Abstract
Wilms' tumor 1-associating protein (WTAP) has been reported to be a ubiquitously expressed nuclear protein. Although a relation to splicing factors has been postulated, its actual physiological function still remains to be elucidated. To investigate the role of WTAP, we generated WTAP-knockout mice and performed small interfering RNA (siRNA)-mediated knockdown analyses in primary cultured cells. In DNA microarrays using human umbilical vein endothelial cells, WTAP-targeted siRNA treatment resulted in markedly reduced expression of cell-cycle-related genes. siRNA-mediated WTAP knockdown down-regulated the stability of cyclin A2 mRNA through a nine-nucleotide essential sequence in cyclin A2 mRNA 3′ UTR. WTAP knockdown induced G2 accumulation, which is partially rescued by adenoviral overexpression of cyclin A2. Moreover, WTAP-null mice exhibited proliferative failure with death resulting at approximately embryonic day 6.5, an etiology almost identical to cyclin A2-null mice. Collectively, these findings establish WTAP as an essential factor for the stabilization of cyclin A2 mRNA, thereby regulating G2/M cell-cycle transition.
Keywords: mRNA stability, cell-cycle regulation
Wilms' tumor 1-associating protein (WTAP) was identified as a protein that specifically interacts with Wilms' tumor 1 (WT1) in both in vitro and in vivo assays (1). The Wilms tumor suppressor gene WT1 is essential for the normal development of the bipotential gonad, and the primordial kidney (2–4) and is associated with a common form of pediatric kidney cancer (5). WTAP and WT1 are present together throughout the nucleoplasm as well as in nuclear speckles and partially colocalize with splicing factors (1). WTAP was also identified as the mammalian homologue of the Drosophila female-lethal-2-D [fl(2)D]. fl(2)D is required for the female developmental pathway because of its activation of female-specific patterns of alternative splicing on SXL and transformer (tra) premRNA (6, 7). The presence of the lethal phenotype of the fl(2)D mutation in both sexes suggests an additional function for the gene (8). In vitro splicing assays demonstrated that depletion of WTAP from HeLa nuclear extracts affects tra but not AdML (a model adenovirus RNA) splicing, suggesting a biochemical function in female-specific splicing regulation (9). Furthermore, proteomic studies isolated WTAP as one of 145 spliceosomal proteins assembled on two distinct premRNAs, adenovirus major late and Fushi tarazu (10). While investigating GATA function in vascular endothelial cells, we serendipitously identified WTAP, in addition to GATA-associating proteins and hematopoietically expressed homeobox, from a yeast two-hybrid screen (11). In spite of these refined studies, the exact function of WTAP remains unknown.
In the present study, we investigated the function of WTAP in vitro by using RNAi in primary cultured cells and in vivo by generating WTAP-knockout mice. Here, we demonstrate that WTAP is a factor essential for cyclin A2 mRNA stabilization and G2/M transition, the understanding of which should yield significant insight into posttranscriptional regulation, as well as cell- cycle regulation in normal and/or tumor cells.
Results
WTAP-Knockdown Cells Display Markedly Reduced Expression of Cyclin A2 and B and Cell Division-Related Genes.
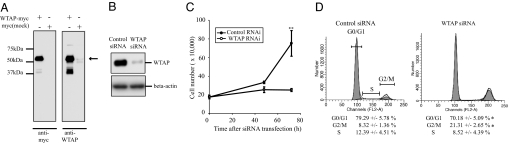
To investigate the physiological function of WTAP, we examined the relative influence of WTAP on gene expression in human umbilical vein endothelial cells (HUVEC) by using an RNAi-based approach. To examine WTAP expression, we first generated an anti-WTAP antibody raised in rabbits by using bacterially expressed GST-WTAP protein. Its specificity for WTAP was tested by Western blot analysis (Fig. 1A). A ≈50-kDa band corresponding to myc-tagged WTAP was detectable by both anti-myc and -WTAP antibodies (Fig. 1A, arrow). Additionally, low levels of endogenous WTAP were also detectable by anti-WTAP antibody. In HUVEC cells, after WTAP siRNA transfection, WTAP protein levels were reduced to <20% of that in control siRNA-transfected cells (Fig. 1B). Subsequently, RNAs from WTAP or control siRNA-treated cells were subjected to microarray analysis. Of 38,500 genes present on a U133 Plus 2.0 array, 329 genes were ≥2-fold down-regulated and 324 genes ≥2-fold up-regulated by WTAP knockdown confirmed in two independent experiments (Tables 3 and 4, which are published as supporting information on the PNAS web site). The top 30 down- or up-regulated genes are listed in Tables 5 and 6, which are published as supporting information on the PNAS web site, in which genes with unknown functions have been excluded. Interestingly, the major genes exhibiting decreased expression are all related to the cell division cycle, including cyclins A2, B1, B2, and CDC20, whereas the up-regulated genes are related to metabolism, inflammation, or cell adhesion.
