology to this RPP5-interaction domain. The central portion of At-RSH1 (residues 160–625) shows a high level of similarity to the central regions (≈450 residues) of bacterial RelA and SpoT proteins (≈30% identity, ≈58% similarity; Figs. 1 B and C and 3). Database searches using At-RSH1 identified uncharacterized amino acid sequences derived from rice (GenBank accession no. D48993), human (THC205397), mouse (AA475394 and AA473095), nematode (Z82096), and two additional Arabidopsis sequences (see next section) that appear to be homologous to (parts of) the RelA/SpoT (p)ppGpp synthetase domain.
Figure 3.
Sequence relationships between the (p)ppGpp synthetase domains of At-RSH1 and At-RSH2, and those of E. coli (Ec) RelA and SpoT and S. coelicolor (Sc) RelA. At-RSH1 and S. coelicolor RelA contain ATP/GTP-binding site motifs (P-loop; boxed) that are absent in other RelA/SpoT proteins.
Two Other Arabidopsis RelA/SpoT Homologs: At-RSH2 and At-RSH3.
Two unlinked and expressed Arabidopsis genes with significant homology to At-RSH1 were identified by using tblastn (30). We designated these genes At-RSH2 on chromosome 3 (GenBank accession nos. AB019229; ESTs N38487, W43725, H76717, AA713029) and At-RSH3 on chromosome 1 (AC006577; EST W43807). At-RSH2 and At-RSH3 are highly similar (75% overall nucleotide identity) but share little DNA homology with At-RSH1; even the regions encoding the putative (p)ppGpp synthetase domains of At-RSH1 and AtRSH2 and 3 share only ≈38% nucleotide identity, whereas these domains of At-RSH2 and At-RSH3 share 84% nucleotide identity. DNA gel blot analysis (2× SSC, 50°C) using At-RSH2 EST N38487 as probe revealed both At-RSH2 and At-RSH3 fragments. Hybridization to poly(A)-enriched leaf RNA (0.2× SSC, 65°C) showed a single transcript of ≈2.7 kb (Fig. 2A). Full-length At-RSH2 and partial At-RSH3 cDNAs were obtained by 5′ rapid amplification of cDNA ends. The At-RSH2 cDNA is 2,605 bp in length with an ORF of 2,130 bp. The At-RSH2 and At-RSH3 genes have five introns at identical positions (Fig. 2B). The low DNA homology between At-RSH1 and At-RSH2 and 3 and the different positions of the introns in the (p)ppGpp synthetase domains indicate an ancient divergence or an independent origin.
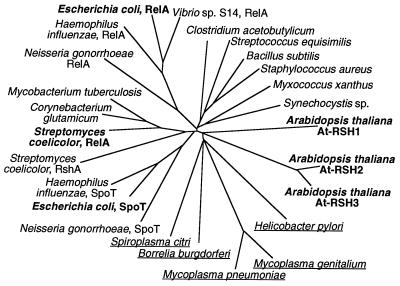
At-RSH2 and At-RSH3 encode 710- and 715-aa residues, respectively, with a molecular size of ≈80 kDa. Their central (p)ppGpp synthetase domains (318 residues) share 90% identity (94% similarity) and are 147 residues (32%) shorter than the same domain of At-RSH1 (465 residues; Figs. 1C and 3). The (p)ppGpp synthetase domains of At-RSH2 and At-RSH3 are markedly more similar to bacterial RelA/SpoT (≈46% identity, ≈66% similarity) than is At-RSH1 (≈30% identity, ≈58% similarity) and share 38% identity and 59% similarity with the corresponding region of At-RSH1 (Fig. 3). At-RSH2 and At-RSH3 do not contain predicted transmembrane-spanning regions, cleavable signal peptides, or hydrophilic C-terminal regions and are predicted to be located in the cytoplasm. The N- and C-terminal portions of At-RSH2 and At-RSH3, excluding the (p)ppGpp synthetase domains, display little or no homology to the corresponding portions of At-RSH1 or to other proteins. Phylogenetic analysis of the (p)ppGpp synthetase domains of At-RSH1, 2, and 3 together with a wide range of bacterial RelA/SpoT proteins grouped the Arabidopsis sequences with homologs from a number of intracellular pathogens (Fig. 4), perhaps indicative of lateral gene transfer early in evolution.
Figure 4.
Phylogenetic analysis of the (p)ppGpp synthetase domains of members of the RelA/SpoT family. The At-RSH1, 2, and 3 homologs and the E. coli and S. coelicolor RelA/SpoT proteins are shown in boldface type. At-RSH1, 2, and 3 cluster with RelA/SpoT proteins from intracellular pathogens (underlined).
