Abstract
The restriction mapping of a massive number of individual DNA molecules by optical mapping enables assembly of physical maps spanning mammalian and plant genomes; however, not through computational means permitting completely de novo assembly. Existing algorithms are not practical for genomes larger than lower eukaryotes due to their high time and space complexity. In many ways, sequence assembly parallels map assembly, so that the overlap–layout–consensus strategy, recently shown effective in assembling very large genomes in feasible time, sheds new light on solving map construction issues associated with single molecule substrates. Accordingly, we report an adaptation of this approach as the formal basis for de novo optical map assembly and demonstrate its computational feasibility for assembly of very large genomes. As such, we discuss assembly results for a series of genomes: human, plant, lower eukaryote and bacterial. Unlike sequence assembly, the optical map assembly problem is actually more complex because restriction maps from single molecules are constructed, manifesting errors stemming from: missing cuts, false cuts, and high variance of estimated fragment sizes; chimeric maps resulting from artifactually merged molecules; and true overlap scores that are “in the noise” or “slightly above the noise.” We address these problems, fundamental to many single molecule measurements, by an effective error correction method using global overlap information to eliminate spurious overlaps and chimeric maps that are otherwise difficult to identify.
Keywords: whole-genome shotgun optical mapping, map assembler
The optical mapping system developed by Schwartz and colleagues (1, 2) constructs genome-wide ordered restriction maps through assembly of individual DNA molecules (genomic) cleaved by a restriction enzyme. Cleavage events on single DNA molecules are imaged by fully automated fluorescence microscopy as visible gaps (≈1 μm) on elongated DNA molecules. The combination of a charged glass surface and fluid flow guided by a microfluidic device (3) simultaneously elongates and deposits DNA random chains as well defined stripes within the device. Because a critical density of charge is maintained on these surfaces, absorbed and elongated molecules under tension uniquely “flag” restriction enzyme cleavage sites as visible gaps formed due to relaxation of adjacent DNA. The distance, or mass of each consecutive restriction fragment is determined by integrated fluorescence intensity measurements against an internal standard. Collectively, these actions produce oriented, labeled molecules that work in concert with downstream image processing, yielding a massive set of restriction maps as relatively compact data files. Due to the enormous throughput of this system, a genome is redundantly spanned by individual restriction maps supporting “shotgun” assembly techniques for whole genome analysis. However, genome assembly is inherently complicated by the fact that measurements are made on random individual DNA molecules, which cannot benefit from averaging steps intrinsic to bulk measurement techniques used by common DNA sequencing platforms; no amplification step is used during optical mapping. This finding places another level of complexity within the genome assembly step, where further error reduction must be must be performed after acquisition. Such errors are characterized as: (i) spurious, or false restriction sites, (ii) partial digestion, or missing cuts (where restriction sites are not observed in optical maps), (iii) small fragments (<2 kb) are underrepresented in maps, (iv) sizing error, and (v) chimeric maps that result from images of ambiguously overlapping DNA molecules.
Ordered restriction maps reveal structural detail across a genome in ways that are only surpassed by DNA sequence data, and recent findings show prevalent structural variations in human populations (4, 5) with many loci linked to diseases. Also, cancer genomes are notoriously rife with aneuploidy and structural aberrations fostered by unchecked genomic instability, which when fully characterized at high-resolution present new routes for diagnostics (6) and treatment. Our current techniques for discovery of structural alterations are somewhat bound by the limitations imposed by DNA hybridization or cost (sequencing). For example, genomic microarrays do reveal deletions (7), but are confounded by common genomic repeats and cannot discern inversions (8). Furthermore, insertions and other genomic events not represented on a chip array cannot be assayed and go undiscovered. As such, ordered restriction maps broadly reveal genome structural events potentiating their discovery and physical characterization in one step. Accordingly, the scalable de novo assembly approach presented here will greatly facilitate the construction of physical maps for this emerging field of human and tumor biology.
Prior Optical Mapping Algorithms
The optical map assembly problem proved to be a challenging task because optical mapping employs measurements performed on individual molecules. Several research groups have worked on restriction map reconstruction algorithms (9–12).
Some formulations of this problem were demonstrated to be NP-hard (12), whereas others allowed polynomial time algorithms (11). These algorithms were designed for reconstruction of short restriction maps using cloned DNA substrates. These methods produced accurate consensus restriction maps but could not be applied to “shotgun” optical mapping data, which is most typical form of data in current optical mapping system. In “shotgun” optical mapping, maps of DNA molecules are produced by random shearing of genomic DNA. This implies that optical maps represent random parts of genome rather than identical DNA molecules.