Fig. 1.
WTAP siRNA-treated cells display growth inhibition in G2/M phase. (A) The specificity of anti-WTAP polyclonal antibody. COS7 cells transiently transfected with a pcDNA3.1(−)-WTAP-myc-His or empty vector were subjected to Western blot analysis by using anti-WTAP or -myc antibody. Arrows indicate the band corresponding to WTAP (≈50 kDa), detected by an anti-myc antibody. (B) Efficient reduction of WTAP protein was confirmed by Western blots in WTAP siRNA-transfected HUVEC. HUVEC were transfected with WTAP siRNA or control siRNA. Total proteins were extracted 72 h after transfection. (C) Cell growth rates in WTAP siRNA or control siRNA-treated HUVEC. Cell numbers were determined by using a hemacytometer. (D) Cell-cycle analysis was carried out by using siRNA-treated HUVEC. Forty-eight hours after siRNA transfection, cells were harvested and stained with propidium iodide and then analyzed for DNA content with a FACScalibur. ∗, P < 0.05; ∗∗, P < 0.001 vs. contol.
siRNA-Mediated WTAP Reduction Results in G2-Phase Accumulation.
To examine the effect of WTAP reduction on cell cycle, proliferation assays and FACS analysis were performed. As shown in Fig. 1C, HUVEC in the control group exhibited moderate growth for the first 48 h after siRNA transfection, then increased 3.5-fold in cell number by 72 h. In contrast, WTAP siRNA-treated cells underwent growth inhibition, as evidenced by the lack of an increase in cell number over time. Moreover, FACS analysis revealed that WTAP siRNA-treated cells exhibited a significantly higher proportion in G2 (21%) compared with that of control cells (8%; Fig. 1D). These data demonstrate that WTAP reduction leads to growth arrest in G2 or reduction in progression through G2. To determine whether this effect is specific to HUVEC, we next performed the same experiments in primary human fibroblasts. G2 accumulation was also induced in human neonatal dermal fibroblasts (Fig. 7, which is published as supporting information on the PNAS web site). This finding demonstrates that the growth inhibition caused by WTAP reduction is not specific to endothelial cells but can be induced in other cell types as well.
WTAP Regulates Cyclin A2 mRNA Stability Through 3′UTR.
