Abstract
A modified susceptible–infected–recovered (SIR) host–pathogen model is used to determine the influence of plant mating system on the outcome of a host–pathogen interaction. Unlike previous models describing how interactions between mating system and pathogen infection affect individual fitness, this model considers the potential consequences of varying mating systems on the prevalence of resistance alleles and disease within the population. If a single allele for disease resistance is sufficient to confer complete resistance in an individual and if both homozygote and heterozygote resistant individuals have the same mean birth and death rates, then, for any parameter set, the selfing rate does not affect the proportions of resistant, susceptible or infected individuals at equilibrium. If homozygote and heterozygote individual birth rates differ, however, the mating system can make a difference in these proportions. In that case, depending on other parameters, increased selfing can either increase or decrease the rate of infection in the population. Results from this model also predict higher frequencies of resistance alleles in predominantly selfing compared to predominantly outcrossing populations for most model conditions. In populations that have higher selfing rates, the resistance alleles are concentrated in homozygotes, whereas in more outcrossing populations, there are more resistant heterozygotes.
Keywords: inbreeding, mathematical model, disease resistance, pathogen
1. Introduction
Broad patterns in nature indicate a relationship between plant mating system and plant–pathogen interactions. There is a positive correlation between the number of fungal pathogen species known to infect a plant host and the outcrossing rate of the host (Busch et al. 2004). In addition, species with higher outcrossing rates tend to occur in less disturbed, biologically complex habitats, where disease and other ‘natural enemies’ are likely to be more prevalent (see Levin (1975), although alternative explanations for this pattern exist). These ecological correlations provide evidence that pathogen pressure may influence mating system evolution and vice versa. Given that the mating system of the host controls the genetic diversity of its progeny (Holsinger 2000) and the degree of similarity between parent and progeny, the mating system of the host could affect the evolution of resistance to infection and the prevalence of disease in populations. For example, biological control of weedy plants is more effective for asexually reproducing species than for sexually reproducing species (reviewed in Burdon & Marshall 1981), indicating that recombination in host species may offer protection from natural enemies and limit the spread of enemies within populations.
Most relevant existing theory investigates the potential for disease to select for increased outcrossing by contrasting sexual and asexual reproduction in the context of the Red Queen hypothesis (e.g. Jaenike 1977; Hamilton 1980; Hamilton et al. 1990; Howard & Lively 1998). Two previous studies have contrasted self-fertilization (‘selfing’) and outcrossing, both of which are forms of sexual reproduction. One model found that parasites can select for a mixed mating system in a haploid host with diallelic matching alleles governing infection (Lively & Howard 1994). Another study used simulations to investigate the possibility that pathogens can select against complete selfing in a host (Agrawal & Lively 2001). The simulations showed that parasites select for outcrossing over a wide range of parameters, but that the results were sensitive to genetic assumptions mediating infection. Both of these models consider individual fitness, but do not scale up to address the potential consequences of varying mating system on the spread of resistance alleles and the transmission of pathogens within populations.
The question of the effect of inbreeding on the prevalence of disease in a population is of broad interest. Humans are causing loss and fragmentation of many species' habitats. This is likely to increase the amount of inbreeding in plants. This paper addresses the possible effects of inbreeding on disease resistance through a modified SIR host–pathogen model (Anderson & May 1981) to determine the influence of mating system on the outcome of the host–pathogen interaction. This model makes the link between host mating system an influential determinant of genotype frequencies and prevalence of infection at a population level, rather than focusing on individual fitness effects. The following specific questions are addressed.
(a) Question 1: will the degree of host selfing affect the incidence of infection in the host population?
We examined this in the model by looking at the effect of the inbreeding coefficient (F) on the number of resistant, susceptible and infected individuals at equilibrium. We assumed that host resistance is determined by a single dominant allele (R). Therefore, in this model, resistant individuals may be either homozygous resistant (RR) or heterozygous resistant (Rr). The genetic effect of selfing alters the distribution of alleles in the next generation by increasing the proportion of homozygotes and decreasing the proportion of heterozygotes without altering the frequency of alleles (Hartl & Clark 1997). Thus, with increasing selfing, there should be fewer resistant heterozygotes produced and, as a result, a greater proportion of individuals susceptible to infection in the population. We therefore expected higher levels of endemic disease in selfing than in outcrossing populations.