Ananthraman et al. designed a Bayesian method that could accommodate shotgun optical mapping data by searching over a large space of order assignments, but their algorithm had deficiencies in terms of scalability to large genomes.¶ Consequently, application of this algorithm to genome assemblies more complex than bacteria required additional extensive ad hoc approaches.
As such, there is a need for new algorithms that are specifically designed for handling many computational issues inherent to the assembly of large genomes, such as plant and mammalian. Here, we approach optical map assembly problem by using an approach that is quite different from existing restriction map reconstruction algorithms. Our method utilizes an overlap–layout–consensus strategy commonly used in existing DNA sequence assemblers because it proved to be practical even for assembling sequence reads from very large genomes. As a key step of our assembly method, we use a highly efficient error correction method to eliminate false positive overlaps and chimeric maps that otherwise render assembly problem highly ambiguous.
DNA Sequence Assemblers
Most existing DNA assembly methods use a three-step computational framework termed overlap–layout–consensus. In this framework, sequence read connectivity is established by the overlap step; then local and global connectivity is combined into assembly contigs and scaffolds and their relative order/orientation is assigned in the layout step; finally, finished sequence contigs are computed in the consensus step. Such an approach is used in the Celera assembler (13), CAP (14), and ARACHNE (15). All of these assemblers also incorporate a number of sophisticated error correction techniques to ensure high accuracy of the finished sequence. Alternative sequence assembly tools such as Euler (16) rely heavily on tuple matches underlying multiple sequence alignment and infer the consensus sequence from the tuple graph.
An Optical Map Assembly Method
Here, we describe an algorithm for the whole-genome de novo assembly of optical maps. The general idea behind the method is to represent significant overlaps of optical maps as a connectivity graph and then apply a three-type error correction method to eliminate errors ubiquitous in that overlap graph. Components of the graph corresponding to genomic regions represented by connected optical maps are explored to construct a draft consensus map with approximate positions of most restriction sites. We then employ a refinement procedure to correct draft map inaccuracies and report a consensus restriction map. To our knowledge, this is the first whole-genome map assembly tool with feasible computational complexity and space requirement. Here we report on the details of the method as well as some assemblies. To date, we have performed assembly of several bacterial genomes (≈5 Mb), one microbial genome (34.5 Mb), one plant genome (430 Mb), and one human genome. Comparison of the two bacterial maps produced by our map assembly method to the known DNA sequences confirms the high accuracy of our method. The overlap structure that we employ for representation of region connectivity is capable of accommodating assemblies from polymorphic genomic regions such as those found in diploid organisms and populations of tumor cells with highly aberrant genomes.
Optical Mapping Measurements
Typically, we use restriction enzymes with a 6-bp cognate recognition sequence or, for mammalian genomes, enzymes that are CpG methylation insensitive. Each optical map is represented by an array of fragment sizes in the order they are determined on a given molecule. Individual optical maps range from 350 kb to 4 Mb in total size, typically bearing 30 restriction fragments. About 20% of restriction sites are not observed in a given optical map due to imperfect digestion. Also, about three false cuts per 1 Mb of DNA are usually present at random positions. Most fragments <500 bp are not observed in our data, and fragments under 2 kb are generally underrepresented. Sizes of restriction fragments X typically have normal distribution (X ≈ N(Y, σ2Y) for σ2 ≈ 0.3), where Y represents the true genomic size of corresponding region of DNA (17). This finding implies, for example, that for a 20-kb DNA fragment, 80% of the measurements are within 3.3 kb of 20 kb.
Results
We evaluated the capabilities of our optical map assembly method, by performing unsupervised de novo assembly of several genomes using experimental results from a series organisms of increasing genomic complexity and size (Table 1).
Table 1.