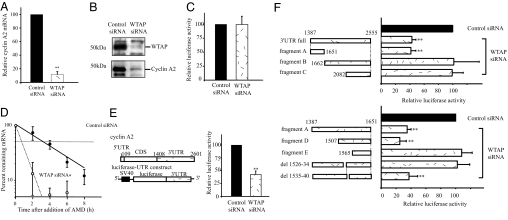
Among the genes down-regulated by the knockdown of WTAP expression, cyclin A2 was the most prominently reduced. Interestingly, it has been reported that cyclin A mutant Drosophila embryos undergo cell-cycle arrest in G2 (12), and somatic mammalian cells are blocked at G2 on ablation of cyclin A (13). Hence, we hypothesized that the G2 accumulation brought about by WTAP reduction was because of the loss of cyclin A2 expression and further investigated how WTAP affects cyclin A2 mRNA expression. We first confirmed that WTAP knockdown led to a decrease in cyclin A2 at the mRNA and protein levels. Quantitative real-time PCR analysis revealed that the cyclin A2 mRNA level was decreased to 10% that of controls within 24 h after siRNA transfection and, as a result, reduction in the cyclin A2 protein level was observed by Western blot analysis (Fig. 2A and B). To investigate the mechanism responsible for the decrease in cyclin A2 mRNA levels, we examined the effect of WTAP on cyclin A2 promoter activity and mRNA stability in HUVEC. By using a cyclin A2 promoter-luciferase reporter construct, the effect of WTAP knockdown on cyclin A2 promoter activity was examined. The full promoter region of cyclin A2, corresponding to −737 to +108 bp (start site, +1), was inserted into a pGL3-basic plasmid. As shown in Fig. 2C, RNAi of WTAP did not result in any substantial change in luciferase activity, suggesting that WTAP has no obvious effect on transcription from the cyclin A2 promoter. Next, we studied the half-life of cyclin A2 mRNA by using standard actinomycin D-based methods to investigate whether its low expression level was because of enhanced degradation of its mRNA. At 24 h after siRNA transfection, 5 μg/ml actinomycin D was added to the medium, and RNA was isolated at each indicated time point. The cyclin A2 mRNA remaining was measured by quantitative real-time PCR normalized against the stable rpL32 mRNA (T1/2 ≥ 25 h). In the absence of WTAP, the cyclin A2 mRNA half-life significantly declined from 2.8 h, in control siRNA-treated cells, to 1.3 h (Fig. 2D). To further examine cyclin A2 mRNA stability, we performed a reporter assay by using a firefly luciferase-cyclin A2 3′UTR chimeric plasmid constructed by conjugating a 3′UTR fragment of cyclin A2 into a pGL3-control vector. The reporter plasmids were cotransfected with a CMV-Renilla luciferase reporter plasmid into HUVEC 12 h after siRNA transfection. Eighteen hours later, dual-luciferase assays were performed. The relative luciferase activity (firefly/Renilla) was normalized to the basal relative luciferase activity obtained from the pGL3-control vector alone. A significant decrease in luciferase activity was observed in WTAP siRNA-treated cells (≈40%) compared with that in control cells (Fig. 2E). These findings suggest that WTAP regulates cyclin A2 mRNA stabilization through its effect on cyclin A2 3′UTR.
Fig. 2.
WTAP knockdown leads to the reduction of cyclin A2 mRNA and protein levels because of destabilization of cyclin A2 mRNA. (A) Quantitative real-time PCR analysis of cyclin A2 expression. Cyclin A2 mRNA levels were decreased to 10% that of controls 24 h after siRNA transfection. Values are WTAP RNAi vs. control RNAi samples normalized to cyclophilin mRNA levels. (B) Western blot analysis of WTAP and cyclin A2. Cells were harvested 72 h after siRNA transfection. (C) The effect of WTAP knockdown on cyclin A2 promoter activity was studied by using a cyclin A2 promoter-luciferase reporter assay. The full promoter region of cyclin A2, corresponding to −737 to +108 bp (start site, +1), was inserted into the pGL3-basic plasmid. (D) Actinomycin D (AMD) was added at 24 h after siRNA transfection, and total RNA was prepared at each indicated time point. The remaining cyclin A2 mRNA was measured by quantitative real-time PCR and normalized to rpL32 mRNA, which has a half-life of >25 h. (E) Influence of the 3′UTR of cyclin A2 mRNA on WTAP-mediated mRNA stability. A luciferase-cyclin A2 3′UTR chimeric plasmid was constructed by subcloning in the 3′UTR fragment of cyclin A2 immediately downstream of the firefly luciferase ORF in a pGL3-control vector. At 12 h after siRNA transfection, HUVEC were transiently transfected with the chimeric luciferase-cyclin A2 3′UTR plasmid. Eighteen hours later, dual-luciferase assays were performed. Relative luciferase activity (firefly/Renilla) was normalized to the relative basal luciferase activity obtained from a pGL3-control vector. (F) Responsible region for WTAP-mediated mRNA stability was analyzed by using deletion constructs of cyclin A2 mRNA 3′UTR. *, P < 0.05; **, P < 0.001 vs. control.
