Abstract
Ecological and mutational explanations for the evolution of sexual reproduction have usually been considered independently. Although many of these explanations have yielded promising theoretical results, experimental support for their ability to overcome a twofold cost of sex has been limited. For this reason, it has recently been argued that a pluralistic approach, combining effects from multiple models, may be necessary to explain the apparent advantage of sex. One such pluralistic model proposes that parasite load and synergistic epistasis between deleterious mutations might interact to create an advantage for recombination. Here, we test this proposal by comparing the fitness functions of parasitized and parasite-free genotypes of Escherichia coli bearing known numbers of transposon-insertion mutations. In both classes, we failed to detect any evidence for synergistic epistasis. However, the average effect of deleterious mutations was greater in parasitized than parasite-free genotypes. This effect might broaden the conditions under which another proposed model combining parasite–host coevolutionary dynamics and mutation accumulation can explain the maintenance of sex. These results suggest that, on average, deleterious mutations act multiplicatively with each other but in synergy with infection in determining fitness.
Keywords: Escherichia coli, evolution of sex, experimental evolution, mutational load, parasitic load
1. Introduction
An understanding of the selective forces underlying the maintenance of sexual reproduction remains one of the most important unanswered questions in evolutionary biology (Maynard Smith 1978; Bell 1982). This slow progress is caused not by a lack of theories about the advantages of sexual reproduction (Kondrashov 1993), but instead by a lack of compelling data. These theories can be broadly classified into two main groups that emphasize either ecological or mutational processes.
Ecological models hypothesize that sex accelerates adaptation to changing environments by creating new gene combinations. For example, the Red Queen hypothesis predicts that sexual recombination is advantageous for populations engaged in antagonistic biotic interactions, such as a coevolutionary arms race between hosts and parasites (Maynard Smith 1978; Bell 1982; Lively & Howard 1994; Howard & Lively 1998). In this case, sex can accelerate the production of new multi-locus genotypes that allow the host to escape infection by parasites that have adapted to the prior generation of hosts, owing to the large populations and rapid generations typical of parasites. However, the Red Queen hypothesis has two key limitations: (i) it requires that parasites have severe effects on host fitness, and (ii) parasites might select for clonal diversity in the host population instead of for recombination (Howard & Lively 1994, 1998). These limitations may constrain the ability of the Red Queen hypothesis to provide a general explanation for the evolution and maintenance of sex (Kondrashov 1993).
Mutational models propose that sexual recombination is advantageous because it breaks apart unfavourable associations between alleles within populations (Peters & Lively 1999). In recent years, the mutational deterministic hypothesis (Kondrashov 1982, 1984, 1985, 1988, 1993) has become one of the most cited explanations for sexual reproduction. According to this hypothesis, if the genomic deleterious mutation rate is high (say, greater than 1), and if deleterious mutations interact synergistically (such that each additional mutation tends, on average, to cause a greater proportional decline in fitness than the previous mutation), then sexual reproduction provides an advantage over asexual reproduction by enabling populations to eliminate deleterious mutations more efficiently. This effect occurs because a disproportionately small number of selective deaths are required in sexual populations to eliminate those low-quality offspring that, by chance, have particularly high numbers of deleterious mutations. The mutational deterministic hypothesis has attracted attention, in part, because its predictions are based on key parameters that, in principle, are experimentally tractable. In practice, however, it has proved difficult to obtain accurate estimates of deleterious genomic mutation rates (Drake et al. 1998; Keightley & Eyre-Walker 2000; Keightley & Lynch 2003). Moreover, most experiments have so far failed to demonstrate a predominance of synergistic epistasis among deleterious mutations (de Visser et al. 1996, 1997a,b; Elena & Lenski 1997; de Visser & Hoekstra 1998; West et al. 1998; Elena 1999; de la Peña et al. 2000; Keightley & Eyre-Walker 2000; Wloch et al. 2001). This failure also reflects considerable variation in the intensity and direction of epistasis between mutations (de Visser et al. 1997a; Elena & Lenski 1997), which further reduces the parameter space in which the mutational deterministic hypothesis predicts an advantage to sexual reproduction (Otto & Feldman 1997).
It has recently been argued that a pluralistic approach combining several hypotheses may be necessary to develop a more complete understanding of the forces that favour sexual reproduction (Lively & Howard 1994; Howard & Lively 1998; West et al. 1999). Several advantages for such an approach have been suggested: (i) there may be different advantages of sex relevant to different species and even to different time points within an evolutionary lineage; (ii) multiple selective forces may act simultaneously to maintain sex even within the same population; and (iii) the emphasis of experimental work shifts from searching for unique predictions to parameter estimation (West et al. 1999). This approach also removes the hidden assumption that a single selective mechanism is responsible for the origin and maintenance of sexual reproduction.