(b) Question 2: will the degree of host selfing affect the frequency of a dominant resistance allele (R) in the population?
Although selfing alone will not alter the frequencies of any allele, increased selfing will lead to an ever greater production of homozygous individuals, which may leave a greater proportion of the population susceptible to infection. Will more susceptibility to infection lead to a higher gene frequency of the resistance allele in later generations? We examined this in the model by looking at the effect of the inbreeding coefficient (F) on the frequency of a dominant resistance allele (R) at equilibrium.
2. The model
Using differential equations, we created a continuous time model of host population dynamics with both disease and host genetics. These epidemiological equations were based on Anderson & May's formulation (1981), but we added a genetic component of host resistance to infection and a host mating system that can be varied. Many host-specific pathogens are able to infect only particular host genotypes (Burdon 1987; Thompson & Burdon 1992). In addition, polymorphism for resistance is common within natural populations (Simms & Triplett 1994) and may be maintained by costs of resistance genes (Bergelson & Purrington 1996) or even by gene flow in structured metapopulations (Thrall & Burdon 1997). We assumed a diploid host with a single locus for resistance to infection. A dominant resistance allele (R) confers complete resistance. Although many factors can contribute to overall pathogen resistance, single dominant alleles of major phenotypic effect have been found to confer resistance in both natural and agricultural systems (Flor 1955; Thompson & Burdon 1992), and thus represent a tractable way to model resistance. For simplicity, the pathogen was assumed to be genetically uniform. Explicit numbers of individual pathogens were not quantified because the pathogen was a microparasite that could reproduce within the host. The pathogen caused similar levels of disease in the host whether the infection was caused by a single individual or many individuals of the pathogen, due to the rapid reproductive rate of the pathogen inside of a susceptible host (Anderson & May 1981). Infection affected host fitness by increasing the death rate of infected hosts. The model is suitable for pathogens that spread through horizontal, density-dependent pathogen transmission, where the host is a perennial. It was assumed that the pathogen had no alternative hosts, and that the offspring were all healthy, regardless of the infection status of the parent(s). We assumed a cost of resistance in the form of lower fecundity for any individual with the resistance allele.
The model considered three genotypes, one of which could be either in the infected state or in the uninfected (susceptible) state. Table 1 is a complete listing of the variables and parameters in the following equations. Following Anderson & May (1981), the set of differential equations for the system is:
| 2.1a |
| 2.1b |
| 2.1c |
| 2.1d |
where XRR, XRr and Xrr are the numbers of healthy individuals carrying two R alleles, one R and one r allele and two r alleles, respectively, while Yrr is the number of infected individuals with two r alleles. We assumed that individuals with an R allele are completely resistant, or immune, to infection, so there is no need for equations for YRR and YRr. The model contains several additional assumptions. Once an individual host was infected it either remained infected or died because, unlike the assumption in many traditional models and vertebrate systems, there was no recovery from the disease, following the observation that plants often do not recover from pathogen infection (for analysis of a more general model in which recovery can occur, see electronic supplementary material, part A). The pathogen was transmitted directly and equally from any infected individual. An Xrr individual's chance of getting the pathogen depended on the number of infected individuals in the population, with an infection rate coefficient β (for analysis of more general assumptions on pathogen transmission, see electronic supplementary material, part B). For healthy individuals (Xrr), the death rate was b, whereas for infected individuals (Yrr), the death rate was (b+α). New individuals were figured as the number of gametes produced for resistant and susceptible genotypes, such that the birth rates for the two gametes were
| 2.2a |
| 2.2b |
Here, aRR, aRr and arr are the birth rates of the three genotypes. The infected individuals generally had no reproductive cost to infection. In the analyses that follow, we considered three cases: (i) aRR=aRr<arr; (ii) aRR<aRr<arr; and (iii) assign a reproductive cost to infection by setting the fecundity of infected individuals to some value (ainf) equal to or less than the fecundity of resistant individuals. The term ρN represents density-dependent self-limitation on reproduction, where
| 2.3 |
Table 1.