To determine the responsible region for WTAP-mediated stabilization, we generated 3′UTR deletion constructs and used reporter assays (Fig. 2F). There are three AUUUA motifs, sequences known to be involved in mRNA decay, in the 3′UTR of cyclin A2 mRNA at +1,686, +2,297, and +2,417. Notably, a 1,387/1,651 construct (fragment A) with no AUUUA motif gave a similar or greater decrease in relative luciferase activity (≈40%) as the 3′UTR full construct in WTAP siRNA-treated cells. Deletion of this region (fragment B) completely abolished the loss in luciferase activity induced by WTAP reduction. As expected, fragment C showed no effect after WTAP RNAi.
To identify the responsible region for stabilization by WTAP within fragment A, we undertook a luciferase assay using the deletion constructs of fragment A (Fig. 2F). Construct (1,565/1,651; fragment E) eliminated the destabilizing activity of WTAP knockdown, as evidenced by an increase in luciferase activity to control levels (Fig. 2F). These results suggest that the responsible element confirming cyclin A2 mRNA stability is present between 1,507 and 1,565 bp of the 3′UTR. Deletion of 1526–34 (del 1526–34) abolished the destabilizing activity, whereas the adjacent deletion of 1535–40 (del 1535–40) showed similar activity to fragment A. Taken together, these findings suggest that ACAAAUUAU, which corresponds to the 3′UTR 1526–34, is an essential element required for the WTAP-mediated stabilization of cyclin A2 mRNA, in a manner independent of AUUUA motifs.
Adenoviral-Mediated Expression of Cyclin A2 Protein Rescued the G2 Accumulation Caused by Knockdown of WTAP.
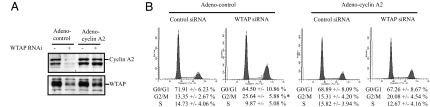
To confirm the contribution of cyclin A2 reduction to the G2 accumulation induced by WTAP knockdown, FACS analysis was performed by using HUVEC infected with IRES-containing adenoviruses expressing both cyclin A2 and EGFP (adeno-cyclin A2) or EGFP alone (adeno-control) at a multiplicity of infection of 20. As shown in Fig. 3A, cells infected with adeno-cyclin A2 showed cyclin A2 expression both in the absence or presence of WTAP siRNA. Adeno-cyclin A2 carries the cDNA encoding the human cyclin A2 coding region. These findings are consistent with WTAP-mediated regulation of cyclin A2 expression through the cyclin A2 mRNA 3′UTR. Adenovirus-mediated cyclin A2 expression had no effect on the cell-cycle profile (Fig. 3B). The G2 accumulation was partially rescued by the overexpression of cyclin A2, whereas the knockdown of WTAP still led to the G2 accumulation of adeno-control-infected cells.
Fig. 3.
Influence of adenovirus-mediated expression of cyclin A2 on cell-cycle profile in WTAP siRNA or control siRNA-treated HUVEC. HUVEC were infected with adeno-control or adeno-cyclin A2 at a multiplicity of infection of 20. (A) Forty-eight hours after siRNA transfection, cells were harvested and subjected to the Western blots for WTAP and cyclin A2. (B) FACS analysis. ∗, P < 0.05 vs. contol.
WTAP-Null Mice Die and Exhibit Proliferative Failure at Approximately Embryonic Day 6.5 (E6.5).
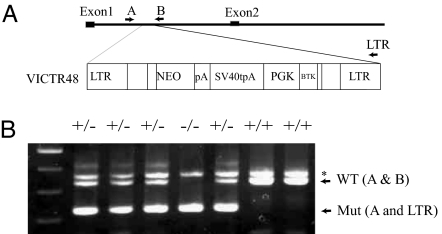
Finally, we extended our studies in vivo using WTAP-null mice. The WTAP-knockout mice were generated by gene trapping by using a retroviral vector integrated between exons 1 and 2 (Fig. 4A). Genotyping by PCR using three primer sets, which lie on opposite sides of the insertion site, and a gene-trap vector (Fig. 4B) revealed that no newborn homozygous mice were yielded from heterozygous intercrosses (data not shown). This finding demonstrates that WTAP is required for embryonic development. To determine the timing of lethality during embryogenesis, we therefore performed genotyping and histological analysis of embryos from the heterozygous matings. Table 1 depicts genotyping by using the yolk sac DNA of embryos from E8.5 to E10.5. Homozygous WTAP−/− embryos are obviously small and without an embryo body (Fig. 5A and B; heterozygous WTAP +/− embryos at E8.5 and homozygous littermate, respectively). Histological analyses of decidua during early stages of development revealed that abnormal embryos were observed at E6.5 (Table 2). Morphologically abnormal embryos are small like blastocysts and exhibit defects in proliferation and an absence of the shapes characteristic of endoderm and ectoderm (Fig. 5 D, E, and H), whereas normal embryos displayed defined layers of embryonic ectoderm and endoderm (Fig. 5 C and F) by E6.5. Interestingly, these features are strikingly similar to those seen in the cyclin A2 null mutant. Cyclin A2 null mutants cease development at approximately E6.5 and present a morphology reminiscent of blastocysts (14).
