Abstract
Strategies for high-throughput analysis of interactions between various hormones and drugs with the estrogen receptor (ER) are crucial for accelerating the understanding of ER biology and pharmacology. Through careful analyses of the crystal structures of the human ER (hER) ligand-binding domain (hER–LBD) in complex with different ligands, we hypothesized that the hER–LBD intramolecular folding pattern could be used to distinguish ER agonists from selective ER modulators and pure antiestrogens. We therefore constructed and validated intramolecular folding sensors encoding various hER–LBD fusion proteins that could lead to split Renilla/firefly luciferase reporter complementation in the presence of the appropriate ligands. A mutant hER–LBD with low affinity for circulating estradiol was also identified for imaging in living subjects. Cells stably expressing the intramolecular folding sensors expressing wild-type and mutant hER–LBD were used for imaging ligand-induced intramolecular folding in living mice. This is the first hER–LBD intramolecular folding sensor suited for high-throughput quantitative analysis of interactions between hER with hormones and drugs using cell lysates, intact cells, and molecular imaging of small living subjects. The strategies developed can also be extended to study and image other important protein intramolecular folding systems.
Keywords: complementation, optical imaging, split reporters
Estrogens are responsible for the growth, development, and maintenance of the reproductive, skeletal, neuronal, and immune systems as well as and other systems. The physiological effects of these hormones are mediated by the estrogen receptor (ER), which is a ligand-inducible nuclear transcription factor (1). In the classical pathway of steroid hormone action, 17β-estradiol (E2), hormones, and a variety of other estrogens bind to the ligand-binding domain (LBD) of ER and lead to its dimerization and subsequent binding to a specific regulatory sequence in the promoters of ER target genes known as the estrogen response elements (2, 3), which then trigger activation or repression of many downstream target genes (4). The deficiency or excess of estrogens can lead to various pathological conditions including osteoporosis and breast carcinomas (5), making ER a major cellular therapeutic target.
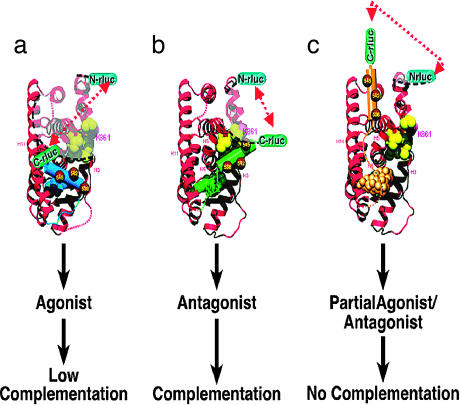
Elegant crystallographic studies with ER–LBD have shown that conformation of helix 12 (H12) is critical in responses observed with various ER ligands (4, 6, 7). The conformation of H12 behaves as a “molecular switch” that either prevents or enhances ER from binding to an array of coactivator proteins, which then activates transcription of many downstream estrogen-regulated genes responsible for cell growth. Given the critical role of H12 in ER signaling, we reasoned that it may be feasible to develop an intramolecular ER folding sensor with specific split reporter complementation patterns to study ligand pharmacology based directly on the conformational changes of H12 in response to different ligands (Fig. 1).
Fig. 1.
Schematic representation of the hypothetical model of ligand-induced intramolecular folding of ER that leads to split RLUC complementation. The N- and C-terminal fragments of split RLUC were fused to the N and C terminus, respectively, of the hERα of various lengths (amino acids 281–549 and 281–595). Binding of ER ligands to the intramolecular folding sensor (N-RLUC-hER-C-RLUC) induces different potential folding patterns in the LBD based on the type of ligand. This folding leads to split RLUC complementation for ER antagonist (b) (H12 and ligands are colored green), low complementation for ER agonist (a) (H12 and ligands are colored blue), and no complementation for partial ER agonist/antagonist (c) (H12 and ligands are colored gold) with the selective folding sensor. Even though the distance between the N- and C-RLUC fragments after binding with partial agonist (c) is smaller than that of agonists (b), this model depicts the importance of the orientations of the split RLUC fragments in complementation. The yellow spheres are hydrophobic amino acids located between helix 3 and helix 5 of LBD.
We used a split synthetic Renilla luciferase (RLUC) and firefly luciferase (FLUC) complementation system, which we previously developed and validated (8, 9), to test this hypothesis by assaying ligand-induced RLUC/FLUC complementation in cell lysates, intact cells, and cell implants in living mice by noninvasive bioluminescence optical imaging. The validated ER intramolecular folding sensors can also be used to distinguish ligand pharmacology in cell culture studies and cell implants in living animals treated with different ER ligands, agonists, selective ER modulators (SERMs), and pure antiestrogens. Moreover, a mutant human ER (hER) with low affinity to E2 was identified and used as an ER sensor for characterization of ER ligand interaction in living mice without significant competition from endogenous circulating estrogens.
Results
RLUC Complementation as Sensors of ER Ligand-Induced Intramolecular Folding.