A specific example of the pluralistic approach is the suggestion that parasites may interact with deleterious mutations to increase the strength of selection maintaining sexual recombination (West et al. 1999). Specifically, if parasites interact with deleterious mutations so as to increase the strength of synergistic epistasis, then the conditions under which the mutational deterministic hypothesis predicts an advantage to sex will be broadened (West et al. 1999). The goal of the present study is to test this hypothesis. To do this, we introduced an intracellular parasite into each member of a collection of Escherichia coli mutants that contained one, two or three transposon-insertion mutations (Elena & Lenski 1997; Elena et al. 1998). We measured the fitness of each mutant by means of direct competitions against the parasitized, unmutated progenitor. These results were then compared with data from competitions between non-parasitized versions of the same mutants to determine the effect of parasites on the nature of the interactions among deleterious mutations. The use of genotypes carrying known numbers of mutations avoids various methodological problems that have complicated the interpretation of many previous studies that have tested for synergistic epistasis (Elena & Lenski 1997).
Overall, we found no tendency for an increase in synergistic epistasis in the parasitized mutants, contrary to the particular pluralistic hypothesis that we sought to test. However, we observed a significant increase in the average severity of deleterious mutations in the presence of the parasites. From this result, we suggest that parasites might broaden the conditions favouring sex by amplifying the adverse effect of mutation accumulation during population bottlenecks (including those that may often occur in host–parasite dynamics), a process that disproportionately harms asexual lineages.
2. Material and methods
2.1 Construction of mini-Tn10 insertion mutants
Insertion mutations were generated using mini-Tn10 constructs, which have certain properties that make them useful for mutagenesis of E. coli (Kleckner et al. 1991). The methods for producing random insertion mutations, and for molecular confirmation of the insertions, are described fully elsewhere (Elena & Lenski 1997; Elena et al. 1998). A subset of 216 mutants from a set of 225 used in a previous study was chosen randomly for this study (Elena & Lenski 1997). Mutants with one, two and three insertion mutations were equally represented in the subset. All mutants were derived from strain REL4548, a clone isolated at generation 10000 from one of the long-term populations described elsewhere (Lenski et al. 1991; Lenski & Travisano 1994; Lenski 2004). REL4548 and its mutant derivatives are unable to use the sugar arabinose (Ara−). We isolated a spontaneous Ara; mutant, designated SFE4548;, that was used as the common competitor in the fitness assays described below. The Ara marker is selectively neutral in the conditions used in thus study, as described below.
2.2 Construction of parasite-bearing strains
As the parasite in this study, we used a horizontally transmissible plasmid derived from the F episome, pTP102 (jP145::Gmr) (Cooper & Heinemann 2005). The F episome is a naturally occurring parasite of E. coli (Stewart & Levin 1977) E. coli strain DH5α/pTP102 was used as the donor to transfer the parasite to the 216 mutants as well as to REL4548 and SFE4548;. Conjugations were performed as previously described (Cooper & Heinemann 2000). Recipients infected with the parasite were identified by plating on Luria-Bertani (LB) agar containing antibiotics streptomycin (Sm) (100μgml−1), to which only recipient cells were resistant, and gentamicin (Gm) (5μgml−1), to which only parasite-infected cells were resistant. Parasite-infected recipients were streaked on LB agar supplemented with Sm and Gm, and a single colony was used to inoculate Davis minimal (DM) medium supplemented with 250μgml−1 glucose. Cultures were grown overnight and then stored at −80°C following addition of glycerol. All strains were checked for the presence of the appropriate antibiotic-resistance markers.
The reduction in fitness associated with pTP102 infection was determined by competitions (see below) between REL4548/pTP102 and SFE4548; performed with 10-fold replication. The fitness of infected cells was 7.96% lower than their uninfected counterparts (t9=4.6208, one-tailed p=0.0006). Therefore, this parasite has a substantial deleterious effect on host fitness.
To ensure that independent parasite infection events had the same effect on host fitness, we performed six separate conjugations between DH5α/pTP102 and REL4548 to obtain six independent REL4548/pTP102 clones. The fitness of each REL4548/pTP102 isolate was measured with fivefold replication against the same SFE4548;/pTP102 clone, as described below. No variation in the cost of independent infection events was detected (one-way ANOVA: F5,24=0.4464, p=0.8116). We also performed a similar experiment with two independent conjugations for each of seven mutant strains, and ran fitness assays with, on average, almost 11-fold replication. Again, there was no significant variation in fitness among independent infection events (nested ANOVA: F7,137=1.7036, p=0.1130).