Summary of de novo assemblies for several organisms
| Genome | Genome size, Mb | Enzyme | Average genomic fragments size, kb | No. of optical maps | Over-sampling | Overlap calculation, h | Layout-consensus calculation, h | Refinement calculation, h | Overlap yield | Contig yield (mass, Mb) | % of genome covered | Contigs aligned to ref. (mass, Mb) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Y. pestis | 4.6 | XhoI | 17.3 | 251 | 49× | 0.1 | 0.01 | 0.1 | 691 | 1 (4.8) | 100 | 1 (4.8) |
| E. coli | 4.6 | XhoI | 26 | 6,750 | 708× | 31 | 1 | 1 | 184,522 | 1 (4.6) | 100 | 1 (4.6) |
| T. pseudonana | 34.5 | NheI | 9.8 | 34,460 | 435× | 2,400 | 1 | 6 | 254,978 | 46 (38) | 100 | 18 (6.4) |
| O. sativa | 430 | NheI | 11.8 | 260 × 103 | 268× | 96 × 103 | 1.2 | 1 | 8 × 106 | 307 (236) | 52 | 288 (225) |
| Human CHM | 3,200 | SwaI | 13.5 | 213 × 103 | 30× | 57 × 103 | 1 | 2 | 12 × 106 | 219 (150) | 4.6 | 171 (109) |
Our first map assembly attempts focused on small bacterial genomes, Yersinia pestis strain KIM genome and Escherichia coli (strain K12), with results closely compared with their reference maps (in silico maps). The reference map was obtained by the restriction digestion, in silico, of the published sequence (18, 19) with the same enzyme used for optical map construction (20). Looking at Table 1, the first step of calculating all pairwise overlaps is the most computationally intensive of all, quadratic in the number of optical maps and requiring extensive computer resources. However, subsequent steps, layout, consensus, and refinement, are fast, requiring no more than 60 min on a single 3-GHz desktop computer. We assessed the quality of the draft and refined consensus maps by their alignments to corresponding DNA sequence for tabulation of errors consisting of missing fragments, false, or missing restriction sites, and restriction fragment size discrepancy. For Y. pestis, the draft assembly contained 30 missing and 12 false cuts, which after the refinement step (using the entire optical map data set) reduced to only one missing cut and no false cuts. Also, only six small restriction fragments (<2 kb) were missing after the refinement, reflecting their known underrepresentation in optical maps (21). The E. coli strain K12 assembly benefited from a large number of optical maps (6,750), yielding 184,522 accurate overlaps that were combined in the overlap graph producing a draft map spanning the entire genome. After the refinement, the final assembly contained 4,352 maps (≈500× coverage), and map to sequence alignments showed no false or missing cuts and only seven very small, missing restriction fragments (0.1–0.7 kb).
In terms of map assembly, the greater genome size and complexity of eukaryotic genomes dictate that significantly larger and more informative sets of optical maps must be considered. Accordingly, assemblies for Thalassiosira pseudonana, Oryza sativa ssp. japonica (rice), and Homo sapiens used only optical maps containing 15 or more restriction fragments as a strict filter for map quality, commesurate with the current throughput of the optical mapping system, to reduce the amount of computation while ensuring sufficient genome coverage. Looking at Table 1, we see very long computational times for calculating the overlaps for T. pseudonana and rice, but subsequent layout-consensus and refinement steps are rapid; taking only ≈1 h on a desktop computer. In terms of assembly accuracy of O. sativa ssp. japonica genome, of maps which aligned with the threshold to the reference genome, 91% of their total length matched by using local and gapped-local alignment. The nonaligned part concentrated most often in the end of the maps due to lower optical map coverage. Details of the alignments revealed that ≈1.4% of contig restriction sites did not align to any restriction site in the sequence (this corresponds to ≈1.4 extra sites per 1 Mb of DNA sequence). Also, 13% of genomic DNA sequence restriction sites did not match to any contig site (in the original optical map data set, 20% of sites are missing). Also, the vast majority of fragments <1 kb was not represented by our consensus maps, and 1,107 of 1,575 fragments between 1 and 3 kb were missing in our consensus maps. Finally, nine of 288 contigs produced alignment patterns consistent with misassemblies (we should note, however, that the published rice genome may also contain misassemblies because it is not entirely finished).
Following the same steps as before, the human genome assembly (from a tumor sample derived from a complete hydatidiform mole; S. Reslewic, personal communication) produced 219 assembled contigs (150 Mb) containing 10 or more optical maps. These were compared through local and gapped alignment to NCBI human build 35 (22) showing 171 (109 Mb) of 219 (124 Mb) contigs aligning with high scores (q score, 11). We attribute a low amount of assembled contig mass to low effective coverage of this human data set. Specifically, even when optical maps are aligned to the reference genome, only 20% of these maps score above threshold. In terms of accuracy of the assembled contigs, we observed 81 extra sites (one extra site per 1.3 Mb of reference sequence), and 210 missing sites of 6,153 reference map sites. Also, vast majority of fragments under 1 kb was not represented by our consensus maps, and 420 of 645 fragments between 1 and 3 kb were not represented. Finally, two assembled contigs showed patterns that appeared to be possible misassemblies.