Fig. 4.
WTAP-null mice exhibited proliferative failure, with death resulting at approximately E6.5. (A) Generation of WTAP-null mice. Retroviral integration and disruption of the WTAP gene. VICTR 48 integrated in the intron between exons 1 and 2. NEO, a functional fusion between the β-gal and neomycin resistance genes; pA, polyadenylation sequence; SV40tpA, SV40 triple polyadenylation sequence; PGK, phosphoglycerate kinase-1 promoter; BTK, Burton's tyrosine kinase. (B) PCR genotyping of embryos from WTAP+/− intercrosses by using three primers, A, B, and LTR, to simultaneously amplify both WT (341 bp) and mutated (mut, 255 bp) alleles. ∗, Nonspecific band.
Table 1.
Genotyping of mouse embryos from heterozygous matings by PCR by using yolk sac DNA
| Stage of development | Numbers of embryos |
Total number | |||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | Reabsorbed | ||
| E8.5 | 7 | 16 | 2 | 1 | 26 |
| E9.5 | 4 | 17 | 5 | 26 | |
| E10.5 | 4 | 12 | 2 | 18 | |
Fig. 5.
Morphology of embryos from WTAP+/− heterozygous matings. (A and B) Heterozygous WTAP+/− embryo at E8.5 (A) and homozygous WTAP−/− littermate (B). Homozygous WTAP−/− embryos are small and present no embryo body. (C–E) Representative H&E stainings of E6.5 normal (C) and abnormal (D and E) embryos cross-sectioned. (E) Some abnormal embryos have a dilated proamniotic canal. (F–I) Representative H&E and WTAP staining of E6.5 embryos sagittally sectioned . (F) Normal embryos exhibit defined layers of cells from embryonic ectoderm and endoderm. (H) Abnormal embryos are smaller and present no defined layers. No specific WTAP stainings were observed in morphologically abnormal embryos (I, compare with G), confirming that these abnormal embryos are homozygous null mutants. (Scale bars: A and B, 500 μm; C–E, 200 μm; F and H, 100 μm.)
Table 2.
Morphology of embryos from heterozygous matings
| Stage of development | Numbers of embryos |
Total number | ||
|---|---|---|---|---|
| Normal | Abnormal | Reabsorbed | ||
| E6.5 | 28 | 4 | 32 | |
| E7.5 | 14 | 2 | 16 | |
| E8.5 | 8 | 1 | 1 | 10 |
| E10.5 | 0 | 1 | 1 | |
Abnormal embryos were not stained by immunostaining with anti-WTAP polyclonal antibodies.
Immunostaining of serial sections with rabbit polyclonal anti-WTAP antibodies revealed that WTAP was detectable in the nuclei of morphologically normal embryos (Fig. 5G). In contrast, no specific WTAP staining was observed in morphologically abnormal embryos, confirming that these embryos are homozygous null mutants (Fig. 5I). In addition to the proliferative failures observed in the WTAP−/− embryos, some presented a dilated proamniotic canal (Fig. 5E).
Cell-Cycle-Dependent WTAP Expression Is Synchronous with Cyclin A2.