A list of the terms in the model equations presented in the order of their appearance in the text.
| F | inbreeding coefficient |
| X RR | number of homozygous resistant individuals |
| X Rr | number of heterozygous resistant individuals |
| X rr | number of susceptible individuals |
| Y rr | number of infected individuals |
| new | rate of creation of new individuals of a given genotype |
| b | death rate for healthy individuals |
| β | pathogen transmission parameter |
| α | additional death rate for infected individuals |
| gamXR | number of resistant gametes |
| gamXr | number of susceptible gametes |
| a RR | fecundity of homozygous resistant individuals |
| a Rr | fecundity of heterozygous resistant individuals |
| a rr | fecundity of susceptible and infected individuals |
| a inf | fecundity of infected individuals |
| ρ | density-dependence factor |
| N | total number of individuals |
| f | frequency of accompanying variable |
| seed | new individual of a given genotype |
In animal-pollinated plants, mating system can be considered a continuous variable from complete selfing to complete outcrossing (Vogler & Kalisz 2001). The mating system of a population can be estimated using the inbreeding coefficient (F), which ranges from 0 to 1 (Hartl & Clark 1997). In comparison to a population composed of randomly mating (i.e. outcrossing) individuals, complete selfing halves the frequency of heterozygotes each generation (Wright 1921). Selfing decreases the frequency of heterozygotes by F, which is the probability that two alleles in the same individual are identical by descent (Hartl & Clark 1997). Therefore, offspring genotype frequencies are determined by the following equations:
| 2.4a |
| 2.4b |
| 2.4c |
The frequencies, fgamXR and fgamXr, of each gamete in the population are simply and , respectively. The total numbers of the three offspring genotypes (newXRR, newXRr and newXrr) used in equations (2.1a)–(2.1c) are determined respectively, by multiplying each of the above functions, fseedXRR, fseedXRr and fseedXrr by the total number of offspring . This completes the development of the model.
3. Results
In order to address the two questions noted above, the model was studied at steady-state equilibrium. Where relevant, we examined the behaviour of the model for a range of model parameters to determine the robustness of the results.
(a) Steady-state equilibrium, case 1: no pathogen present and aRR≤aRr<arr
In the absence of a pathogen, because aRR≤aRr<arr, the R allele disappears from the system at equilibrium. Therefore, XRR=XRr=Yrr=0 and it is easy to show that
| 3.1 |
The loss of the R allele in the absence of a pathogen is predicted in other models with similar assumptions about the cost of resistance (e.g. Thrall & Antonovics 1995). It is important to note that in a spatially explicit metapopulation context, unlike the single population model presented here, gene flow between populations may increase the time for gene fixation or prevent fixation altogether.
(b) Steady-state equilibrium, case 2: pathogen present and aRR=aRr<arr
For this special case, where resistance is dominant and the cost of resistance is associated with the resistant phenotype, it is possible to solve analytically for the steady-state equilibrium by setting the right-hand sides of equations (2.1a)–(2.1d) to zero and using equations (2.2a,b), (2.3) and (2.4a)–(2.4c). Resistant individuals are maintained in the population by the presence of the pathogen. The solutions for , and are:
| 3.2a |
| 3.2b |
| 3.2c |
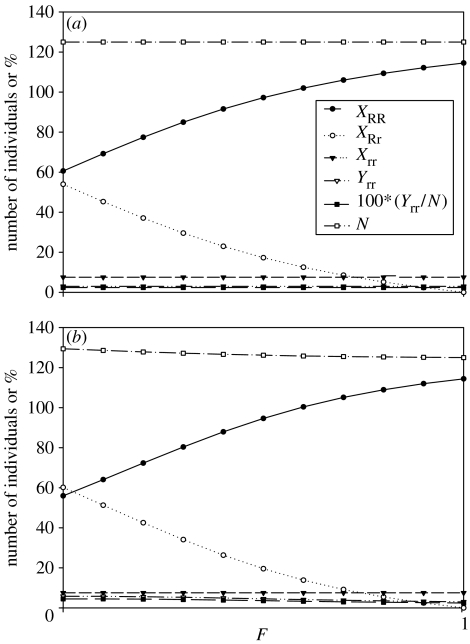
(Analytic formulae can also be found individually for the last two individual variables, and , as quadratic functions of F; see electronic supplementary material, part C.) Note that the total population size, including infected individuals, is in the case where disease is present, as compared with when the disease is absent. An important conclusion is that the level of inbreeding does not affect the above equilibrium values, including , the fraction of the infected individuals in the population (figure 1a), because F, the inbreeding coefficient, does not appear in the solutions. However, although F does not affect the sum, , it does affect the individual values of and (also see electronic supplementary material, part C). When the population is completely outcrossing (F=0) there are more heterozygous individuals, while there are none for complete selfing (F=1). Because the numbers of resistant, susceptible and infected individuals are the same for completely selfing and completely outcrossing populations, the frequency of the R allele is higher in selfing populations than in outcrossing populations due to the absence of heterozygous resistant individuals in completely selfing populations. (It is possible to extend these results to more general infection rates than ; see electronic supplementary material, part B.)