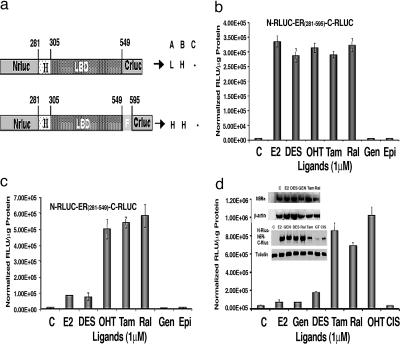
Several factors need to be considered to achieve efficient ligand-induced split RLUC complementation, and also ligand-induced complementation that distinguishes agonists from antagonists. Key factors include (i) the distance between the complementing N- and C-RLUC fragments in the intramolecular folding sensor before and after the binding of ligands, (ii) the orientation of N- and C-RLUC fragments after binding of different ER ligands, and (iii) the position of RLUC fragments in recovering the substrate binding pockets through complementation after ligand binding. By carefully considering these factors along with the crystal structures of different ER ligand complexes (bound with agonists, SERMs, and pure antiestrogens) and the importance of H12 within the LBD, we constructed a series of vectors that expresses fusion proteins with split RLUC fragments and different portions of the hER (Fig. 2a) (4, 10). We also confirmed the orientation of split RLUC fragments needed for efficient complementation (Fig. 6, which is published as supporting information on the PNAS web site). All of these vectors were studied in 293T cells (ER-negative cells, transiently transfected with ER) treated with several ER ligands, including E2, tamoxifen, raloxifene, genistein, diethylstilbestrol (DES), and 4-hydroxytamoxifen (4-OHT) (structures of ligands are in Fig. 7, which is published as supporting information on the PNAS web site). Among the different portions of hER examined, the fusion protein containing hER of amino acids 281–595 [hER281–595: partial hinge (domain D), LBD (domain E), and domain F] showed a significant level of ligand-induced RLUC complementation for both agonists and SERMs. The hERα was used for constructing all vectors in this study. The level of complementation achieved by this fusion protein with both SERMs and agonists was 80 ± 15 times greater than in cells exposed to carrier controls (P < 0.001). The cells exposed with genistein, a very-low-affinity ERα agonist, showed no complementation, similar to untreated cells (Fig. 2b). At the same time, the cells transfected with the vector expressing the fusion protein containing amino acids 281–549 of hER (hER281–549: domains D and E) show complementation that clearly distinguishes agonists from SERMs. In transfected cells treated with SERMs and agonists, 80-fold ± 15-fold and 15-fold ± 5-fold increases in complementation were observed, respectively, relative to untreated cells (P < 0.05) (Fig. 2c).
Fig. 2.
Intramolecular folding sensors and their response to various ER ligands. (a) Schematic diagram of different intramolecular folding sensor constructs with the split RLUC fragments and different flanking sequences (ERα domains) on either side of the ER–LBD were used to identify a vector that leads to ligand-induced intramolecular folding-based RLUC complementation and can distinguish ER agonists (A), antagonists (B), and partial antagonists (C). H, high complementation; L, low complementation. (b) ER ligand-specific intramolecular folding sensor. 293T cells were transiently transfected to express the intramolecular folding sensor N-RLUC-hER281–595-C-RLUC and treated with the indicated ER ligands or carrier control for 18 h. Treatment with ER antagonists and agonists led to similar levels of intramolecular-folding-assisted complementation that was significantly higher than that of carrier control-treated cells (P < 0.001). (c) Antagonist-specific intramolecular folding sensor. 293T cells were transiently transfected to express the intramolecular sensor N-RLUC-hER281–549-C-RLUC and treated with the ER ligands or carrier control as in b. Treatment with ER antagonists and agonists led to relatively high signal and low signal levels of intramolecular-folding-assisted RLUC complementation, respectively, relative to carrier control-treated cells (P < 0.01). (d) Western blot analysis of MCF7 cells using anti-ERα antibody after treatment with different ligands (Inset Upper). Shown are ER ligand antagonist-specific and agonist-specific intramolecular-folding-assisted RLUC complementation in 293T cells and the Western blot analysis of corresponding sample using RLUC antibody (Inset Lower). Error bars of all of the graphs represent the averages of triplicate samples ± SEM.
Up-regulation of the multidrug-resistant P-glycoprotein by ER ligands and associated change in the signal by coelenterazine (11) were ruled out by different experiments in cell lysates and intact cells (Fig. 8, which is published as supporting information on the PNAS web site).
Complementation of ER–LBD Intramolecular Folding Sensor Is Independent of the Levels of Endogenous ERα Expression.