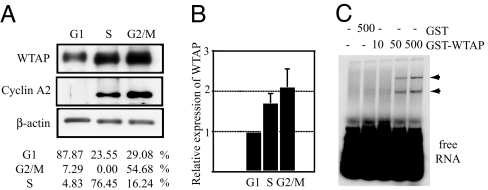
It has been reported that cyclin A2 mRNA stability varies with the cell cycle, which helps account for its cell-cycle-dependent relative abundance (15). Thus, we next examined the expression of WTAP throughout the cell division cycle. To this end, HUVEC were subjected to cell-cycle synchronization experiments by serum starvation for 36 h, so that cells exhibited G1-enriched distribution (≈90%). Then cells were passaged and released from quiescence by exchanging the medium for complete medium EGM2, and cell-cycle progression was monitored by FACS analysis. Protein was harvested at time points when the majority of the cells were in G1 (87%), S (76%), or G2/M (55%). As shown in Fig. 6A, WTAP expression varied with the cell cycle. WTAP was low in G1, elevated to 1.7-fold in S and 2.1-fold in G2. The cell-cycle-dependent expression of cyclin A2 was detected in only the S- and G2-rich phases. The cell-cycle-dependent changes in the expression levels of cyclin A2 and WTAP were similar, even though WTAP expression was maintained at a certain basal expression level throughout the cell cycle.
Fig. 6.
Cell-cycle-dependent expression of WTAP. (A) Western blots of WTAP and cyclin A2 of the G1, S, or G2/M phase. For synchronization studies, HUVEC were serum-starved for 36 h, then released by medium exchange. The cell-cycle profile was monitored by FACS analysis, and proteins were collected at each time point. (B) Relative protein expression of WTAP was normalized to β-actin expression. (C) EMSA using cyclin A2 mRNA 3′UTR full and the indicated concentrations of either GST or GST-WTAP. Arrowheads indicate the RNA–protein complexes.
WTAP Binds to the 3′UTR of Cyclin A2 mRNA.
To investigate the interaction of WTAP protein with cyclin A2 mRNA, we performed RNA EMSAs by using purified GST-WTAP and in vitro synthesized transcripts. GST-WTAP or GST protein alone was incubated with radiolabeled cyclin A2 3′UTR transcripts, and the formation of protein–RNA complexes was examined. As shown in Fig. 6C, GST-WTAP bound to the 3′UTR of the cyclin A2 mRNA, whereas control GST protein did not. No protein–RNA complexes were detected when GST-WTAP was incubated with the coding region of the cyclin A2 (data not shown).
Discussion
In the present study, our data suggested that WTAP regulates cyclin A2 mRNA stability, and WTAP−/− mice suffer embryonic lethality because of the loss of cyclin A2 expression. WTAP knockdown reduced cyclin A2 expression to ≈10% of control cells by decreasing the stability of cyclin A2 mRNA, suggesting that a specific change in mRNA stability is an important component in the maintenance of the cell cycle. The G2 accumulation induced by WTAP RNAi is likely because of cyclin A2 mRNA destabilization, because overexpression of cyclin A2 released WTAP siRNA-treated cells from G2 accumulation. The other mitosis-related genes down-regulated by WTAP RNAi, as detected by microarray analysis, are believed to be secondary effects because of G2 accumulation.
The regulation of mRNA stability has been recognized as an important process in the control of gene expression. The best-characterized cis element mediating mRNA decay is the AU-rich element (ARE) (16), which is present in the 3′UTRs of many mRNAs, including those of growth factors (17), oncogenes (18), cytokines (19, 20), and cell-cycle regulatory proteins (21, 22). It has been reported that the RNA-binding protein HuR selectively binds to AREs and enhances cyclin A and cyclin B1 mRNA stability (23). As shown in Fig. 6C, RNA EMSA analysis revealed a direct interaction of WTAP protein with the 3′UTR of cyclin A2 mRNA, suggesting that WTAP enhances cyclin A2 mRNA stability by binding its 3′UTR. Yet, it remains to be elucidated that other unidentified proteins may function in concert with WTAP to modulate mRNA stability. Based on chimeric luciferase experiments using 3′UTR deletion constructs, we demonstrated that the nine-base sequence ACAAAUUAU was required for the regulation of stability by WTAP. There are no previous reports of this sequence being involved in the regulation of mRNA stability. It remains to be determined whether this nine-base sequence itself or the mRNA secondary structure is responsible for WTAP-mediated stabilization of cyclin A2 mRNA.