Figure 1.
The numbers of homozygous resistant, heterozygous resistant, susceptible and infected individuals, percentage infected in the population and total number of individuals at equilibrium across the range of complete selfing to complete outcrossing. Note that the Yrr symbols are obscured by the 100*(Yrr/N) symbols. Parameters: b=0.2, α=0.1, β=0.04, ρ=0.02; for (a) aRR=aRr=0.7, arr=0.8 (case 2, aRR=aRr<arr) and (b) aRR=0.7, aRr=0.75, arr=0.8 (case 3, aRR<aRr<arr).
The analytic solutions allow us to examine the effects of model parameters on the number of susceptible individuals in the population, , as well as the number of infected individuals, . Increased values of β and decreased values of the ratio arr/aRR (recall aRR<arr) will decrease equilibrium numbers of infected individuals. Increased death rates of infected individuals, α, lead to larger values of , because a larger value of α decreases the steady-state number of the infected individuals in the population, as close examination of equation (3.2b) reveals. A similar effect occurs in a predator–prey population with a prey-dependent functional response; a higher death rate of a predator increases the equilibrium population size of its prey.
(c) Steady-state equilibrium, case 3: pathogen present and aRR<aRr<arr
In case 2 above, in which aRr=aRR, heterozygous individuals bear the same cost of resistance as the resistant homozygote. A possible alternative is for the heterozygote reproductive rate to be intermediate between the two homozygote reproductive rates. For this case, the model could not be solved analytically at equilibrium. However, solutions were possible through numerical evaluation. We examined the effects of F, in combination with each parameter, on the prevalence of disease in the population and summarize the main relationships below.
Consider again the example described in case 2, where reproductive coefficients aRR=aRr=0.7 and arr=0.8. But now let aRr=0.75, meaning that the effect of resistance on reproduction is proportional to the number of resistant alleles in an individual. The first thing to note, in this case, is that the equilibria of all variables except for vary with F (figure 1b). The variable , the number of susceptibles, is held at a fixed level by top-down control by the disease parameters alone and is thus independent of both F and aRr. Any change in the rate of production of Xrr, that is, newXrr, changes the steady-state value of . Second, note that for F=1 the equilibrium values , , and N* are the same as the values in case 2 (aRR=aRr). The reason is that when F=1, so all of the variables are completely independent of the value of aRr, and thus they are the same as if aRr=aRR. Third, note that as F is decreased from 1, N* increases (only slightly in this particular case, but, with other sets of parameters, decreases in F can result in large increases in N*). The increase in N* is due to the higher reproductive rate of the XRr relative to the XRR in conjunction with the greater prevalence of the XRr as outcrossing increases. Fourth, as F is decreased from 1, the sum no longer remains constant, as it did in case 2; the increase in slightly exceeds the decrease in . Fifth, and increase with decreasing F and are almost double in size when F reaches 0.
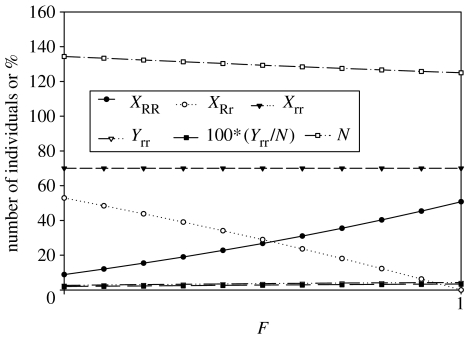
The behaviours of and as functions of F displayed in figure 1b are typical of situations in which the infection rate, β, is relatively large. Under the condition of smaller β, the opposite dependence of and as functions of F occurs, in which case these variables decrease as F decreases (figure 2). In this example, we also increased α, the increment of mortality due to infection, from 0.1 to 0.5, which contributes to the reversed slopes of and as functions of F relative to parameter sets with higher β and lower α. Now decreases by more than 40% as F decreases from 1 to 0.