Discussion
In this paper, we have described an algorithm for whole-genome unsupervised de novo restriction map assembly using optical maps constructed from randomly sheared genomic DNA molecules. To our knowledge, this is the first algorithm capable of producing accurate restriction maps, using single DNA molecules, of very large genomes (such as human or rice) in feasible time, through the leveraging of increasingly available cluster computing resources. The uniqueness of the method is in the application of the overlap–layout–consensus strategy to the assembly of optical maps and in the effective distance-based error-elimination method. Together, these features enable “impossible” assemblies due to ubiquitous false overlaps created by local errors and chimerism found in optical maps. The main application of our method will be in the realm of “structural genomics,” where restriction maps reveal kilobase-sized alterations in test genomes as novel restriction sites, missing restriction sites, large indels (>5 kb), and complex rearrangements. When such alterations are assessed in populations, new structural polymorphisms will emerge; in cancer, new breakpoints will be discovered and characterized at high-resolution.
Methods
Optical maps from a target organism serve as input data for the whole-genome optical map assembly process, which computes a consensus restriction map of the genome. The assembly process consists of seven steps that we outline in this section. The method is designed in such way that the most computationally demanding step (calculation of pairwise overlaps) only needs to be done once, and the other less computationally intense steps use computed overlaps as an input for further calculations. In this way, maps can be fast and easily reassembled if a change in the assembler parameters is required. Below, we briefly describe our map assembly method and in the following sections we discuss the details of the outlined steps.
Calculation of overlaps. We first compute all pairwise alignments (overlaps) of optical maps. These overlaps are screened to identify accurate overlaps by using an alignment score as a proxy for overlap significance. Provided that we have large quantities of optical maps such as those from currently mapped human genomes (0.5 million or more optical maps), the amount of necessary computation can be very impressive. Fortunately, this step can take advantage of the massive parallelization offered by large modern computing clusters. The computation can be performed in a relatively short time provided a large number of available processors available. Furthermore, the memory requirement for this computation is very moderate. Each overlap between a pair of maps with m and n fragments is computed in O(mn) time, equivalent of ≈1/100 of a second on an average PC, and requires O(mn) storage.
Overlap graph construction. Pairwise overlap relations between individual maps are represented by the overlap graph, which is a central object of our analysis. In this graph, optical maps are represented by graph nodes, whereas overlaps between pairs of maps are represented by edges connecting corresponding nodes. This representation is convenient, because we can carry out all calculations in the graph by using simple graph algorithms such as breadth-first search (BFS), depth-first search (DFS), and heaviest path.
Graph correction procedure. Unless the errors present in the overlap graph are addressed, construction of accurate consensus maps is often impossible. Overlap graph errors appear in the form of false edges (due to spurious overlaps) and spurious nodes (due to chimeric maps) that suggest false connectivity of genomic regions. To address this problem, we carry out a graph correction procedure to eliminate such errors. This is accomplished in three steps to account for three error types: (i) orientation consistent false overlaps, (ii) orientation inconsistent false overlaps, and (iii) chimeric maps.
Identification of islands. After the graph correction procedure, the overlap graph breaks into multiple components corresponding to connected genomic regions spanned by overlapping optical maps. Each of these graph components, also termed islands, must be processed independently of each other to yield consensus maps representing these regions.
Contig construction. Within each of the graph components, contigs can be represented by paths connecting sources and sinks. To produce the most extensive map representing the genomic region of a given graph component, we find the heaviest cycle-free path, maximizing the estimated genomic distance spanned by that path. In sequence assembly problems, scaffolding, described orienting and ordering contigs relative to each other and a reference genome, is commonly provided because sequence reads are typically mate-paired. In optical mapping, this is not the case, so sets of assembled contigs are not “scaffolded”, i.e., no additional orientation and/or order between the map contigs is given. However, if the mapping process is parallelled by resequencing, this disadvantage can be overcome to provide more detailed information about the assembled maps.
Construction of draft consensus map. Extracted paths within the overlap graph produce draft consensus maps by merging corresponding optical maps according to their overlaps.
Consensus map refinement. In our approach, draft consensus maps are constructed by concatenating single optical maps. Therefore, errors in the form of missing cuts, false cuts, and fragment size inaccuracies inherently appear in these draft maps. However, if the draft map retains enough accuracy, errors can be corrected by combining information from a large number of optical maps obtained from the corresponding graph component. This refined map is reported as a consensus map representing the relevant genomic region.