To determine whether ligand-induced hER intramolecular-folding-assisted RLUC complementation in transfected 293T cells is due to increased protein expression of the folding sensor N-RLUC-hER281–549-C-RLUC, Western blot analysis was performed by using the RLUC antibody before and after treatment with different ER ligands. Treatment of 293T cells with E2, DES, genistein, and raloxifene did not lead to significant changes in the protein level of the folding sensor (Fig. 2d Inset Lower). Interestingly, even though the levels of sensor protein were significantly reduced in the transfected cells treated with the SERM tamoxifen and two anticancer drugs, epigallocatechin gallate and cisplatin (both non-ER ligands), the RLUC signal in tamoxifen-treated cells was still significantly greater than that in the cells treated with the agonists E2 and DES (P < 0.05) (Fig. 2d). The anticancer drugs did not lead to any RLUC signal, compared with carrier control-treated cells (P > 0.05). These results show that the variations in the RLUC activity in the cells treated with different ligands were not mediated through changes in the level of sensor protein but were more likely to be caused by different complementation patterns within the receptor induced by the ligands. The ERα protein level in transfected MCF-7 cells was also estimated with Western blot analysis before and after the cells were treated with different ER ligands. Treatment of MCF-7 cells with ER ligands did not lead to significant changes in the intracellular ERα protein levels (Fig. 2d Inset Upper). Also, ligand-induced intramolecular folding of ER studied in different ER-positive (MCF-7) and ER-negative (MDA-MB-231 and 293T) cell lines showed no significant relation with the intracellular ER level (Fig. 3d–f).
Fig. 3.
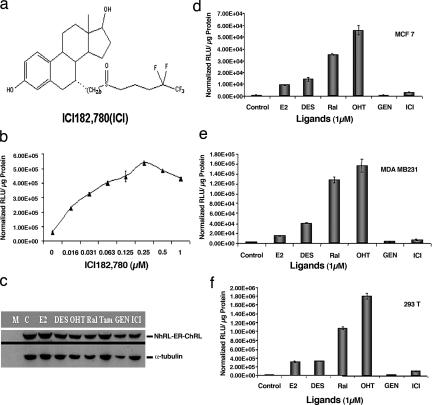
Study of ER-ligand-induced intramolecular folding system with pure ER antagonist ICI182, 780. (a) Structure of the ER ligand ICI used for the study. (b) Dose-dependent RLUC complementation of ICI. 293T cells were transiently transfected to express the intramolecular folding sensor N-RLUC-hER281–549-C-RLUC and treated with various concentrations of ICI for 18 h. RLUC activity was determined as in b. (c) Expression of the intramolecular folding sensors in 293T cells treated with different ER ligands (1 μM). The expression of N-RLUC-hER281–549-C-RLUC in transiently transfected 293T cells treated with different ER ligands or ICI was determined by Western blotting as described in Fig. 4c. (d–f) Ligand-induced intramolecular folding by different ER agonists and antagonists in comparison with ICI. The intramolecular folding sensor was transiently transfected into ER-positive MCF-7 cells and ER-negative MDA-MB-231 and 293T cells and treated with the indicated ligands or carrier control for 18 h. RLUC activity was determined as in b. The error bars represent SEM of triplicate determinations.
Time- and Dose-Dependent Induction of hER–LBD Intramolecular RLUC Complementation.
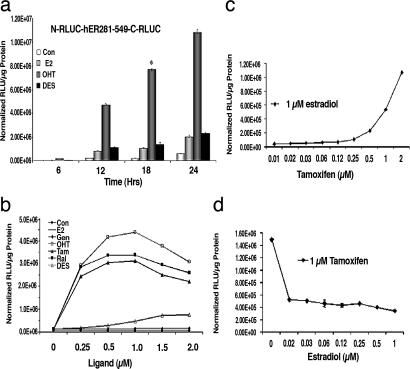
To determine the kinetics of ligand-induced RLUC complementation, 293T cells transiently transfected to express the fusion protein N-RLUC-hER281–549-C-RLUC were exposed to 1 μM of three representative ligands (the agonists E2 and DES and the SERM 4-OHT) and assayed for complemented RLUC activity at 6, 12, 18, and 24 h. The results showed significant levels of RLUC complementation in transfected cells exposed to the 4-OHT at all time points studied (P < 0.002 relative to carrier control-treated/untreated cells). The agonists E2 and DES led to RLUC complementation that was significantly lower than 4-OHT but higher than that of carrier control/untreated cells at all time points studied (P < 0.001). Furthermore, the maximum level of ligand-induced RLUC complementation was achieved after 18 h of exposure to ligands (Fig. 4a). The potency of each ligand to induce RLUC complementation in transiently transfected 293T cells expressing N-RLUC-hER281–549-C-RLUC was studied at different ligand concentrations with RLUC activities determined by luminometer assays. A dose-dependent increase in complemented RLUC activity was observed in all cases, with maximum induction at 1 μM except for the agonist DES, which showed maximum activity at 1.5 μM relative to carrier control-treated/untreated cells (Fig. 4b).
Fig. 4.