Figure 2.
The numbers of homozygous resistant, heterozygous resistant, susceptible and infected individuals, percentage infected in the population and total number of individuals at equilibrium across the range of complete selfing to complete outcrossing. Note that the Yrr symbols are obscured by the 100*(Yrr/N) symbols. Parameters: b=0.2, α=0.5, β=0.01, ρ=0.02, aRR=0.7, aRr=0.75, arr=0.8 (case 3, aRR<aRr<arr).
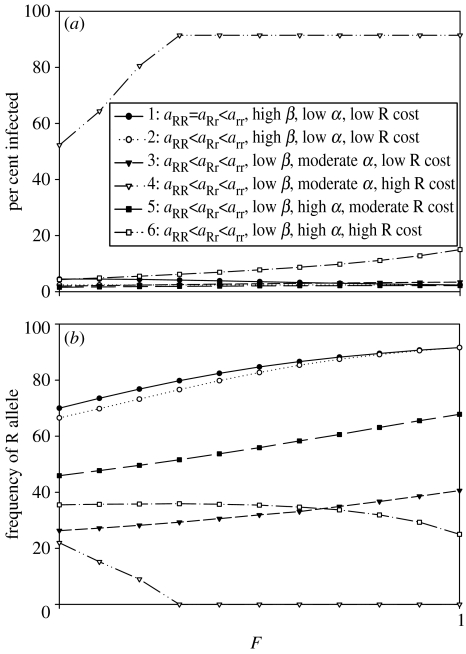
Thus, depending on parameter values, and may increase or decrease as functions of the degree of selfing. For comparison, from the above two cases is plotted together with curves for three additional parameter sets (figure 3a). In one case (curve 5 in figure 3a), has a very slight peak at an intermediate value of F. In a case in which the reproductive rate associated with the resistance allele is half that of the non-resistant allele and the disease mortality increment (α) is small (e.g. curve 4 in figure 3), Yrr is an extremely large fraction of the population when selfing is relatively high, as the R-allele is eliminated from the population. The plots of all of the variables related to curves 4–6 in figure 3a,b are appended in electronic versions (see figures E1–E3 in the electronic supplementary material).
Figure 3.
(a) Summary of percentages infected in the population at equilibrium as a function of F. (b) Summary of the frequency of R alleles in the population at equilibrium as a function of F. Parameters: ‘curve 1’ as in figure 1a; ‘curve 2’ as in figure 1b; ‘curve 3’ as in figure 2; ‘curve 4, high cost of R’ as in curve 2, but with aRR=0.4 and aRr=0.6 (see figure E1 in electronic supplementary material); ‘curve 5, high α, moderately high cost of R’ as in curve 2, but with α=1.0, aRR =0.6 and aRr=0.7 (see figure E2 in electronic supplementary material); ‘curve 6, high α, high cost of R’ as in curve 4, but with α=1.0 (see figure E3 in electronic supplementary material).
The frequency of the R-allele in the population at equilibrium for aRR<aRr<arr also follows a variety of patterns (figure 3b). We examined the relationship under the same sets of model parameters as above. In general, increased inbreeding leads to a greater frequency of the R-allele. However, when there is a high cost of carrying the R-allele, that is, when values of aRR and aRr are low relative to arr (curves 4 and 6 in figure 3), as F increases from zero the frequency of the R-allele in the population will decrease or even disappear from the population. A low cost of resistance with a relatively high β will lead to the highest frequencies of the R-allele at equilibrium (curves 1 and 2).
(d) Steady-state equilibrium, case 4: pathogen present and
We considered the case in which infected individuals have a reproductive rate that is either equivalent to or less than that of the resistant individuals. The equilibrium values of , and were independent of F and decreased with decreasing ainf (please refer to electronic supplementary material, part A). The frequency of the R allele increases with increasing inbreeding as in case 2.