Given the potentially large number of pairwise comparisons required in the overlap step, a question arises as to whether something can be done to reduce the number of candidate pairs by some heuristic method without calculating all pairwise alignments. In sequencing, this problem is addressed by means of k-mer hashing (e.g., k = 23) that allows detection of a high percentage of correct overlaps by finding long word matches and thus avoids the expensive dynamic programming step for many pairs of sequence reads. The reason why this idea works well for sequence reads produced by the Sanger sequencing method is because the errors in sequence reads are usually somewhat rare. This rarity is due to the fact that averaging over a large population of clonal sequences results high sequence read accuracy (>95%). We investigated the possibility of geometric hashing of adjacent optical map fragments for quick elimination of spurious overlaps (23). However, because optical mapping is a single-molecule mapping technology, averaging is not embedded in the primary measurement, as it is in Sanger sequencing, which results in higher error frequency when compared with sequence reads. False cuts and especially missing cuts are ubiquitous in optical maps, thus requiring the tuple size k to be small to ensure a high probability of finding true overlaps. On the other hand, if k is chosen to be small, only a small number of nonoverlapping map pairs can be eliminated before the alignment step, and a large fraction of maps still need to be aligned. Therefore, we abandoned hashing in favor of exhaustive pairwise comparisons for the purpose of finding accurate overlaps.
Overlap Graph Construction.
A directed overlap graph G = (V, E) is defined by a set of nodes V corresponding to individual optical maps taken in a particular orientation, and a set of directed edges E, corresponding to high-quality overlaps between optical maps. Initially, all pairwise overlaps of optical maps are calculated by using our likelihood-based scoring method (17). The quality of reported overlaps is based on the site match measure that we term “q score” (A.V., unpublished data). Overlaps with q scores exceeding a specified threshold are considered to be accurate and are selected for construction of the graph. Specifically, they are sorted according to their q score values and progressively added to the graph in decreasing order of significance. Each map is placed in a particular orientation (normal or reverse), and the graph is grown as more edges are added. At this point, orientation consistency is checked. If an edge, suggested for addition to the graph, connects two nodes within the same component of the graph, orientation of maps represented by these nodes must be consistent with orientation of maps within the suggested overlap. If orientation is inconsistent, the edge is not added to the graph and therefore not considered in further analysis. This step accomplishes elimination of false overlaps with inconsistent orientation. The idea behind this step is to embed correct overlaps into the graph as early as possible. False overlaps generally have low q scores; hence, by the time they are considered for the addition to the graph, we hope that enough accurate overlaps are embedded already to purge the spurious orientation-inconsistent overlaps from further consideration. Unlike the greedy merge procedure used in CAP (14), this step does not create a problem if a false edge is incorporated early in the graph. As will become clear below, our graph correction procedure will eliminate such an edge, so corresponding regions will remain unconnected instead of causing a spurious consensus map to be reported.
Previously, we explained that each map in the overlap graph appears in a particular orientation o ∈ {N, R}, where N stands for normal orientation (fragments appear in the same order as in the map stored in the file) and R stands for reverse (fragments appear in the opposite order compared with the way the map is stored in the file). Furthermore, edges within the overlap graph can be of two types: containment edges and noncontainment edges. A containment edge connects two maps, one of which is contained by another through their pairwise alignment. More precisely, if map M1 represents genomic region G1 and map M2 represents genomic region G2, than map M1 contains map M2 if G1 ⊇ G2. In this case, M1 is called master map, and map M2 is called a slave map. Therefore, noncontainment edges represent overlaps of maps that contain both common and unique regions.
Below we describe how we assign edge directions and weights in the overlap graph. Suppose that maps M1 and M2 appear in orientation o1 and o2 in their pairwise overlap. The edge weights in the overlap graph are given by estimates of genomic distances between optical map midpoints deduced from their overlap, so that weight (M1o1 → M2o2) = dist(M1o1, M2o2). Fig. 1 gives an illustration how this distance can be calculated for maps M1 and M2. Based on the positions of map centers C1 and C2, we can identify the largest common alignment block (u1, u2; v1, v2) that does not contain map centers C1 and C2. We use corresponding map regions with sizes B1 = |u2 − u1|, B2 = |v2 − v1|, A1 = |u1 − C1|, and E2 = |C2 − v2| to estimate the distance: dist(M1o1, M2o2) = (−1) I{u1≤C1, C2 ≤v2} × (A1 + (B1 + B2)/2 + E2). Of course, all of the numbers are defined by the map orientation; for example, v2 = Σi=1k−1 fi for M2N and v2 = ‖M2‖ − Σi=1k−1 fi for M2R. Here fi are fragment sizes of map M2 and k is the index of the site corresponding to v2. It is clear that changing the direction of the edge will change the sign of the edge weight, but not its absolute value. So, when constructing the graph, we choose edge directions so that edge weights are positive.