Antagonists- and agonists-specific induction of split RLUC complementation of the intramolecular folding sensor. (a) 293T cells transiently transfected with the intramolecular folding sensor (N-RLUC-hER281–549-C-RLUC) were assayed for RLUC activity at different time points after exposure to E2, 4-OHT, and DES. The maximum fold ligand induction of split RLUC complementation relative to carrier control-treated cells was achieved at 18 h (∗). (b) Concentration-dependent activation of ligand-induced RLUC complementation. 293T cells were transiently transfected with the intramolecular folding sensor and exposed to increasing concentrations of the indicated ligands for 18 h. RLUC activity was determined by luminometer assays as in a and normalized to that of carrier control-treated cells. RLUC activity increased significantly in transfected cells treated with the ER antagonists 4-OHT (□), tamoxifen (▴), and raloxifene (■). The ER agonist DES (▵) led to maximum induction of complemented RLUC activity at 1.5 μM. All other ligands led to a dose-dependent increase in complemented RLUC activity with maximum induction at 1 μM, relative to carrier control-treated cells. (c) Competitive binding of tamoxifen to the intramolecular folding sensor. 293T cells transiently transfected with the intramolecular sensor N-RLUC-hER281–549-C-RLUC were treated with various concentrations of the ER antagonist tamoxifen in the presence of a fixed concentration of the ER agonist estradiol (1 μM). RLUC activity was determined as in b, and the samples were normalized for transfection efficiency by cotransfecting with FLUC. (d) Competitive binding of E2 to the intramolecular folding sensor. 293T cells stably transfected with the intramolecular sensor N-RLUC-hER281–549-C-RLUC were treated with various concentrations of E2 in the presence of a fixed concentration of tamoxifen (1 μM). RLUC activity was determined as in b.
Competitive Binding of ER Agonists and SERMs in Ligand-Induced Intramolecular-Folding-Assisted RLUC Complementation.
To study ER intramolecular folding in living mice, the 293T cells were stably transfected to express the intramolecular folding sensor N-RLUC-hER281–549-C-RLUC. No significant difference in RLUC activity was observed between the stable and transiently transfected cells with all ligands studied (data not shown). These stable cells were used for studying the competitive binding of an agonist (E2) and a SERM (tamoxifen) in inducing RLUC complementation. The cells were assayed for complemented RLUC activity 18 h after being simultaneously exposed to a fixed concentration of E2 (1 μM) with various concentrations of tamoxifen (0.008–1 μM). Similarly, stable cells expressing the intramolecular folding sensor were exposed to a fixed concentration of tamoxifen (1 μM) and various concentrations of E2 (0.008–1 μM). E2-induced complemented RLUC activity was significantly enhanced in the presence of tamoxifen (P < 0.05) (Fig. 4c), and the tamoxifen-induced complemented RLUC activity was reduced in the presence of E2 (P < 0.05) (Fig. 4d).
The Pure Antiestrogen Fulvestrant [ICI182,780 (ICI)] Leads to RLUC Complementation Signal That Is Neither Like an Agonist nor a SERM.
The drug ICI is a pure antiestrogen (Fig. 3a) that is currently in clinical use for the treatment of both ER-positive and ER-negative breast cancers (12, 13). Crystallographic studies with ERβ/pure antiestrogen complex suggest unique structural features of this receptor complex leading to its interesting pharmacology (14). In our system with 293T cells transfected to express the sensors RLUC-ER (281–549) or RLUC-ER (281–595), ICI led to dose-dependent RLUC complementation with maximum induction at 0.25 μM (Fig. 3b). Furthermore, RLUC complementation induced by ICI was distinct from that of ER SERMs or agonists (Fig. 3f). It is of note that no significant degradation of the fusion sensor protein was observed in transfected 293T cells in the presence of ICI (Fig. 3c) (15).
A Single Amino Acid Change at Glycine-521 with Threonine Selectively Abolishes the E2-Mediated Complementation of Folding Sensor Without Significantly Affecting the Complementation Induced by Other ER Ligands.
Binding of endogenous E2 to the intramolecular folding sensor in living animals will limit its use in tumor models or a transgenic model for studying hER interactions with ligands and drugs. Hence, to overcome this issue, we generated single amino acid mutations at position 521 of hERα that are analogous to the mutation generated in mouse ERα at amino acid position 525 (G525R), which was reported to reduce affinity to E2 by 1,000-fold (16). We constructed 19 different mutants at position 521 within N-RLUC-hER281–595-C-RLUC, which leads to RLUC complementation in the presence of both agonists and SERMs. The results of constructs screened with six different ER ligands are presented in Table 1, which is published as supporting information on the PNAS web site. Among all of the mutations created at position 521, the glycine-to-threonine (G521T) transition led to a 94% reduction in the intramolecular folding-mediated RLUC complementation induced by E2 (P < 0.001); an only 12–22% reduction for DES, 4-OHT, and raloxifene (P < 0.05); and no significant changes in response to genistein and ICI (P < 0.05) (Fig. 9, which is published as supporting information on the PNAS web site). The sensor with mutant hER (N-RLUC-hER281–549/G521T-C-RLUC), which distinguishes ER antagonists from SERMs (Fig. 2d), and the sensor with mutant mouse ER (N-RLUC-mER281–549/G525R-C-RLUC) were transiently transfected into 293T cells and treated with different ER ligands, and RLUC complementation was determined by luminometer assay. Our folding sensor with mutant hER (G521T) appeared less sensitive to the endogenous ligand E2 and retained more of the DES and raloxifene effects compared with the folding sensor with the mutant mER (G525R) (Fig. 10, which is published as supporting information on the PNAS web site).