4. Discussion
According to our model, when a single R allele can confer resistance and the cost of resistance (lower birth rate) is the same for the homozygote and heterozygote carrying that allele (i.e. aRR=aRr, case 2), the host mating system does not affect the fraction of resistant individuals, or the prevalence of infection in a population (figure 1a). This is an apparently new and broad analytic result. Ecological and life-history factors such as the pathogen transmission parameter (β), the birth (aRR, aRr, ainf and arr) and death rates (b and α) and the density dependence factor (ρ) determine the number of susceptible and infected individuals at equilibrium. Only the frequency of the resistance allele varies with F. The population dynamics ‘compensate’ for disease dynamics such that resistant individuals are still born into the population at a rate high enough to maintain their existence despite the potential influence of selfing in creating a higher number of susceptible individuals in the population.
The selfing rate does not affect the sizes of or when aRR=aRr (case 2), because the RR-homozygote and the heterozygote are both resistant and have the same reproductive rate coefficients. However, when the cost of resistance differs between the RR-homozygote and the heterozygote (aRR<aRr<arr, case 3) the situation is different, because the heterozygote has higher fitness than the RR-homozygote. For case 3, for the particular situation of complete selfing (F=1), the fraction of resistant individuals and the prevalence of infection, , in a population are the same as in the case aRR=aRr (case 2; compare figure 1a,b), because there are no heterozygotes at equilibrium for F=1, so the value of aRr has no influence. But, unlike case 2, in case 3 as F decreases from 1, both the absolute number, and the fraction, , of infected individuals in the population can decrease, increase, or display unimodal behaviour, depending on the set of model parameters.
Case 3 had to be explored by computer simulations of the model, so it is hard to draw generalizations, but some patterns emerge. Increases in with increasing F tend to be associated with small values of β and large values of α (hence large values of ), while the reverse trend generally occurs for large values of β and small values of α. This difference appears to stem from two facts. First, XRr is superior to XRR because it has a higher reproductive rate. This alters the balance of XRR and XRr population from the case in which aRr=aRR (compare figure 1a,b). In particular, when aRr=aRR, any decrease in F results in an increase in XRr that is exactly matched by the decrease in XRR (figure 1a), because they are equivalent ecologically. However, when aRR<aRr, the increase in XRr for decreasing F is not matched in magnitude by the decrease in XRR. In all examples under case 3 examined here, the increase in XRr exceeded the decrease in XRR. Thus, XRr+Xrr increases with decreasing F, and there is a higher relative proportion of XRr (compared with XRR) in the population for F<1 in the case when aRr<aRR than when aRr=aRR. Second, the increased size of XRr has a negative density-dependent effect on the reproduction of Xrr by causing N* to increase, while the increased proportion of XRr in the population has a potentially positive effect on Xrr reproduction because mating between the additional Rr-individuals and both other Rr-individuals and rr-individuals contributes to the Xrr reproductive rate. It is difficult to say a priori whether the negative or positive effect will dominate for a particular set of parameters, but the positive effect tends to exceed the negative effect for large values of (small β and large α) and vice versa. Hence our original prediction for question 1 that higher levels of endemic disease should be found in selfing than in outcrossing populations was found not to be true in general.
In question 2 (§1b), we asked if increasing inbreeding would lead to a greater frequency of the resistance allele in subsequent generations. We usually found a greater frequency of the resistance allele with increased inbreeding using numerical evaluations of the model (figure 3b). However, under certain circumstances, such as a high cost of resistance for homozygotes (i.e. low aRR), the resistance allele may actually be present in higher frequencies in populations with more outcrossing, whereas it has smaller frequencies, or is even lost in some selfing populations (curves 4 and 6 in figure 3b).
For case 2, or aRR=aRr, despite the higher R-frequency in a selfing population, selfing and outcrossing populations have the same phenotypic frequencies of resistant and susceptible individuals due to their differing levels of heterozygosity. For case 3, or aRR<aRr<arr, as mentioned earlier the total number of resistant individuals generally seems to increase with decreasing F, though we could not determine in general whether or the frequency always do so. The only study to date to examine whether the selfing rate affected the prevalence of resistance in a population was a study of two Linum marginale–Melampsora lini host–pathogen metapopulations with different outcrossing rates (Burdon et al. 1999). Results of this study showed that the predominantly selfing populations did not have significantly lower levels of resistance to a variety of fungal isolates than predominantly outcrossing populations.