At-RSH1 Restores Growth of an E. coli relA Mutant but Not of a relA, spoT Double Mutant.
RelA and SpoT play central roles in the bacterial stringent response, allowing prompt physiological responses to rapidly changing environmental conditions (18). The primary functions of RelA and SpoT are to synthesize and to degrade (p)ppGpp, respectively, and SpoT also is capable of (p)ppGpp synthesis under conditions of energy limitation. (p)ppGpp functions to regulate the transcription of a large number of genes, both positively and negatively. E. coli relA mutants are unable to grow on minimal agar medium supplemented with the amino acids Ser, Met, and Gly (SMG medium), a phenotype that is complemented through engineered (p)ppGpp synthesis (27). Moreover, because a relA, spoT mutant lacks (p)ppGpp phosphohydrolase activity, induced synthesis of (p)ppGpp in the double mutant abolishes growth, presumably through cessation of rRNA and tRNA synthesis, a primary characteristic of the stringent response (18).
To examine whether Arabidopsis At-RSH1 might function as a (p)ppGpp synthetase, the 1.6-kb 5′ region of At-RSH1 containing the putative (p)ppGpp synthetase domain was cloned in the temperature-inducible pT7–7 vector (26) and introduced into E. coli CF1648 (relA+, spoT+), CF1652 (relA−, spoT+), and CF1693 (relA−, spoT−). None of the At-RSH1-containing strains grew on SMG agar under inducing conditions (37°C and 42°C), indicating that high-level expression of At-RSH1 was toxic, potentially reflecting levels of (p)ppGpp synthesis sufficient to prevent growth of even a spoT+ strain on supplemented minimal medium agar. However, growth of the relA mutant was restored on SMG agar under noninducing conditions (30°C; Fig. 5A), consistent with a low level of expression of At-RSH1 from the T7 promoter that would give rise to sufficient (p)ppGpp to suppress the SMG phenotype, but not to prevent growth (growth of E. coli on SMG medium requires only low steady-state levels of (p)ppGpp). Growth of the double mutant was not restored on SMG agar at 30°C (not shown), consistent with the inability of the relA, spoT double mutant to degrade At-RSH1-derived (p)ppGpp.
Figure 5.
Functional analysis of At-RSH1 in bacteria. (A) At-RSH1 complements an E. coli relA− mutant. The 1.6-kb 5′ region of At-RSH1 restores growth of the E. coli relA mutant (CF1652) on SMG medium at 30°C. (B) At-RSH1 expression is lethal in an E. coli relA−, spoT− double mutant. The 1.6-kb 5′ region of At-RSH1 was cloned in the temperature-inducible pT7–7 vector (26). Expression was induced by rapidly transferring rich L-broth cultures from 30°C to 42°C (denoted by arrows). Induced expression of At-RSH1 (1.6 kb) in the E. coli relA mutant (CF1652) slightly reduces growth rate (Upper), whereas in the E. coli relA, spoT double mutant (CF1693) growth is abolished (Lower). (C) At-RSH1 expression in S. coelicolor M600. At-RSH1 was expressed from the thiostrepton-inducible tipA promoter of pIJ8600 (28). Spores of each strain were dropped on SMMS medium (28) and allowed to dry. Twelve microliters of 1 mg/ml thiostrepton in 2% DMSO was added to induce tipA expression (+), and 12 μl of 2% DMSO was added to the control cultures (−). The plates were incubated at 30°C for 4 days. Induced expression of the S. coelicolor relA gene and the full-length and 1.6-kb 5′ region of At-RSH1 results in precocious antibiotic production (the blue pigment, actinorhodin) and precocious aerial hyphae formation, giving a white appearance to the mycelium.
There was no difference in growth rate between the At-RSH1-containing derivatives and their vector controls when the strains were grown in rich L-broth under noninducing conditions (30°C); any basal level of expression of At-RSH1 under these conditions presumably produces insufficient (p)ppGpp to impair growth in this nutrient-rich liquid medium. In contrast, induction of the truncated At-RSH1 at 42°C in rapidly dividing L-broth cultures reduced the growth rate of CF1652 (relA−, spoT+) and essentially abolished growth of CF1693 (relA−, spoT−) (Fig. 5B), consistent with high levels of (p)ppGpp synthesis. Thus, expression at two different levels of the 5′ portion of At-RSH1 containing the (p)ppGpp synthetase domain in E. coli confers two distinct phenotypes associated with (p)ppGpp synthesis: restoration of growth of a relA mutant on minimal SMG agar when expressed at a low level, and abolition of growth of a relA, spoT double mutant in rich L-broth when expressed at a high level.
At-RSH1 Confers Phenotypes Associated with (p)ppGpp Synthesis in S. coelicolor.