The ER Intramolecular Folding System with LBDs ER281–549/G521G, ER281–549/G521T, and ER281–595/G521G Using Split FLUC Enzyme Fragments.
To evaluate the ER intramolecular folding system's utility and generalizability, we also constructed vectors with improved split FLUC fragments (our unpublished data) with different ER constructs and various ligands (Fig. 11, which is published as supporting information on the PNAS web site). The results show similar patterns as achieved when using the corresponding system with split RLUC fragments (Fig. 12, which is published as supporting information on the PNAS web site).
Imaging of ER Intramolecular Folding in Living Mice.
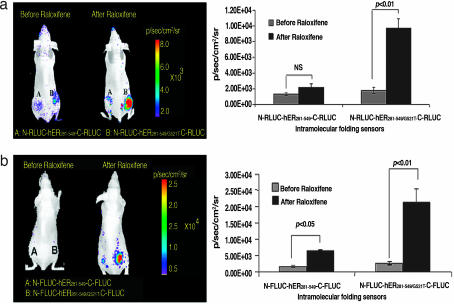
We studied the interactions between hER with ligands in living mice using the ER intramolecular folding-mediated RLUC complementation system, by implanting 293T cells stably expressing wild-type (N-RLUC-hER281–549-C-RLUC) or mutant (N-RLUC-hER281–549/G521T-C-RLUC) hER sensor on either side in the lower back of female nude mice (n = 3). Bioluminescence imaging of RLUC activity was performed immediately after cell implantations and 18 h after i.p. injection of raloxifene (0.5 mg). There was no significant difference in RLUC complementation induced by the SERMs 4-OHT and raloxifene in different experiments; we used raloxifene for many of our animal experiments. Significant RLUC activity was observed only in the site implanted with the stable cells expressing the mutant hER sensor (wild-type hER, 2.16 ± 0.52 × 103 photons per second per square centimeter per steradian; mutant hER, 9.7 ± 1.2 × 103 photons per second per square centimeter per steradian) (P < 0.01 relative to the site with wild-type hER) (Fig. 5a). Similar FLUC activity pattern was observed with wild-type and mutant ER constructs (Figs. 11 and 12). The lower level of signal produced from implanted cells expressing the wild-type hER sensor is most likely due to competitive binding of endogenous estrogen before the availability of raloxifene. The split FLUC system showed significantly more signal in living animals than the same system with the RLUC fragments, so this sensor was used for studying the agonist- and antagonist-specific induction of complementation in living mice.
Fig. 5.
Bioluminescence imaging of ER antagonist-induced intramolecular folding in a mouse model. (a) Shown is optical CCD camera imaging of 293T cells stably expressing intramolecular folding sensors N-RLUC-hER281–549-C-RLUC and N-RLUC-mutant-hER281–549/G521T-C-RLUC in living female nude mice before and after treatment with antagonist raloxifene (0.5 mg per mouse) and the corresponding quantitative graph. (b) Similar imaging conducted by using the same sensors with the split FLUC fragment system (N-FLUC-hER281–549-C-FLUC and N-FLUC-mutant-hER281–549/G521T-C-FLUC). The site implanted with the cells expressing the intramolecular folding sensor with the mutant hER (G521T) shows a higher RLUC complementation signal after raloxifene treatment compared with that of the wild-type hER.
Imaging of ER Intramolecular Folding Sensor to Distinguish ER Ligands in Living Mice.
We show the utility of our system in differentiating pharmacological classes of ER ligands in living animals based on receptor conformations by implanting 293T cells stably expressing the wild-type ER sensor (281–549) with FLUC fragments and the mutant form of ER–LBD (N-RLUC-hER281–549/G521T-C-RLUC) on either side of the lower back of the male nude mice (n = 2; two implants in each animal) and imaged 24 h after implantation (before ligand administration) and every 24 h after administration of 20 μg of DES or 4-OHT with adjuvant (sesame oil) or adjuvant in a 50-μl volume (17). These results show a significantly greater level of complementation (P < 0.05) from animals that received the SERM than the animals that received the agonist or adjuvant only (Fig. 13, which is published as supporting information on the PNAS web site).
Discussion
We have developed and validated two hER intramolecular folding sensors that can be used to distinguish ER ligand pharmacology. These receptor sensors can be directly translated from cell culture studies to molecular imaging in small living subjects. In this study we used an ER-based split reporter complementation strategy to follow the position of H12 within the ER–LBD to detect changes in the receptor structural folding in response to ligand binding. The longer construct with the F domain (281–595) appears ligand-pharmacology-independent (Fig. 2b), whereas the shorter construct without the F domain (281–549) leads to highest levels of split luciferase complementation in response to SERMs, moderate levels for agonists, and minimal levels for pure antiestrogens (Fig. 2c). We validated these intramolecular folding sensors with various ER ligands in both transiently and stably transfected 293T cells and transiently transfected MDA-MB-231 (ER-negative) and MCF-7 (ER-positive) cells. To extend the folding sensor for applications in living animals, we incorporated a previously undescribed mutant of hER (G521T) into the folding sensor that was insensitive to circulating endogenous estrogen but retained its ability to distinguish SERMs from synthetic agonists. Alternatively, ovariectomized mice can likely be used with the wild-type hER with minimal competition from endogenous estrogens while retaining the ability to study estrogen-like drugs. Future studies should be performed to compare strategies with the wild-type and mutant systems.