Fig. 1.
The distance between optical map midpoints. The distance between maps M1N and M2N is given by the distance between map centers. In this case, dist(M1N, M2N) = dist(C1, C2) = (−1)I{u1≤C,1v2≥C2} × (A1 + (B1 + B2)/2 + E2), where u1 represents the closest to C1 matching site of the largest alignment block (u1, u2; v1, v2) that does not contain center points C1 and C2.
Graph Correction Procedure.
Although we only embed high quality overlaps in the overlap graph, some of them can be spurious. Such overlaps introduce false edges. Chimeric maps are another type of error in which a single map represents a concatenate of two or more different genomic regions. Thus, in the overlap graph, these maps bridge unrelated genomic regions and can cause errors in the construction of consensus maps. Fig. 2 gives examples of what such errors look like when they arise in the overlap graph. Below we describe in more detail how we eliminate false edges and chimeric maps from the overlap graph. Note that false edges and chimeric maps can look very similar to regions with low coverage and poor overlap alignment quality. In our method, however, we require that genomic regions connected in the overlap graph provide multiple evidence of connectivity. In other words, any two connected regions must be connected by at least two different paths within the overlap graph with no common intermediate nodes. If such evidence cannot be found in the overlap graph, our procedure will disconnect the graph at the corresponding node. Although this implies that genomic regions must be deep in terms of map coverage, there is a significant benefit in terms of reduction of overlap graph errors, which allows for accurate map assemblies.
Fig. 2.
Errors in the overlap graph. (i) Cycles in graphs from linear genomes. False edge (red) connects two nodes within the same component and creates a cycle. In graphs built for circular genomes, cycles arise naturally, but graphs from linear genomes should not contain cycles. (ii and iii) False edges. Orientation inconsistent false edge (ii, red edge) creates an orientation conflict when placing a map in the graph in a particular orientation. False edges that do not introduce orientation conflict (iii, red edge) can spuriously connect maps from unrelated genomic regions. (iv) Chimeric maps. Chimeric maps (D) combine maps from at least two different genomic regions (shown in blue and orange), resulting in falsely connected regions.
Elimination of False Edges with Inconsistent Orientation.
This step is accomplished at the graph construction stage because edges added to the overlap graph must connect maps consistently with the orientation of maps already placed in the graph. We first sort all overlaps in decreasing order of significance and then add them to the overlap graph. Suppose an edge suggested for addition to the graph connects two maps M1 and M2 in orientation o1 and o2. If these maps belong to the same component, they are already assigned orientations r1 and r2. Hence, the overlap is consistent if either r1 = o1 and r2 = o2 or r̄1 = o1 and r̄2 = o2. If the edge is consistent, it is included in the graph, otherwise it is skipped.
Elimination of False Edges with Consistent Orientation.
We can use the proposed genomic distance between optical maps to our advantage to eliminate orientation consistent false edges still present in the overlap graph. For every node Ni in the graph, we perform a depth-first search of specified depth taking only outgoing edges and collect all nodes Nj such that there exist multiple independent paths through the graph connecting nodes Ni and Nj. For each of those paths Pα we compute its spanning genomic distance Dα by adding weights of edges taken along the edges of each path. Distances from correct paths must be distributed according to the distance error model. In our previous analysis (17), we have established that for genomic region of size y, its size X, estimated from the optical map, is normally distributed with mean EX = y and variance Var(X) = σ2y. Naturally, we adopt the same error model for the genomic distances between optical maps, because they are calculated by adding sizes of fragments within optical maps. Unfortunately, for a given pair of maps Mi and Mj, represented by nodes Ni and Nj, their true genomic distance is generally unknown. To overcome this limitation, we want to find the path Pα with distance Dα connecting Ni and Nj, that maximizes the size of the cluster of paths connecting Ni and Nj with distances found within χ standard deviations χ σ √Dα of Dα. If there is more than one path within such a cluster (including the path corresponding to Dα), then all of the edges within those paths are marked as “confirmed” (because we found multiple evidence of connectivity with agreeable distance). Unconfirmed edges are then eliminated from the overlap graph along with all isolated nodes.