To analyze further the potential (p)ppGpp synthetase activity of At-RSH1, it was expressed in S. coelicolor. S. coelicolor relA null mutants do not produce (p)ppGpp (A. Hesketh and M.J.B., unpublished data); consequently, they are deficient in antibiotic production and show delayed morphological differentiation, i.e., delayed formation of aerial hyphae and spores (19, 29). Induced expression of the endogenous relA gene in the relA+ S. coelicolor M600 strain provokes the precocious production of the blue-pigmented antibiotic actinorhodin and of aerial hyphae (Fig. 5C).
Expression of At-RSH1 or a 5′ portion containing the (p)ppGpp synthetase domain in M600 using the thiostrepton-inducible pIJ8600 vector (28) yielded phenotypes that were indistinguishable from those observed upon induction of the endogenous relA gene (Fig. 5C). Thus, expression of At-RSH1 in S. coelicolor again induces two distinct phenotypes associated with (p)ppGpp synthesis: precocious antibiotic production and the early onset of morphological differentiation.
Discussion
We identified an Arabidopsis RelA/SpoT homolog, At-RSH1, that specifically interacts with the NB-ARC domain of RPP5 in yeast. We showed that At-RSH1 confers disparate phenotypes associated with (p)ppGpp synthetase activity in both S. coelicolor and E. coli. In addition to At-RSH1, we characterized two other Arabidopsis RelA/SpoT homologs, At-RSH2 and At-RSH3. The At-RSH genes are the first eukaryotic homologs of bacterial relA or spoT genes described to date. RelA and SpoT play a central role in the bacterial stringent response (18). Both enzymes are made constitutively and are allosterically activated under sudden nutritional and environmental stress conditions such as amino acid, carbon, nitrogen, or phosphate starvation, as well as upon abrupt increases in temperature and osmolarity. RelA and, under certain conditions, SpoT transfer pyrophosphate groups from ATP to the 3′ positions of GDP and GTP, resulting in the rapid accumulation of ppGpp and pppGpp. In bacteria, (p)ppGpp induces and represses transcription of genes involved in a wide variety of processes (18).
The striking amino acid similarity and the ability of At-RSH1 to complement such disparate phenotypes in two evolutionary distinct bacteria strongly suggest that At-RSH1 is capable of (p)ppGpp synthesis. Although several nucleotide derivatives such as cAMP, cGMP, and cADP-ribose have been implicated in plants as intracellular secondary signaling molecules (31, 32), ppGpp and pppGpp have not been described unambiguously in plants or other eukaryotes. By analogy to its role in bacteria, it is conceivable that (p)ppGpp functions in plants as a rapidly activated transcription cofactor. Rapid cellular stress responses are required in plants subject to pathogen infection and after wounding, sudden drought or flooding conditions, or osmotic shock. Stress-induced (p)ppGpp-mediated transcriptional repression of genes involved in normal cellular metabolic processes could rapidly prevent loss of compounds and energy and would nutritionally deprive invading pathogens. In addition, (p)ppGpp-mediated transcriptional activation may rapidly induce stress- and defense-related genes and compounds.
The yeast two-hybrid interaction between At-RSH1 and RPP5 is intriguing, but a function for At-RSH1 has not yet been established in planta. We have postulated that NB-LRR proteins may “guard” host proteins for interference from pathogen Avr products (33). In line with this model is the idea that RPP5 has evolved to specifically recognize the physical association of P. parasitica (a)virulence factors with At-RSH1 and subsequently activate defense mechanisms. The C-terminal LRR domain of RPP5 could, like the WD40 repeats in Apaf-1 (34), ensure that activation of the protein complex is signal-dependent. This model implies that plants do not adapt to pathogens producing a virtually unlimited number of Avr effectors, but rather produce a restricted number of R proteins that guard a finite number of host targets.
Acknowledgments
We thank Bart Feys and Jane Parker for the Arabidopsis RNA preparations and Andy Hesketh for help with the HPLC assays. E.v.d.B. was supported in part by a fellowship from the European Community. This project was funded in part by grants from Zeneca and from the Biotechnology and Biological Sciences Research Council. The Sainsbury Laboratory is supported by the Gatsby Charitable Foundation.
Abbreviations
- Avr
avirulence
- NB
nucleotide-binding
- ARC
Apaf-1, R proteins, CED-4
- LRR
leucine-rich repeat
- R
resistance
- ppGpp
guanosine tetraphosphate
- pppGppp
guanosine pentaphosphate
- SMG medium
minimal agar medium supplemented with Ser, Met, and Gly
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AF225702, AF225703, and AF225704 (At-RSH1, At-RSH2, and At-RSH3, respectively)].