To date, several in vitro assays have been developed for screening ER ligands by using either purified ERα protein or ER isolated from cell lysates (18–21). Limited fluorescence-based assays (22) have been developed to measure receptor conformational changes (23) and recruitment of coactivator peptides (22, 24, 25) in the full-length hERα within cell culture (26). Other assays have been designed to study the effects of synthetic ligands on ER transcription through the activation of downstream target genes (27). However, most of these reported assays are not suitable for quantitative, high-throughput screening of ER ligands in intact cells and especially in living subjects through noninvasive molecular imaging.
A nontranscriptional assay containing fusion chimeras of either Flp recombinase (28) or Cre recombinase (29) with a truncated mouse ERα (amino acids 281–599) has been reported and used for regulating the recombination of reporter genes in cells and living animals. This system demonstrates high background activity even before the addition of ER ligands, mainly through enzymatic amplification, thus limiting its dynamic range in response to different ER ligands. We developed an analogous fusion chimera by fusing a truncated version of hER (amino acids 281–595) with FLUC, which leads to luciferase activity that is 104-fold greater than background (mock-transfected cells) even before the addition of ligands (unpublished data). The addition of ligands generates FLUC activity that is only 5- to 6-fold higher than that of carrier control-treated cells (unpublished data). These results clearly support the notion that these nontranscriptional chimeras containing ER–LBD are not optimal for studying concentration-dependent interactions between ER and their ligands.
To our knowledge, only one study has reported the construction of mutant versions of hER (G521R and G521V) for selective ER ligand binding using a fusion chimera containing hER251–595 with Flp recombinase enzyme (28). Incorporation of the same mutation into our intramolecular folding sensor (N-RLUC-hER281–595-C-RLUC) led to nearly complete abolishment of signal for all ER ligands (hERG521R) and a significant reduction in signal (77–89%) for all agonist activities (hERG521V) relative to hERG521G (P < 0.05) (Table 1). We constructed intramolecular folding sensors using the hERG521 mutants with 19 different possible amino acids. We found that the replacement of hERG521 with threonine leads to nearly complete abolishment of the E2-induced RLUC complementation and to only a 10–20% reduction for all other ER ligands studied. Subsequently, 293T cells stably expressing this intramolecular folding sensor (N-RLUC-hER281–549/G521T-C-RLUC) were generated for imaging hERα/ligand complexes in living animals.
The advantages of the intramolecular folding sensor strategy developed and validated include the following: (i) it is real-time (because RLUC exhibits flash kinetics) and quantitative; (ii) it can be used to distinguish binding of agonists, SERMs, and pure antiestrogens; (iii) it can be adapted for studying ligand binding to hER in living animal models by molecular imaging, and thus pharmacokinetic properties of each drug/ligand can be examined; (iv) it allows for a high-throughput strategy for screening/comparing different ER ligands and drugs in multiple cell lines; (v) it allows direct transition from cell culture studies to small living subjects because it is based on a bioluminescence split reporter strategy; and (vi) it will allow for applications using transgenic models that incorporate the intramolecular folding sensor. In addition, the availability of other split reporters with different properties and substrate specificity should allow multiplexing with other reporter assays.
The limitations with using split RLUC as the reporter gene regarding efflux of its substrate coelenterazine were resolved by showing experiments that resulted in no significant relation between the RLUC complementation and the multidrug resistance systems (Fig. 8) (11). In addition, the intramolecular folding system was also studied with the improved split FLUC fragments by replacing RLUC fragments. Both systems showed equal sensitivity in different cell culture experiments. The FLUC fragments showed more detectable signal in mouse experiments than RLUC because of more light penetration through tissues due to the more red-shifted wavelengths of FLUC. Also, the FLUC-based folding system showed greater efficiency in differentiating ER ligands in living mice. It is also possible that the exact locations (cytosolic vs. nuclear) of our fusion reporter proteins may affect the results obtained, and this will need to be explored in future studies. In addition, for some applications in vivo, the developed strategies may have difficult distinguishing agonists from background, and this will have to be investigated with testing of additional drugs.
The strategies developed in this study can also be extended to FRET and bioluminescence resonance energy transfer (30) by replacing the split RLUC/FLUC reporter fragments with the appropriate choice of donors and acceptors. This system will help to validate a new class of molecular “switch” for imaging drug–receptor interactions in living subjects that was previously not feasible. This study will eventually translate to improved methods for understanding the underlying estrogen biology, preclinical drug development, and target validation, as well as investigation of other important intramolecular folding systems.