2.3 Culture conditions and fitness assays
The fitness of each non-parasitized mutant genotype was measured in competition against the unmutated reference strain SFE4548;. Competition conditions were the same as those in the long-term evolution experiment except that, in this study, competitions were performed in unshaken 96-well plates, whereas the long-term evolution experiment used shaken flasks. The fitness of each infected mutant was measured against the parasitized derivative of the reference strain, SFE4548;/pTP102. Before each fitness assay, the two competitors (a mutant and the appropriate reference strain) were acclimatized to the competition environment by growing them separately under the same conditions as used for the competition. Each competitor was then diluted 200-fold into fresh DM medium supplemented with glucose at 25μgml−1 (Lenski et al. 1991), and a sample immediately plated on tetrazolium arabinose (TA) agar to estimate the initial densities of the competing strains. Ara− and Ara; cells are distinguished by their red and white colonies, respectively, on TA agar (Lenski et al. 1991). After each day of competition, the population was diluted 100-fold into fresh DM25, allowing the mixed population to grow 100-fold, which is equivalent to 6.64 generations per day. After t days of competition (usually t=2, but some competitions were performed over 1 or 4 days for genotypes with very low or high fitness values, respectively), a sample was plated on TA agar to obtain the final densities of the competitors. The fitness of the mutant relative to its Ara; competitor is calculated as W=ln(100tMt/M0/ln(100tPt/P0), where M and P represent the densities of the Ara− mutant and the Ara; progenitor at the start of the competition and after t days, respectively. Relative fitness is thus defined as the ratio of the population growth rates achieved by two genotypes during their direct competition. All fitness assays were replicated fourfold. Five additional replicates were performed for those mutants that had a coefficient of variation among the first four replicates above 5%. Finally, for each numerical class of mutants (carrying 1, 2 or 3 mutations), the five genotypes with the lowest and highest values were further replicated six- and fourfold, respectively. To minimize the influence of any outlying measurements, the median value of all the replicate assays for a particular mutant was used as the fitness estimate for that mutant when performing the overall regression analyses; this conservative approach is appropriate because the 216 mutant genotypes (and not the assays) are the fundamental units of replication in our study.
At the end of some competitions, strains infected with the parasite were tested on TA agar plates supplemented with Gm (to which the plasmid–parasite confers resistance) to check for retention of the parasite during the fitness assays. The parasite was retained in all 400 colonies thus screened.
Previous work showed that fitness effects of insertions were caused by disruption of specific genes at the insertion site, rather than by the addition of the antibiotic-resistance gene encoded by the mini-Tn10 (Elena et al. 1998). We also confirmed that the Ara marker was neutral in both the uninfected and infected states: the fitness of REL4548 relative to SFE4548; was 1.0036±0.0058 s.e.m. (H0: W=1; t9=0.6303, p=0.5442); and the fitness of REL4548/pTP102 relative to SFE4548;/pTP102 was 0.9846±0.0083 s.e.m. (H0: W=1; t9=1.8434, p=0.0984). Therefore, any fitness differences between the mutants and their non-mutated progenitor can be attributed to the insertion mutations.
2.4 Statistical analyses
A simple way to quantify the average strength and direction of epistasis is by fitting the power function lnWk=−skβ to the data (Lenski et al. 1999; Wilke & Adami 2001; You & Yin 2002), where Wk is the mean fitness of genotypes with k mutations. For deleterious mutations, s>0 and is proportional to average severity. The form and strength of epistasis is defined by β. If mutations interact synergistically (β>1), then the average marginal effect of each additional mutation increases with mutation number. In that case, the equilibrium load of deleterious mutations is lower in sexual than in asexual populations, providing an advantage for sex (Kimura & Maruyama 1966; Crow 1970; Otto & Feldman 1997). If deleterious mutations interact multiplicatively (β=1) or antagonistically (0<β<1), then sex is not effective in reducing the equilibrium mutational load. The intercept is fixed at zero because fitness values are expressed relative to the unmutated progenitor. To maximize statistical power, it would be desirable to perform the regressions using each mutant as an independent observation. In practice, this approach has two technical difficulties: (i) zero-fitness mutants cannot be ln-transformed; and (ii) the variance in fitness is expected to increase with the number of mutations, violating an assumption of standard least-squares regression. To circumvent these difficulties, while retaining the statistical power corresponding to a sample of 216 mutant genotypes, we used Tukey’s jackknife procedure to obtain 216 quasi-independent estimates of s and β (Elena & Lenski 1997; Manly 1997).