Murphy et al. (14) demonstrated that a targeted deletion of the murine cyclin A2 gene is embryonically lethal at approximately E6.5, with a blastocyst like morphology, although homozygous-null embryos develop normally until postimplantation, approximately E5.5. It has also been demonstrated that cyclin A2 is not required for cellular progression during the preimplantation nongrowth period of mouse embryogenesis (24). The timing of lethality and abnormal phenotype reported in cyclin A2-null mice are strikingly similar to those observed in WTAP-null mice, providing further evidence of their functional relationship.
A previous report demonstrated that WTAP has a ubiquitous expression pattern in mouse tissues, as confirmed by RT-PCR experiments (1). We also confirmed its ubiquitous expression in our gene chip database (Fig. 8, which is published as supporting information on the PNAS web site). Additionally, WTAP was present in nonmitotic potential tissues where cyclin A2 expression is barely detected. Importantly, in “normal” tissues, abundant expression of WTAP was observed. Adenoviral overexpression of WTAP in HUVEC did not affect cell proliferation or the cell-cycle profile in HUVEC (data not shown). Thus, we hypothesize that the role of WTAP may be more to maintain cell-cycle progression through the stabilization of cyclin A2 mRNA than to promote it. We cannot exclude the possibility that WTAP may have additional roles requisite in many, if not all, cell types. A recent report demonstrated that the WT-1 (+KTS) isoform is involved in translational enhancement of an unspliced RNA with a retained intron (25). Further studies are needed to clarify the role of WTAP in cell- cycle regulation and should yield novel insights into the posttranscriptional mechanisms involved in cell-cycle regulation.
Materials and Methods
Cell Culture.
HUVEC (Clonetics, La Jolla, CA) were cultured in EGM2 medium (Clonetics). HUVEC were used within the first six passages. For synchronization studies, HUVEC were serum-starved in EBM2 (Clonetics) for 36 h, then released by exchange medium to EGM2. Normal human dermal fibroblasts (Clonetics) were cultured in FGM-2 medium (Clonetics).
RNAi.
For WTAP RNAi analysis, siRNA duplexes targeting the sequences AAG CTT TGG AGG GCA AGT ACA (Qiagen, Valencia, CA) or Qiagen control siRNA were transfected with lipofectamine 2000 (Invitrogen, Carlsbad, CA), according to the manufacturer's protocol. Growth medium was replaced 6 h after transfection.
Generation of the WTAP-Knockout Mice.
WTAP-knockout mice were obtained from Lexicon Genetics (The Woodlands, TX). Genomic DNA was extracted from mouse tail or embryo yolk sac by using a DNeasy kit (Qiagen). Genotyping was done by PCR by using the following primers to simultaneously amplify both WT (341 bp) and mutated (mut, 255 bp) alleles; A, 5′-AAATGCGGTCTGGAGAGAGA-3′; B, 5′-CCTAACTTGCCCCAGCAGTA-3′; LTR, 5′-AAATGGCGTTACTTAAGCTAGCTTGC-3′. The original founder mice were 129 SvEvBrd × C57BL/6 heterozygotes from the F2 generation. Heterozygous mice were maintained by backcross with C57BL/6 mice until the N7 generation. Similar results were observed in mice from earlier generations and the N7 generation.
Statistics.
All experiments were performed a minimum of three times. Data points represent the mean ± SD calculated from multiple independent experiments. Statistically significant differences for data points and half-life curves represent P < 0.05 and were calculated by using either the unpaired t test or analysis of variance [Fisher's Least Significant Difference (LSD) test]. Statistical analyses were performed by using Statview version 5 software (Abacus Concepts, Berkeley, CA).
Further Details.
Further details of the experimental procedures used in this study are provided in Supporting Text, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Dr. K. Boru of Pacific Edit for review of the manuscript, Drs. Yoshifumi Yamaguchi and Norihiko Ohbayashi for helpful discussion, and Naomi Saito for helpful technical assistance. This work is supported in part by grants from the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation; from the Special Coordination Fund for Science and Technology from the Ministry of Education, Culture, Sports, Science, and Technology; and from the New Energy and Industrial Technology Development Organization Bionanochip Project (P03023), Ministry of Economy, Trade, and Industry, Japan.