Chimeric Map Elimination.
Even after elimination of false edges, some chimeric maps may be present in the overlap graph. Chimeric maps have a very distinctive appearance in the overlap graph (Fig. 2), namely, a set of maps L(M) overlapping with the left part of chimeric map M and a set of maps R(M) overlapping with the right part of M must not be locally connected in the overlap graph other than through that chimeric map. This observation is based on the fact that right and left parts of map H must belong to different regions. Therefore, potential chimeric maps are identified as local articulation nodes, removal of which disconnects the local subgraph (in this case, edge direction is not important, because finding articulation points suffices). Provided enough optical map coverage, we can use this as a strategy for finding chimeric maps. From every node adjacent to map M, we perform a breadth first search of specified depth without taking paths through M to discover all immediate neighbors of M. If we fail, the node corresponding to M is removed from the graph.
Genomic distances between map centers as we described in this section provide simple, yet powerful measures that can be used to filter out overlap errors that make assembly problematic. Conveniently, this method can also be extended into assembly of sequences, where distance would represent nucleotide count between centers of sequence reads based on their overlap. A corresponding distance error model would be derived from parameters of the sequencing instrument, where the amount of sequencing errors will depend on the DNA content of a particular genomic region. Naturally, this distance would allow detection of false overlaps between sequence reads and aid the sequence assembly process.
Layout and Consensus Map Reconstruction.
After false edges and chimeric maps are eliminated from the overlap graph, the graph should break into components representing islands of overlapping maps. For each component of the graph, we want to extract a contig representing the genomic region corresponding to this island. In other words, we must find a cycle-free path through a subgraph maximizing the genomic distance spanned by this path. To do this, we first identify sources within the component and perform depth-first search from each source assigning the longest distance accumulated along the paths to each of the discovered nodes. Consider node Ni with weight wi corresponding to the longest spanning distance of the incoming path ending at that node. If Oi is a set of edges incoming to Ni, then Si gives the set of nodes for which edges from Oi are outgoing. The maximization recursion for Ni outgoing is given by
where wj is the the weight stored at the node Nj ∈ Si and wi→j = dist(Ni, Nj) is the distance between maps Ni and Nj. The heaviest path is found by locating the node with the largest weight and the heaviest path ending at that node. The prescribed merging of optical maps within a corresponding island is thus given by the set of maps as they appear along this heaviest path starting from a relevant source node.
Each graph component yields a draft map by combining portions of corresponding optical maps based on their pairwise overlap relations (Fig. 3). Although the draft map is a concatenate of multiple optical maps, each fragment in it is given by a fragment from a single optical map. Therefore, a draft map inherently contains errors in the form of missing cuts, false cuts, and fragment size inaccuracies. These are subject to further correction, which we accomplish through a map refinement procedure (24). More specifically, maps from the corresponding overlap graph component are used to improve draft map accuracy by (i) removing false cuts, (ii) adding missing cuts, and (iii) reestimating fragment sizes (Fig. 4). To accomplish this, we align optical maps to the draft map and perform hypothesis testing to identify positions of draft map where sites need to be added and/or deleted. Fragment size reestimation is accomplished by taking average of fragments of optical maps corresponding to fragments of the draft map. This procedure is repeated until no further corrections can be made. During each iteration of the refinement procedure, optical maps are realigned to yield more accurate corrections. The refinement process converges rapidly and equilibrium is usually reached within 10−13 iterations. The corrected consensus map is reported as a map representing the corresponding island.
Fig. 3.
Draft map construction based on the path from the overlap graph. Draft map is constructed by concatenating regions of optical maps based on their pairwise overlaps.
Fig. 4.
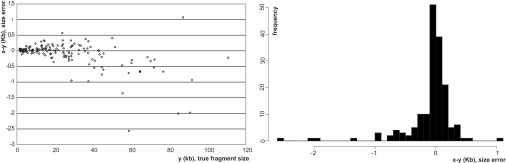
Fragment size discrepancy for map assembly of XhoI digest of E. coli K12 strain. For the restriction fragment sizes x of XhoI E. coli consensus map, y gives the true fragment sizes inferred from the published sequence; x − y gives the sizing error of consensus map fragments produced by our optical map assembler. (Left) The scatter plot shows the size discrepancy depending on the size of the underlying fragment. (Right) The histogram illustrates the marginal distribution of the size discrepancy for the finished restriction map.