3. Results
3.1 Test of synergistic epistasis
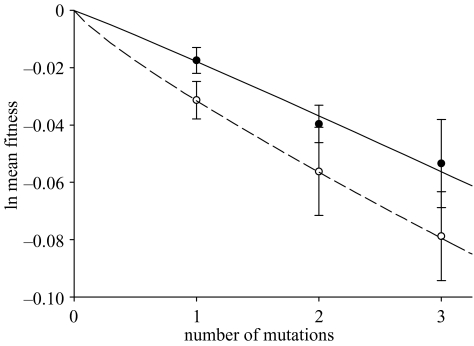
A requirement of the mutational deterministic hypothesis is that, on average, interactions between deleterious mutations are synergistic (Kondrashov (1982, 1984, 1985) but see also Otto & Feldman (1997)). To test this hypothesis, we therefore measured the effect on fitness of introducing known numbers of mutations into a reference genotype. The solid line in figure 1 shows the relationship between mutation number and average fitness. The power-function model explains 98.4% of the variation in sample means across the three mutational classes (F2,1=219.7465, p=0.0476). The jackknifed estimate of s is 0.0178±0.0045 s.e.m., a value significantly above zero (t215=3.9556, one-tailed p < 0.0001), which confirms that the mutations are deleterious on average. The corresponding estimate of β is 1.0481±0.3875 s.e.m., which does not differ significantly from 1 (t215=0.1241, p=0.9014). These results are consistent with previous work, also carried out in the absence of parasites, which found no overall tendency for deleterious mutations to interact synergistically in this system (Elena & Lenski 1997).
Figure 1.
Decline in ln-transformed mean fitness with increasing numbers of mutations. The filled circles and solid curve are parasite-free mutants relative to the non-mutated reference strain. The open circles and dashed curve are parasitized mutants relative to the parasitized non-mutated reference strain. Each curve shows the best fit of a power model to the data, with the intercept constrained to a ln-transformed fitness of zero; however, neither curve differs significantly from a log-linear model (see § 3a). Error bars show s.e.m. calculated using the jackknife method (Elena & Lenski 1997).
Next, we sought to test whether the presence of parasites affected the pattern of epistasis among deleterious mutations. The relationship between mutation number and average fitness for parasitized mutants is shown by the dashed line in figure 1. The power-function regression explains more than 99.9% of the variation in sample means among the mutational classes (F2,1=620 234.2165, p=0.0009). The jackknifed estimate of s is 0.0315±0.0067 s.e.m., which is again significantly greater than zero (t215=4.7015, one-tailed p < 0.0001). The estimate of the epistatic term β is 0.8428±0.2909 s.e.m. which does not differ significantly from 1 (t215=0.5404, p=0.5895). In fact, the direction of epistasis tends towards the opposite direction of that required by the hypothesis that parasites promote synergistic epistasis of deleterious mutations. Of course, it is possible that our experiment lacked sufficient statistical power to detect synergistic epistasis (β>1). In this regard, our conclusion is limited by the fact that the maximum number of mutations was three. However, contributing to statistical power, our sample sizes were large, with 72 mutants in each of the three mutational classes.
3.2 Synergy between parasitism and mutational load
The results in § 3a indicate that there is no overall tendency for synergistic epistasis among deleterious mutations in either the presence or absence of the parasite. Nonetheless, the parasite had a large influence on the fitness effects associated with the mutations. The average severity of the deleterious mutations was ca. 77% greater in parasitized than in non-parasitized cells, and this difference is highly significant (paired t-test: t215=3.8773, one-tailed p < 0.0001). It is important to emphasize that this difference does not simply reflect the burden of infection by the parasite, because the fitness of parasitized mutants was measured in competition with the parasitized progenitor, whereas the fitness of non-parasitized mutants was measured in competition with the non-parasitized progenitor. This result therefore shows that the parasite interacts synergistically with the harmful mutations to increase their average severity.
The increased average severity of deleterious mutations in the presence of the parasite could arise in two different ways. One possibility is that the parasite might cause a consistent change in the severity of all deleterious mutations. Alternatively, parasites might interact inconsistently with the various mutations, such that the severity of some mutations increases in the presence of the parasite, while the severity of other mutations does not change or perhaps is even reduced by the parasite. To distinguish between these possibilities, we first performed a two-way ANOVA examining the joint effects of the parasite and mutant genotype. The interaction term is highly significant (table 1: p < 0.0001), which indicates that the effect of the parasite varied among the mutants. To examine further the pattern of this variation, we then performed t-tests comparing the fitness of parasitized and non-parasitized derivatives of each of the 216 mutants relative to the parasitized and non-parasitized forms of the progenitor. In 45 cases (20.8%), mutational effects were significantly aggravated by the parasite. More surprisingly, in 14 cases (6.5%) the mutational effects were significantly alleviated in the presence of the parasite. (Again, it must be emphasized that such alleviation does not imply that the parasite increased fitness, but rather that the parasite reduced the severity of the fitness decrement associated with deleterious mutations.) With 216 tests performed, one would expect ca. 5% of them, or ca. 11 cases, to be ‘significant’ by chance alone, whereas the observed number of significant effects greatly exceeded that level. Even the 14 cases in which the parasite significantly alleviated the harmful mutational effect exceed the 2.5% of cases that, by chance, might appear ‘significant’ in that one direction (binomial test, p=0.0012). Thus, the increased average severity of deleterious mutations in the presence of the parasite was caused by a combination of many cases in which the parasite had a large aggravating effect, many cases in which the parasite had little or no effect, and a few cases in which the parasite alleviated the mutational effects.