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.060392397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.060392397
References
- 1.Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar S P. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 2.Galán J E, Collmer A. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 3.Hammond-Kosack K E, Jones J D G. Annu Rev Plant Physiol Mol Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- 4.Kearney B, Staskawicz B J. Nature (London) 1990;346:385–386. doi: 10.1038/346385a0. [DOI] [PubMed] [Google Scholar]
- 5.Ritter C, Dangl J L. Mol Plant Microbe–Interact. 1995;8:444–453. doi: 10.1094/mpmi-8-0444. [DOI] [PubMed] [Google Scholar]
- 6.Bogdanove A J, Kim J F, Wei Z, Kolchinsky P, Charkowski A O, Conlin A K, Collmer A, Beer S V. Proc Natl Acad Sci USA. 1998;95:1325–1330. doi: 10.1073/pnas.95.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laugé R, Joosten M H, Haanstra J P, Goodwin P H, Lindhout P, De Wit P J G M. Proc Natl Acad Sci USA. 1998;95:15849–15854. doi: 10.1073/pnas.95.15.9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H-W, Lucy A P, Guo H-S, Li W X, Ji L-H, Wong S-M, Ding S W. EMBO J. 1999;18:2683–2691. doi: 10.1093/emboj/18.10.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammond-Kosack K E, Jones J D G. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parker J E, Coleman M J, Szabò V, Frost L N, Schmidt R, Van der Biezen E A, Moores T, Dean C, Daniels M J, Jones J D G. Plant Cell. 1997;9:879–894. doi: 10.1105/tpc.9.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van der Biezen E A, Jones J D G. Curr Biol. 1998;8:R226–R227. doi: 10.1016/s0960-9822(98)70145-9. [DOI] [PubMed] [Google Scholar]
- 12.Aravind L, Dixit V M, Koonin E V. Trends Biochem Sci. 1999;24:47–53. doi: 10.1016/s0968-0004(98)01341-3. [DOI] [PubMed] [Google Scholar]
- 13.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Alnemri E S. Mol Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 14.Yang X, Chang H Y, Baltimore D. Science. 1998;281:1355–1357. doi: 10.1126/science.281.5381.1355. [DOI] [PubMed] [Google Scholar]
- 15.Zou H, Li Y, Wang X. J Biol Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
- 16.Noël L, Moores T L, Van der Biezen E A, Parniske M, Daniels M J, Parker J E, Jones J D G. Plant Cell. 1999;11:2099–2111. [PMC free article] [PubMed] [Google Scholar]
- 17.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1996. [Google Scholar]
- 18.Cashel M, Gentry D R, Hernandez V J, Vinella D. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 1458–1496. [Google Scholar]
- 19.Chakraburtty R, Bibb M. J Bacteriol. 1997;179:5854–5861. doi: 10.1128/jb.179.18.5854-5861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Botella M A, Parker J E, Frost L N, Bittner-Eddy P D, Beynon J L, Daniels M J, Holub E B, Jones J D G. Plant Cell. 1998;10:1847–1860. doi: 10.1105/tpc.10.11.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant M R, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes R W, Dangl J L. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 22.Gassmann W, Hinsch M E, Staskawicz B J. Plant J. 1999;20:265–277. doi: 10.1046/j.1365-313x.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]
- 23.Whitham S, Dinesh-Kumar S P, Choi D, Hehl R, Corr C, Baker B. Cell. 1994;78:1101–1115. doi: 10.1016/0092-8674(94)90283-6. [DOI] [PubMed] [Google Scholar]
- 24.Thompson J D, Higgens D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Tabor S, Richardson C C. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- 28.Sun J, Kelemen G H, Fernández-Abalos J M, Bibb M J. Microbiology. 1999;145:2221–2227. doi: 10.1099/00221287-145-9-2221. [DOI] [PubMed] [Google Scholar]
- 29.Chakraburtty R, White J, Takano E, Bibb M. Mol Microbiol. 1996;19:357–368. doi: 10.1046/j.1365-2958.1996.390919.x. [DOI] [PubMed] [Google Scholar]
- 30.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 31.Walden R. Curr Opin Plant Biol. 1998;1:419–423. doi: 10.1016/s1369-5266(98)80266-5. [DOI] [PubMed] [Google Scholar]
- 32.Durner J, Wendehene D, Klessig D J. Proc Natl Acad Sci USA. 1998;95:10328–10333. doi: 10.1073/pnas.95.17.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van der Biezen E A, Jones J D G. Trends Biochem Sci. 1998;23:454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- 34.Hu Y, Benedict M A, Ding L, Núñez G. EMBO J. 1999;18:3586–3595. doi: 10.1093/emboj/18.13.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]