Acknowledgments
We thank Susan Reslewic and Gene Ananiev for experimental results and critical feedback on genomic applications; Miron Livny for generous CONDOR support; Dan Forrest, Yu-Chi Liu, and John Nguyen for stimulating conversations concerning the details of our assembly method; Juan Nunez-Iglesias for excellent comments and critical reading of the manuscript; and anonymous reviewers for their excellent comments that helped us improve the presentation of the material. This research was supported by National Institutes of Health Grants P50 HG002790, CA119333, and HG000225; National Science Foundation Grant DBI-0501818; and a Preuss Foundation Fellowship (to A.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Ananthraman, T., Schwartz, D, Mishra, B. The Seventh International Conference on Intelligent Systems for Molecular Biology, 1999.
References
- 1.Zhou S, Kile A, Bechner M, Place M, Kvikstad E, Deng W, Wei J, Severin J, Runnheim R, Churas C, et al. J Bacteriol. 2004;186:7773–7782. doi: 10.1128/JB.186.22.7773-7782.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwartz DC, Li X, Hernandez L, Ramnarain SP, Huff EJ, Wang Y-K. Science. 1993;262:110–114. doi: 10.1126/science.8211116. [DOI] [PubMed] [Google Scholar]
- 3.Dimalanta ET, Lim A, Runnheim R, Lamers C, Churas C, Forrest DK, dePablo JJ, Graham MD, Coppersmith SN, Schwartz DC. Anal Chem. 2004;76:5293–5301. doi: 10.1021/ac0496401. [DOI] [PubMed] [Google Scholar]
- 4.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 5.Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, Baker A, Jonasdottir A. Nat Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 6.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, et al. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 7.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, Dallaire S, Gabriel SB, Lee C, Daly MJ, Altshuler DM. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 8.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 9.Ananthraman T, Mishra B, Schwartz D. J Comp Biol. 1997;4:91–118. doi: 10.1089/cmb.1997.4.91. [DOI] [PubMed] [Google Scholar]
- 10.Lee JK, Dancik V, Waterman MS. J Comp Biol. 1997;5:505–516. doi: 10.1089/cmb.1998.5.505. [DOI] [PubMed] [Google Scholar]
- 11.Karp R, Shamir R. Proceedings of the 2nd ACM Conference on Computational Molecular Biology; New York: Assoc Comput Machinery; 1998. pp. 117–124. [Google Scholar]
- 12.Muthukrishnan S, Parida L. Proceedings of the Annual International Conference onComputational Molecular Biology (RE-COMB 1997); New York: Assoc Comput Machinery; 1997. pp. 209–219. [Google Scholar]
- 13.Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KHJ, Remington KA, et al. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 14.Huang X, Maddan A. Genome Res. 1999;9:868–977. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batzoglou S, Jaffe BD, Stanley K, Butler J, Gnerre S, Mauceli E, Berger B, Mesirov JP, Lander ES. Genome Res. 2002;12:177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pevzner PA, Tang H, Waterman MS. Proc Natl Acad Sci USA. 2001;98:9748–9753. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valouev A, Li L, Liu YC, Schwartz DC, Yang Y, Zhang Y, Waterman M. J Comp Biol. 2006;13:442–462. doi: 10.1089/cmb.2006.13.442. [DOI] [PubMed] [Google Scholar]
- 18.Deng W, Burland V, Plunkett G, III, Boutin A, Mayhew GF, Liss P, Perna NT, Rose DJ, Mau B, Zhou S, et al. J Bacteriol. 2002;184:4601–4611. doi: 10.1128/JB.184.16.4601-4611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 20.Zhou S, Deng W, Anantharaman TS, Lim A, Dimalanta ET, Wang J, Wu T, Chunhong T, Creighton R, Kile A, et al. Appl Environ Microbiol. 2002;68:6321–6331. doi: 10.1128/AEM.68.12.6321-6331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou S, Kvikstad E, Kile A, Severin J, Forrest D, Runnheim R, Churas C, Hickman JW, Mackenzie C, Choudhary M, et al. Genome Res. 2003;13:2142–2151. doi: 10.1101/gr.1128803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International Human Genome Sequencing Consortium. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y. PhD thesis. Los Angeles: University of Southern California; 2005. [Google Scholar]
- 24.Valouev A, Zhang Y, Schwartz DC, Waterman MS. Bioinformatics. 2006;22:1217–1224. doi: 10.1093/bioinformatics/btl063. [DOI] [PubMed] [Google Scholar]