Methods
The methods used for constructing different plasmid vectors and the procedures used for cell culture, transfection, and luciferase assays for RLUC and FLUC are in Supporting Materials and Methods, which is published as supporting information on the PNAS web site (Figs. 2a and 6a).
The Ligand-Concentration-Dependent Intramolecular-Folding-Assisted Complementation Study.
To determine the concentration of different agonists and antagonists of ERα required for inducing efficient intramolecular folding of the ER–LBD sensors, 293T cells transiently transfected with pcDNA-N-RLUC-hER281–549-C-RLUC were exposed to different ligands including E2, raloxifene, tamoxifen, DES, 4-OHT, and genistein at six different concentrations (0–1,500 μM). The transfected cells were assayed for RLUC activity after 18 h of incubation and normalized as mentioned in Supporting Materials and Methods.
Kinetics of Ligand-Induced Intramolecular Folding of hER281–549 and Split RLUC Complementation.
To determine the time point for maximum ER ligand-induced split RLUC complementation, 293T cells transiently transfected with pcDNA-N-RLUC-hER281–549-C-RLUC were exposed to E2, 4-OHT, and DES (1 μM). The cells were assayed for RLUC activities at 6, 12, 18, and 24 h after exposure to ligands as described in Supporting Materials and Methods.
Competitive Binding of ER Agonists and Antagonists in Induction of Intramolecular-Folding-Assisted RLUC Complementation.
To determine the effect of competitive binding on ER ligand-mediated split RLUC complementation, 293T cells transiently transfected to express the fusion protein N-RLUC-hER281–549-C-RLUC were exposed to agonist E2 (1 μM) with different concentrations of antagonist tamoxifen (0.008–2 μM) or to tamoxifen (1 μM) with different concentrations of E2 (0–1 μM) for 18 h. RLUC activities were determined as described in Supporting Materials and Methods.
The Ligand Agonist- and Antagonist-Specific Intramolecular Folding in ER-Positive and ER-Negative Cell Lines.
To determine the specificity of ligand agonists and antagonists in induction of intramolecular folding, ER positive (MCF-7) and negative (MDA-MB-231) cell lines were transfected with pcDNA-N-RLUC-hER281–549-C-RLUC and immediately treated with different ER ligands dissolved in DMSO (1 μM) or carrier control (DMSO). Complemented RLUC activities and expression of the folding sensor were determined 18 h after transfection as described in Supporting Materials and Methods.
Selection of 293T Cells Stably Expressing N-RLUC-hER281–549-C-RLUC and N-RLUC-mutant hER281–549-C-RLUC for in Vivo Imaging Studies.
293T cells stably expressing the intramolecular folding sensor with mutant (G521T) and wild-type hERα were selected by transfection of respective vectors with Lipofectamine 2000 and selected by using puromycin hydrochloride (1.5 μg/ml). Stable clones were propagated in MEM containing puromycin hydrochloride and used for imaging studies in living mice.
Optical CCD Imaging of ER Ligand-Induced Intramolecular Folding in Living Mice.
All animal handling was performed in accordance with Stanford University Animal Research Committee guidelines. For imaging in living nude mice (nu/nu), 293T cells stably expressing one of the fusion proteins N-RLUC-hER281–549-C-RLUC, N-RLUC-hER281–549/G521T-C-RLUC, N-FLUC-hER281–549-C-FLUC, or N-FLUC-hER281–549/G521T-C-FLUC were used. In vivo imaging for luciferase and RLUC were performed as per refs. 9 and 31 (see Supporting Materials and Methods for details).
Supplementary Material
Acknowledgments
We acknowledge Anobel Tamrazi and Carmel Chan for help in improving the manuscript and Tim Doyle and Shay Keren for the help with instrumentation. This work was supported by National Cancer Institute In Vivo Cancer Molecular Imaging Centers Grant P50 CA114747, National Cancer Institute Small Animal Imaging Resource Program Grant R24 CA92865, and National Institutes of Health Grant R01 CA82214 (to S.S.G.).
Abbreviations
- RLUC
Renilla luciferase
- FLUC
firefly luciferase
- N-RLUC
N-fragment of RLUC gene
- C-RLUC
C-fragment of RLUC gene
- ER
estrogen receptor
- hER
human ER
- LBD
ligand-binding domain
- SERM
selective ER modulator
- DES
diethylstilbestrol
- 4-OHT
4-hydroxytamoxifen
- E2
17β-estradiol
- H12
helix 12
- ICI
ICI182,780.
Footnotes
The authors declare no conflict of interest.