Table 1.
Two-way ANOVA examining the fitness effects of parasite infection, mutant genotype and their interaction.
(Mixed-model ANOVA, with parasite infection a fixed effect and mutant genotype a random effect. The fixed effect was tested over the mean squares (MS) interaction, and the random and interaction effects were tested over the MS error. The error d.f. reflect unequal sample sizes for fitness assays performed for the various mutants, as explained in § 2c.)
| source | d.f. | MS | F | p |
| parasite infection | 1 | 0.1310 | 8.5955 | 0.0037 |
| mutant genotype | 215 | 0.0494 | 19.0361 | <0.0001 |
| interaction | 215 | 0.0152 | 5.8760 | <0.0001 |
| error | 1843 | 0.0026 | — | — |
4. Discussion
We measured the fitness of 216 genotypes bearing one, two or three mutations in both the absence and presence of a plasmid parasite. We found no evidence for an overall excess of synergistic epistasis in either case (figure 1). On average, the parasite tended to aggravate the harmful effects of deleterious mutations (figure 1). However, the effect of the parasite depended on the particular genotype (table 1), with infection ameliorating the severity of some mutations while aggravating the harmful effects of others.
The finding that parasite infection does not promote synergistic epistasis between deleterious mutations in this system fails to support one particular pluralistic hypothesis for the evolution of sex, in which it has been proposed that parasites might interact with synergistically epistatic deleterious mutations to increase the strength of selection favouring sexual recombination (West et al. 1999). Nevertheless, our finding that parasite infection increases the average severity of deleterious mutations is relevant to another model in which the advantage to sex accrues as a consequence of the ameliorating effect of recombination on mutation accumulation. The basis of this model is genetic drift, which can cause the stochastic loss of the fittest genotype from a finite population via a process that has become known as Muller’s ratchet. In the absence of back-mutation, the fittest genotype can be regained only by genetic recombination, which provides a potential advantage to sex in opposing the detrimental advance of the ratchet (Muller 1964; Felsenstein 1974). A key parameter determining the influence of Muller’s ratchet is the average severity of deleterious mutations (Haigh 1978; Gabriel et al. 1993; Stephan et al. 1993). Therefore, if parasites increase the average severity of deleterious mutations, then the ratchet should cause a greater reduction in mean fitness of parasitized populations, thereby broadening the conditions over which opposing the ratchet provides an advantage for sex (Howard & Lively 1994; West et al. 1999). Also, the observation that parasites seem to disproportionately aggravate the fitness effects of a subset of deleterious mutations may accelerate the rate at which the ratchet would operate. In a theoretical study, Gordo & Charlesworth (2001) found that, in the absence of recombination, mildly deleterious mutations accumulated more rapidly in the presence of strongly deleterious mutations; this effect was attributed to a reduction in effective population size and, consequently, an increase in the rate at which the ratchet drives the accumulation of mildly deleterious mutations (Charlesworth et al. 1993).
Howard & Lively (1994) proposed a model that encompasses the interaction of Red Queen coevolutionary dynamics of host and parasite populations with Muller’s ratchet. Considering these factors together overcame certain limitations inherent in the separate models by increasing the strength of Muller’s ratchet in larger populations, and by increasing the advantage of sexual reproduction under the Red Queen model (Howard & Lively 1994, 1998). An increase in the average severity of deleterious mutations extended the parameter space in which sex was predicted to be advantageous (Howard & Lively 1994). The results of our study indicate that such an increase in mutational severity for the host can occur as a consequence of parasite infection.