One assumption of this model is that selfers do not experience inbreeding depression. This assumption makes the model a more conservative test of the effect of inbreeding on pathogen transmission and the frequency of resistance in a population. Inbreeding depression could cause disadvantages to selfers in that their offspring would have a generalized poor condition that could increase the probability of pathogen infection. Inbreeding depression would create an inherent advantage to outcrossing that may alter the conclusions of this model. The results of this model are still applicable to populations that do not experience significant inbreeding depression.
An important assumption that affects the results of this model is a cost of resistance to pathogen infection. Is there such a cost? The prevalence of polymorphism for resistance in natural plant populations implies that resistance must be costly; otherwise, all plants would be resistant (Parker 1992; Simms 1992). An extensive review of plants resistant to pathogens found a cost of resistance in 50% of the studies surveyed (Bergelson & Purrington 1996), although costs are difficult to measure and may occur only in certain circumstances. Another study created transgenic Arabidopsis thaliana that differed from control plants by possessing a single resistance gene (Tian et al. 2003). These plants suffered a 9% cost of resistance in the absence of disease.
In conclusion, this model links the genetic effects of host selfing to the prevalence of disease in a population and the frequency of the resistance allele. In this paper, we have considered only one epidemiological model, albeit an important one. We show in the electronic supplementary material, part B, that analytic solution is possible in a broad array of assumed infection rate functions, including dependence on F. When costs of resistance are associated with the number of resistance alleles an individual possesses (aRR<aRr), the relationship between increased inbreeding and both the prevalence of disease and the frequency of the resistance allele is complex. Some sets of parameters lead to more disease, while others lead to less disease with increasing inbreeding. Likewise, the frequency of the resistance allele can increase or decrease with increasing inbreeding depending on other model parameters. On the other hand, although disease has direct negative effects on individual fitness through increased death rate, in the case that costs of resistance are associated with the resistance phenotype (aRR=aRr), the selfing rate does not affect the proportion of resistant, susceptible, or infected individuals in a population. This surprising result exemplifies the difficulty in translating effects on individuals to a population level, an important step when considering density-dependent processes such as pathogen transmission.
Acknowledgments
We thank Stewart Schultz and Jim Bever for discussions and ideas. Janis Antonovics was helpful during the original development of the model. We thank Jeremiah Busch, Doug Scofield and members of the Clay lab for feedback. J.M.K. was supported by the University of Miami Biology Department and the Indiana University Biology Department. D.L.D. was supported by the USGS's Florida Integrated Science Centers.
Supplementary Material
An alternative model including reduced fecundity of infected individuals and recovery. Appendix B: Generalized expression for the infection rate. Appendix C: Calculation of XRr☆ and XRR☆. Additional figures showing all variables in model conditions described in figure 3.