References
- 1.Tsai MJ, O'Malley BW. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 2.Schuur ER, Loktev AV, Sharma M, Sun Z, Roth RA, Weigel RJ. J Biol Chem. 2001;276:33554–33560. doi: 10.1074/jbc.M105555200. [DOI] [PubMed] [Google Scholar]
- 3.Gronemeyer H. Annu Rev Genet. 1991;25:89–123. doi: 10.1146/annurev.ge.25.120191.000513. [DOI] [PubMed] [Google Scholar]
- 4.Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 5.Beck V, Rohr U, Jungbauer A. J Steroid Biochem Mol Biol. 2005;94:499–518. doi: 10.1016/j.jsbmb.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 6.Pike AC, Brzozowski AM, Walton J, Hubbard RE, Bonn T, Gustafsson JA, Carlquist M. Biochem Soc Trans. 2000;28:396–400. [PubMed] [Google Scholar]
- 7.Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 8.Paulmurugan R, Ray P, De A, Chan CT, Gambhir SS. Trends Anal Chem. 2005;24:446–458. [Google Scholar]
- 9.Paulmurugan R, Umezawa Y, Gambhir SS. Proc Natl Acad Sci USA. 2002;99:15608–15613. doi: 10.1073/pnas.242594299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moras D, Gronemeyer H. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 11.Pichler A, Prior JL, Piwnica-Worms D. Proc Natl Acad Sci USA. 2004;101:1702–1707. doi: 10.1073/pnas.0304326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buzdar AU. Oncologist. 2003;8:335–341. doi: 10.1634/theoncologist.8-4-335. [DOI] [PubMed] [Google Scholar]
- 13.Cheung KL, Owers R, Robertson JF. Endocr Relat Cancer. 2006;13:251–255. doi: 10.1677/erc.1.01108. [DOI] [PubMed] [Google Scholar]
- 14.Pike AC, Brzozowski AM, Walton J, Hubbard RE, Thorsell AG, Li YL, Gustafsson JA, Carlquist M. Structure (London) 2001;9:145–153. doi: 10.1016/s0969-2126(01)00568-8. [DOI] [PubMed] [Google Scholar]
- 15.Fan M, Bigsby RM, Nephew KP. Mol Endocrinol. 2003;17:356–365. doi: 10.1210/me.2002-0323. [DOI] [PubMed] [Google Scholar]
- 16.Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacLusky NJ, Luine VN, Hajszan T, Leranth C. Endocrinology. 2005;146:287–293. doi: 10.1210/en.2004-0730. [DOI] [PubMed] [Google Scholar]
- 18.Nichols JS, Parks DJ, Consler TG, Blanchard SG. Anal Biochem. 1998;257:112–119. doi: 10.1006/abio.1997.2557. [DOI] [PubMed] [Google Scholar]
- 19.Inoue A, Yamakawa J, Yukioka M, Morisawa S. Anal Biochem. 1983;134:176–183. doi: 10.1016/0003-2697(83)90280-4. [DOI] [PubMed] [Google Scholar]
- 20.Nasir MS, Jolley ME. Comb Chem High Throughput Screen. 1999;2:177–190. [PubMed] [Google Scholar]
- 21.Krey G, Braissant O, L'Horset F, Kalkhoven E, Perroud M, Parker MG, Wahli W. Mol Endocrinol. 1997;11:779–791. doi: 10.1210/mend.11.6.0007. [DOI] [PubMed] [Google Scholar]
- 22.Zhou G, Cummings R, Li Y, Mitra S, Wilkinson HA, Elbrecht A, Hermes JD, Schaeffer JM, Smith RG, Moller DE. Mol Endocrinol. 1998;12:1594–1604. doi: 10.1210/mend.12.10.0176. [DOI] [PubMed] [Google Scholar]
- 23.Schaufele F, Carbonell X, Guerbadot M, Borngraeber S, Chapman MS, Ma AA, Miner JN, Diamond MI. Proc Natl Acad Sci USA. 2005;102:9802–9807. doi: 10.1073/pnas.0408819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bai Y, Giguere V. Mol Endocrinol. 2003;17:589–599. doi: 10.1210/me.2002-0351. [DOI] [PubMed] [Google Scholar]
- 25.Weatherman RV, Chang CY, Clegg NJ, Carroll DC, Day RN, Baxter JD, McDonnell DP, Scanlan TS, Schaufele F. Mol Endocrinol. 2002;16:487–496. doi: 10.1210/mend.16.3.0813. [DOI] [PubMed] [Google Scholar]
- 26.Michalides R, Griekspoor A, Balkenende A, Verwoerd D, Janssen L, Jalink K, Floore A, Velds A, van't Veer L, Neefjes J. Cancer Cell. 2004;5:597–605. doi: 10.1016/j.ccr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 27.Awais M, Sato M, Sasaki K, Umezawa Y. Anal Chem. 2004;76:2181–2186. doi: 10.1021/ac030410g. [DOI] [PubMed] [Google Scholar]
- 28.Logie C, Nichols M, Myles K, Funder JW, Stewart AF. Mol Endocrinol. 1998;12:1120–1132. doi: 10.1210/mend.12.8.0155. [DOI] [PubMed] [Google Scholar]
- 29.Kemp R, Ireland H, Clayton E, Houghton C, Howard L, Winton DJ. Nucleic Acids Res. 2004;32:e92. doi: 10.1093/nar/gnh090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De A, Gambhir SS. FASEB J. 2005;19:2017–2019. doi: 10.1096/fj.05-4628fje. [DOI] [PubMed] [Google Scholar]
- 31.Bhaumik S, Gambhir SS. Proc Natl Acad Sci USA. 2002;99:377–382. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.