Our finding that parasite infection tends to exacerbate the severity of deleterious mutations is consistent with some previous studies on the interaction between parasites and mutational load (Stevens et al. 1997; Coltman et al. 1999), although other studies have reported variable effects (Wiehn et al. 2002; Haag et al. 2003; Salathé & Ebert 2003). In a broader context, our findings also tend to support previous studies that the average severity of deleterious mutations is greater in stressful than in non-stressful environments, as reported for Saccharomyces cerevisiae (Szafraniec et al. 2001), Caenorhabditis elegans (Vassilieva et al. 2000) and Drosophila melanogaster (Shabalina et al. 1997). However, a recent study using E. coli found variable results, including alleviation as well as aggravation of average mutational severity, depending on the particular stress that was imposed (Kishony & Leibler 2003). The reason for the differences between environmental stresses in the direction of their effects on mutational severity remains unclear at present (Elena & de Visser 2003). In any case, our finding that the magnitude and even direction of the fitness effect of a particular parasite varies among mutations indicates that many mutant genotypes must be tested to understand the pattern of interaction between deleterious mutations and even a single parasite. Also, it has been suggested that synergistic epistasis may predominate only under conditions of severe competition (Peck & Waxman 2000). The bacteria in our experiments clearly competed for limiting glucose, but whether the competition was sufficiently severe might be debated. Finally, we have performed experiments with an asexual organism to test hypotheses concerning the evolution of sex. Our justification for using an asexual organism is threefold: (i) sex must originally have evolved from an asexual ancestral state; (ii) we are broadly interested in the form of mutational interactions, especially in relation to parasitism; and (iii) E. coli offers many important advantages for performing precise and rigorous experiments.
In summary, we tested the hypothesis that the interaction between infection by a parasite and deleterious mutations would exacerbate synergistic epistasis among the deleterious mutations, thereby extending the conditions under which the mutational–deterministic model predicts an advantage to sex. We found that, on average, there was no tendency towards an excess of synergistic epistasis among deleterious mutations in either parasitized or non-parasitized E. coli cells. However, the average mutational severity was significantly greater in the presence of the parasite, indicating synergy between the harmful effects of parasitism and mutations. This result provides support for an alternative model in which the advantage for sexual reproduction derives from combining its effect in opposing Muller’s ratchet with Red Queen coevolutionary dynamics between hosts and parasites (Howard & Lively 1994).
Acknowledgments
We thank A. de Visser, Y. Michalakis, J. Peck, D. Rozen and G. Velicer for helpful comments on the manuscript, and N. Hajela for excellent technical support. This research was supported by a USA NSF grant to R.E.L. and by a Spanish MEC-FEDER grant to S.F.E.
References
- Bell G. The masterpiece of nature. University of California Press; San Francisco, CA: 1982. [Google Scholar]
- Charlesworth B., Morgan M.T., Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coltman D.W., Pilkington J.G., Smith J.A., Pemberton J.M. Parasite-mediated selection against inbred soay sheep in a free-living, island population. Evolution. 1999;53:1259–1267. doi: 10.1111/j.1558-5646.1999.tb04538.x. [DOI] [PubMed] [Google Scholar]
- Cooper T.F., Heinemann J.A. Transfer of conjugative plasmids and bacteriophage λ occurs in the presence of antibiotic that prevent de novo gene expression. Plasmid. 2000;43:171–175. doi: 10.1006/plas.1999.1450. [DOI] [PubMed] [Google Scholar]
- Cooper, T. F. & Heinemann, J. A. 2005 Selection for plasmid postsegregational killing depends on multiple infection: evidence for selection of more virulent parasites through parasite-level competition. Proc. R. Soc. B272 (In the press.) (doi:10.1098/rspb.2004.2921) [DOI] [PMC free article] [PubMed]
- Crow J.F. Genetic load and the cost of natural selection. In: Kijima K.I., editor. Mathematical topics in population genetics. Springer; New York: 1970. pp. 128–177. [Google Scholar]
- de la Peña M., Elena S.F., Moya A. Effect of deleterious mutation-accumulation on the fitness of RNA bacteriophage MS2. Evolution. 2000;54:686–691. doi: 10.1554/0014-3820(2000)054[0686:eodmao]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- de Visser J.A.G.M., Hoekstra R.F. Synergistic epistasis between loci affecting fitness: evidence in plants and fungi. Genet. Res. Camb. 1998;71:39–49. [Google Scholar]