References
- Agrawal A.F, Lively C.M. Parasites and the evolution of self-fertilization. Evolution. 2001;55:869–879. doi: 10.1554/0014-3820(2001)055[0869:pateos]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Anderson R.M, May R.M. The population dynamics of microparasites and their invertebrate hosts. Phil. Trans. R. Soc. B. 1981;291:451–524. [Google Scholar]
- Bergelson J, Purrington C.B. Surveying patterns in the cost of resistance in plants. Am. Nat. 1996;148:536–558. doi: 10.1086/285938. [DOI] [Google Scholar]
- Burdon J.J. Cambridge University Press; Cambridge, UK: 1987. Diseases and plant population biology. [Google Scholar]
- Burdon J.J, Marshall D.R. Biological control and the reproductive mode of weeds. J. Appl. Ecol. 1981;18:649–658. [Google Scholar]
- Burdon J.J, Thrall P.H, Brown A.H.D. Resistance and virulence structure in two Linum marginale–Melampsora lini host–pathogen metapopulations with different mating systems. Evolution. 1999;53:704–716. doi: 10.1111/j.1558-5646.1999.tb05365.x. [DOI] [PubMed] [Google Scholar]
- Busch J, Neiman M, Koslow J. Evidence for maintenance of sex by pathogens in seed plants. Evolution. 2004;58:2584–2590. doi: 10.1111/j.0014-3820.2004.tb00886.x. [DOI] [PubMed] [Google Scholar]
- Flor H.H. Host–parasite interaction in flax rust—its genetics and other implications. Phytopathology. 1955;45:680–685. [Google Scholar]
- Hamilton W.D. Sex versus non-sex versus parasite. Oikos. 1980;35:282–290. [Google Scholar]
- Hamilton W.D, Axelrod R, Tanese R. Sexual reproduction as an adaptation to resist parasites (a review) Proc. Natl Acad. Sci. USA. 1990;87:3566–3573. doi: 10.1073/pnas.87.9.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D.L, Clark A.G. Sinauer Associates, Inc; Sunderland, MA: 1997. Principles of population genetics. [Google Scholar]
- Holsinger K.E. Reproductive systems and evolution in vascular plants. Proc. Natl Acad. Sci. USA. 2000;97:7037–7042. doi: 10.1073/pnas.97.13.7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard R.S, Lively C.M. The maintenance of sex by parasitism and mutation accumulation under epistatic fitness functions. Evolution. 1998;52:604–610. doi: 10.1111/j.1558-5646.1998.tb01658.x. [DOI] [PubMed] [Google Scholar]
- Jaenike J. An hypothesis to account for the maintenance of sex within populations. Evol. Theory. 1977;3:191–194. [Google Scholar]
- Levin D.A. Pest pressure and recombination systems in plants. Am. Nat. 1975;109:437–451. doi: 10.1086/283012. [DOI] [Google Scholar]
- Lively C.M, Howard R.S. Selection by parasites for clonal diversity and mixed mating. Phil. Trans. R. Soc. B. 1994;346:271–280. doi: 10.1098/rstb.1994.0144. [DOI] [PubMed] [Google Scholar]
- Parker M.A. Disease and plant population genetic structure. In: Fritz R.S, Simms E.L, editors. Plant resistance to herbivores and pathogens. The University of Chicago Press; Chicago, IL: 1992. pp. 345–362. [Google Scholar]
- Simms E.L. Costs of plant resistance to herbivory. In: Fritz R.S, Simms E.L, editors. Plant resistance to herbivores and pathogens. The University of Chicago Press; Chicago, IL: 1992. pp. 392–425. [Google Scholar]
- Simms E.L, Triplett J. Costs and benefits of plant-responses to disease—resistance and tolerance. Evolution. 1994;48:1973–1985. doi: 10.1111/j.1558-5646.1994.tb02227.x. [DOI] [PubMed] [Google Scholar]
- Thompson J.N, Burdon J.J. Gene-for-gene coevolution between plants and parasites. Nature. 1992;360:121–125. doi: 10.1038/360121a0. [DOI] [Google Scholar]
- Thrall P.H, Antonovics J. Theoretical and empirical studies of metapopulations: population and genetic dynamics of the Silene–Ustilago system. Can. J. Bot. Revue Canadienne De Botanique. 1995;73:S1249–S1258. [Google Scholar]
- Thrall P.H, Burdon J.J. Host–pathogen dynamics in a metapopulation context: the ecological and evolutionary consequences of being spatial. J. Ecol. 1997;85:743–753. [Google Scholar]
- Tian D, Traw M.B, Chen J.Q, Kreitman M, Bergelson J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature. 2003;423:74–77. doi: 10.1038/nature01588. [DOI] [PubMed] [Google Scholar]
- Vogler D.W, Kalisz S. Sex among the flowers: the distribution of plant mating systems. Evolution. 2001;55:202–204. doi: 10.1111/j.0014-3820.2001.tb01285.x. [DOI] [PubMed] [Google Scholar]
- Wright S. Systems of mating. II. The effects of inbreeding on the genetic composition of a population. Genetics. 1921;6:124–143. doi: 10.1093/genetics/6.2.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An alternative model including reduced fecundity of infected individuals and recovery. Appendix B: Generalized expression for the infection rate. Appendix C: Calculation of XRr☆ and XRR☆. Additional figures showing all variables in model conditions described in figure 3.