Abbreviations
- WT1
Wilms' tumor 1
- siRNA
small interfering RNA
- HUVEC
human umbilical vein endothelial cells
- En
embryonic day n.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo/(accession no. GSE2327).
References
- 1.Little NA, Hastie ND, Davies RC. Hum Mol Genet. 2000;9:2231–2239. doi: 10.1093/oxfordjournals.hmg.a018914. [DOI] [PubMed] [Google Scholar]
- 2.Haber DA, Buckler AJ, Glaser T, Call KM, Pelletier J, Sohn RL, Douglass EC, Housman DE. Cell. 1990;61:1257–1269. doi: 10.1016/0092-8674(90)90690-g. [DOI] [PubMed] [Google Scholar]
- 3.Buckler AJ, Pelletier J, Haber DA, Glaser T, Housman DE. Mol Cell Biol. 1991;11:1707–1712. doi: 10.1128/mcb.11.3.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- 5.Hastie ND. Cell. 2001;106:391–394. doi: 10.1016/s0092-8674(01)00469-x. [DOI] [PubMed] [Google Scholar]
- 6.Granadino B, Campuzano S, Sanchez L. EMBO J. 1990;9:2597–2602. doi: 10.1002/j.1460-2075.1990.tb07441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Granadino B, Penalva LO, Sanchez L. Mol Gen Genet. 1996;253:26–31. doi: 10.1007/s004380050292. [DOI] [PubMed] [Google Scholar]
- 8.Granadino B, San Juan A, Santamaria P, Sanchez L. Genetics. 1992;130:597–612. doi: 10.1093/genetics/130.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ortega A, Niksic M, Bachi A, Wilm M, Sanchez L, Hastie N, Valcarcel J. J Biol Chem. 2003;278:3040–3047. doi: 10.1074/jbc.M210737200. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Z, Licklider LJ, Gygi SP, Reed R. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 11.Minami T, Murakami T, Horiuchi K, Miura M, Noguchi T, Miyazaki J, Hamakubo T, Aird WC, Kodama T. J Biol Chem. 2004;279:20626–20635. doi: 10.1074/jbc.M308730200. [DOI] [PubMed] [Google Scholar]
- 12.Lehner CF, O'Farrell PH. Cell. 1990;61:535–547. doi: 10.1016/0092-8674(90)90535-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy M, Stinnakre MG, Senamaud-Beaufort C, Winston NJ, Sweeney C, Kubelka M, Carrington M, Brechot C, Sobczak-Thepot J. Nat Genet. 1997;15:83–86. doi: 10.1038/ng0197-83. [DOI] [PubMed] [Google Scholar]
- 15.Maity A, McKenna WG, Muschel RJ. Cell Growth Differ. 1997;8:311–318. [PubMed] [Google Scholar]
- 16.Chen CY, Shyu AB. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 17.Shaw G, Kamen R. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 18.Miller AD, Curran T, Verma IM. Cell. 1984;36:51–60. doi: 10.1016/0092-8674(84)90073-4. [DOI] [PubMed] [Google Scholar]
- 19.Henics T, Sanfridson A, Hamilton BJ, Nagy E, Rigby WF. J Biol Chem. 1994;269:5377–5383. [PubMed] [Google Scholar]
- 20.Wodnar-Filipowicz A, Moroni C. Proc Natl Acad Sci USA. 1990;87:777–781. doi: 10.1073/pnas.87.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin S, Wang W, Wilson GM, Yang X, Brewer G, Holbrook NJ, Gorospe M. Mol Cell Biol. 2000;20:7903–7913. doi: 10.1128/mcb.20.21.7903-7913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. EMBO J. 2000;19:2340–2350. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winston N, Bourgain-Guglielmetti F, Ciemerych MA, Kubiak JZ, Senamaud-Beaufort C, Carrington M, Brechot C, Sobczak-Thepot J. Dev Biol. 2000;223:139–153. doi: 10.1006/dbio.2000.9721. [DOI] [PubMed] [Google Scholar]
- 25.Bor YC, Swartz J, Morrison A, Rekosh D, Ladomery M, Hammarskjold ML. Genes Dev. 2006;20:1597–1608. doi: 10.1101/gad.1402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.