- de Visser J.A.G.M., Hoekstra R.F., Van den Ende H. The effect of sex and deleterious mutations on fitness in Chlamydomonas. Proc. R. Soc. B. 1996;263:193–200. [Google Scholar]
- de Visser J.A.G.M., Hoekstra R.F., Van den Ende H. Test of interaction between genetic markers that affect fitness in Aspergillus nidulans. Evolution. 1997a;51:1499–1505. doi: 10.1111/j.1558-5646.1997.tb01473.x. [DOI] [PubMed] [Google Scholar]
- de Visser J.A.G.M., Hoekstra R.F., Van den Ende H. An experimental test for synergistic epistasis and its application in Chlamydomonas. Genetics. 1997b;145:815–819. doi: 10.1093/genetics/145.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J.W., Charlesworth B., Charlesworth D., Crow J.F. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena S.F. Little evidence for synergism among deleterious mutations in a non-segmented RNA virus. J. Mol. Evol. 1999;49:703–707. doi: 10.1007/pl00000082. [DOI] [PubMed] [Google Scholar]
- Elena S.F., de Visser J.A.G.M. Environmental stress and the effects of mutations. J. Biol. 2003;2:12. doi: 10.1186/1475-4924-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena S.F., Lenski R.E. Test of synergistic interactions among deleterious mutations in bacteria. Nature. 1997;390:395–398. doi: 10.1038/37108. [DOI] [PubMed] [Google Scholar]
- Elena S.F., Ekunwe L., Hajela N., Oden S.A., Lenski R.E. Distribution of fitness effects caused by random insertion mutations in Escherichia coli. Genetica. 1998;102–103:349–358. [PubMed] [Google Scholar]
- Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974;78:737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel W., Lynch M., Burger R. Muller’s ratchet and mutational meltdowns. Evolution. 1993;47:1744–1757. doi: 10.1111/j.1558-5646.1993.tb01266.x. [DOI] [PubMed] [Google Scholar]
- Gordo I., Charlesworth B. The speed of Muller’s ratchet with background selection, and the degeneration of Y chromosomes. Genet. Res. Camb. 2001;78:149–161. doi: 10.1017/s0016672301005213. [DOI] [PubMed] [Google Scholar]
- Haag C.R., Sakwińska O., Ebert D. Test of synergistic interaction between infection and inbreeding in Daphnia magna. Evolution. 2003;57:777–783. doi: 10.1111/j.0014-3820.2003.tb00289.x. [DOI] [PubMed] [Google Scholar]
- Haigh J. The accumulation of deleterious genes in a population: Muller’s ratchet. Theor. Popul. Biol. 1978;14:251–267. doi: 10.1016/0040-5809(78)90027-8. [DOI] [PubMed] [Google Scholar]
- Howard R.S., Lively C.M. Parasitism, mutation accumulation and the maintenance of sex. Nature. 1994;367:554–557. doi: 10.1038/367554a0. [DOI] [PubMed] [Google Scholar]
- Howard R.S., Lively C.M. The maintenance of sex by parasitism and mutation accumulation under epistatic fitness functions. Evolution. 1998;52:604–610. doi: 10.1111/j.1558-5646.1998.tb01658.x. [DOI] [PubMed] [Google Scholar]
- Keightley P.D., Eyre-Walker A. Deleterious mutations and the evolution of sex. Science. 2000;290:331–333. doi: 10.1126/science.290.5490.331. [DOI] [PubMed] [Google Scholar]
- Keightley P.D., Lynch M. Toward a realistic model of mutations affecting fitness. Evolution. 2003;57:683–685. doi: 10.1111/j.0014-3820.2003.tb01561.x. [DOI] [PubMed] [Google Scholar]
- Kimura M., Maruyama T. The mutational load with epistatic gene interactions in fitness. Genetics. 1966;54:1337–1351. doi: 10.1093/genetics/54.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishony R., Leibler S. Environmental stress can alleviate the average deleterious effect of mutations. J. Biol. 2003;2:14. doi: 10.1186/1475-4924-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Bender J., Gottesman S. Uses of transposons with emphasis on Tn10. Meth. Enzymol. 1991;204:139–181. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- Kondrashov A.S. Selection against harmful mutations in large sexual and asexual populations. Genet. Res. Camb. 1982;40:325–332. doi: 10.1017/s0016672300019194. [DOI] [PubMed] [Google Scholar]
- Kondrashov A.S. Deleterious mutations as an evolutionary factor. I. The advantage of recombination. Genet. Res. Camb. 1984;44:199–217. doi: 10.1017/s0016672300026392. [DOI] [PubMed] [Google Scholar]
- Kondrashov A.S. Deleterious mutations as an evolutionary factor. II. Facultative apomixis and selfing. Genetics. 1985;111:635–653. doi: 10.1093/genetics/111.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov A.S. Deleterious mutations and the evolution of sexual reproduction. Nature. 1988;336:435–440. doi: 10.1038/336435a0. [DOI] [PubMed] [Google Scholar]
- Kondrashov A.S. Classification of hypotheses on the advantage of amphimixis. J. Heredity. 1993;84:372–387. doi: 10.1093/oxfordjournals.jhered.a111358. [DOI] [PubMed] [Google Scholar]
- Lenski R.E. Phenotypic and genomic evolution during a 20 000-generation experiment with the bacterium Escherichia coli. Pl. Breeding Rev. 2004;24:225–265. [Google Scholar]
- Lenski R.E., Travisano M. Dynamics of adaptation and diversification: a 10 000-generations experiment with bacterial populations. Proc. Natl. Acad. Sci. USA. 1994;91:6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski R.E., Rose M.R., Simpson S.C., Tadler S.C. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2000 generations. Am. Nat. 1991;138:1315–1341. [Google Scholar]
- Lenski R.E., Ofria C., Collier T.C., Adami C. Genome complexity, robustness and genetic interactions in digital organisms. Nature. 1999;400:661–664. doi: 10.1038/23245. [DOI] [PubMed] [Google Scholar]
- Lively C.M., Howard R.S. Selection by parasites for clonal diversity and mixed mating. Phil. Trans. R. Soc. B. 1994;346:271–281. doi: 10.1098/rstb.1994.0144. [DOI] [PubMed] [Google Scholar]
- Manly B.F.J. Randomization, bootstrap and Monte Carlo methods in biology. Chapman & Hall; London: 1997. [Google Scholar]
- Maynard Smith J. The evolution of sex. Cambridge University Press; 1978. [Google Scholar]
- Muller H.J. The relation of recombination to mutational advance. Mut. Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Otto S.P., Feldman M.W. Deleterious mutations, variable epistatic interactions, and the evolution of recombination. Theor. Popul. Biol. 1997;51:134–147. doi: 10.1006/tpbi.1997.1301. [DOI] [PubMed] [Google Scholar]
- Peck J.R., Waxman D. Mutation and sex in a competitive world. Nature. 2000;406:399–404. doi: 10.1038/35019055. [DOI] [PubMed] [Google Scholar]
- Peters A.D., Lively C.M. The Red Queen and fluctuating epistasis: a population genetic analysis of antagonistic coevolution. Am. Nat. 1999;154:393–405. doi: 10.1086/303247. [DOI] [PubMed] [Google Scholar]
- Salathé P., Ebert D. The effect of parasitism and inbreeding on the competitive ability of Daphnia magna: evidence for synergistic epistasis. J. Evol. Biol. 2003;16:976–985. doi: 10.1046/j.1420-9101.2003.00582.x. [DOI] [PubMed] [Google Scholar]
- Shabalina S.A., Yampolsky L.Y., Kondrashov A.S. Rapid decline of fitness in panmictic populations of Drosophila melanogaster maintained under relaxed natural selection. Proc. Natl. Acad. Sci. USA. 1997;94:13034–13039. doi: 10.1073/pnas.94.24.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W., Chao L., Smale J.G. The advance of Muller’s ratchet in a haploid asexual population: approximate solutions based on diffusion. Genet. Res. Camb. 1993;61:225–231. doi: 10.1017/s0016672300031384. [DOI] [PubMed] [Google Scholar]
- Stevens L., Guiyun Y., Pray L.A. Consequences of inbreeding on invertebrate host susceptibility to parasitic infections. Evolution. 1997;51:2032–2039. doi: 10.1111/j.1558-5646.1997.tb05126.x. [DOI] [PubMed] [Google Scholar]
- Stewart F.M., Levin B.R. The population biology of bacterial plasmids: a priori conditions for the existence of conjugationally transmitted factors. Genetics. 1977;87:209–228. doi: 10.1093/genetics/87.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafraniec K., Borts R.H., Korona R. Environmental stress and mutational load in diploid strains of the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2001;98:1107–1112. doi: 10.1073/pnas.021390798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilieva L.L., Hook A.M., Lynch M. The fitness of spontaneous mutation in Caenorhabditis elegans. Evolution. 2000;54:1234–1246. doi: 10.1111/j.0014-3820.2000.tb00557.x. [DOI] [PubMed] [Google Scholar]
- West S.A., Peters A.D., Barton N.H. Testing for epistasis between deleterious mutations. Genetics. 1998;149:435–444. doi: 10.1093/genetics/149.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S.A., Lively C.M., Read A.F. A pluralistic approach to sex and recombination. J. Evol. Biol. 1999;12:1003–1012. [Google Scholar]
- Wiehn J., Kopp K., Rezzonico S., Karttunen S., Jokela J. Family-level covariation between parasite resistance and mating system in a hermaphroditic freshwater snail. Evolution. 2002;56:1454–1461. doi: 10.1111/j.0014-3820.2002.tb01457.x. [DOI] [PubMed] [Google Scholar]
- Wilke C.O., Adami C. Interaction between directional epistasis and average mutational effects. Proc. R. Soc. B. 2001;268:1469–1474. doi: 10.1098/rspb.2001.1690. doi:10.1098/rspb.2001.1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloch D.M., Borts R.H., Korona R. Epistatic interactions of spontaneous mutations in haploid strains of the yeast Saccharomyces cerevisiae. J. Evol. Biol. 2001;14:310–316. [Google Scholar]
- You L., Yin J. Dependence of epistasis on environment and mutation severity as revealed by in silico mutagenesis of phage T7. Genetics. 2002;160:1273–1281. doi: 10.1093/genetics/160.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]