Abstract
In metazoans, cell-cycle-dependent histones are produced from poly(A)-lacking mRNAs. The 3′ end of histone mRNAs is formed by an endonucleolytic cleavage of longer precursors between a conserved stem–loop structure and a purine-rich histone downstream element (HDE). The cleavage requires at least two trans-acting factors: the stem–loop binding protein (SLBP), which binds to the stem–loop and the U7 snRNP, which anchors to histone pre-mRNAs by annealing to the HDE. Using RNA structure-probing techniques, we determined the secondary structure of the 3′-untranslated region (3′-UTR) of mouse histone pre-mRNAs H4–12, H1t and H2a–614. Surprisingly, the HDE is embedded in hairpin structures and is therefore not easily accessible for U7 snRNP anchoring. Probing of the 3′-UTR in complex with SLBP revealed structural rearrangements leading to an overall opening of the structure especially at the level of the HDE. Electrophoretic mobility shift assays demonstrated that the SLBP-induced opening of HDE actually facilitates U7 snRNA anchoring on the histone H4–12 pre-mRNAs 3′ end. These results suggest that initial binding of the SLBP functions in making the HDE more accessible for U7 snRNA anchoring.
INTRODUCTION
Histones are among the most conserved proteins in the eukaryotic kingdom. Their amino acid sequences are highly conserved as well as the coding sequence suggesting a strong selection pressure on the secondary structure of the corresponding mRNAs (1,2). In contrast, sequences of the 5′- and 3′-untranslated regions (5′- and 3′-UTRs) are more divergent. They are believed to be modern acquisitions that restrict the histone biosynthesis to the S phase of the cell cycle. Different strategies have been developed by eukaryotes for that purpose. In yeast and plants, histones are produced from classical polyadenylated mRNAs. In metazoans, a sophisticated machinery involving many factors and ribonucleoprotein particles (RNPs) is used for the histone synthesis (3,4). In the latter case, histone mRNAs are not polyadenylated but instead end in a highly conserved hairpin structure. The histone mRNAs are synthesized as precursor mRNAs with a 3′ extension. The mature histone mRNAs are generated by a single endonucleolytic cleavage occurring between two cis-acting elements: a highly conserved hairpin structure upstream of the cleavage site and a so-called histone downstream element (HDE also called spacer element; see Figure 1). These two sequences are the scaffold for the assembly of the whole processing machinery. The highly conserved hairpin structure is bound by the stem–loop binding protein (SLBP, also called HBP for hairpin binding protein) (5,6). Downstream of the cleavage site, the HDE anneals to the 5′ end of the minor U7 snRNP. Then, the Zinc-Finger rich Protein (ZFP100) bridges the two cis-elements by interacting on one hand with the U7-specific protein Lsm11 and on the other hand with SLBP bound to its target hairpin (7–9). Results from recent UV-crosslinking studies suggest that CPSF-73, a known component of the cleavage/polyadenylation machinery, is the cleavage factor, which is recruited in a U7-dependent manner (10). CPSF-73 would act in a larger processing complex comprising eight proteins of the cleavage/polyadenylation machinery including the heat labile factor (HLF) also called Symplekin (11). The mature histone mRNAs are exported to the cytoplasm for translation. Therefore, the 3′ end processing of histone pre-mRNAs is the cornerstone of histone biosynthesis. Early in vitro processing experiments have suggested that the hairpin for SLBP binding is required for efficient 3′ end processing. However, the effect of loss of binding mutants of the hairpin can be compensated by optimizing the base pairing between the HDE and the U7 snRNP (12). Furthermore, the distance between the hairpin and the HDE is critical for efficient processing (13–15). In vitro experiments also showed that efficient processing of mouse H2a-614 mRNA can be observed in absence of the SLBP whereas processing of mouse H1t mRNA is fully dependent on the presence of the SLBP on its target hairpin because of weak base-pairing with U7 (16). In vivo experiments on mouse histone mRNA H2a-614 showed that an intact hairpin target for the SLBP is required for efficient 3′ end processing (17). Recently, this has been validated using the nowadays-available knockout techniques and indeed the SLBP is an essential protein in Caenorhabditis elegans (18,19) and in HeLa cells (20,21). On the other hand, Drosophila histone genes are special in that they contain polyadenylation sites downstream to the normal 3′ end site. Therefore, the SLBP depletion by RNAi in Drosophila is not lethal but only causes infertility because the histone mRNAs become polyadenylated. This allows in vivo examination of 3′ processing reaction, which shows that the reaction is strongly affected in the absence of the SLBP (22). Altogether, these data suggest that SLBP is required for efficient histone 3′ end processing, in facilitating U7 snRNP binding by protein–protein contacts mediated through ZFP100 (8).
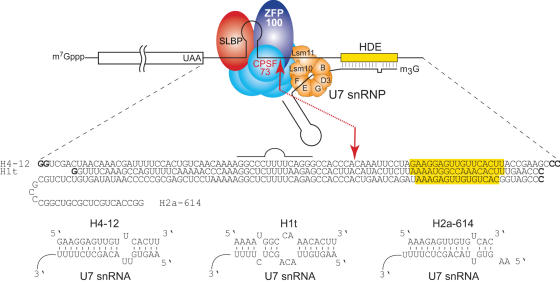
Figure 1.
Schematic representation of the 3′ end processing of replication-dependent histone pre-mRNAs. Cleavage of the RNA occurs at the red arrow between a highly conserved stem–loop structure and the HDE. Trans-acting factors are SLBP, which binds to the hairpin structure, U7 snRNP which anchors the pre-mRNAs by annealing to HDE and the cleavage complex including CPSF-73 which is believed to be the cleavage factor. U7 snRNP contains two U7-specific proteins named Lsm10 and Lsm11. ZFP100 bridges U7 to histone pre-mRNAs by interacting with SLBP and Lsm11. The sequences of the 3′-UTRs used in this work are indicated below the cartoon. Nucleotides shown in bold were added to optimize transcription reactions. Base-pairing of H4-12, H1t and H2a-614 with mouse U7 snRNA is shown at the bottom of the figure.
The rationale of the present study was to investigate whether the effect of the SLBP on the 3′ end processing could also be mediated by other mechanisms involving RNA structure. For this, we investigated the secondary structure of the 3′-UTR of histone H4-12 pre-mRNAs, which is processed in vitro in a SLBP-dependent manner (6,12). Our results show that SLBP induces conformational rearrangements of the HDE that enables U7 snRNP anchoring. This is also observed for the histone gene H1t, which displays SLBP-dependent processing but not for H2a-614 (16). The implications for histone biosynthesis in metazoan are discussed.
MATERIALS AND METHODS
Construction of templates
Templates for T7 in vitro transcription of the 3′-UTRs and U7 snRNA have been cloned into the SmaI site of pUC19. Sequences of the 3′-UTRs of mouse H4–12 (accession no. X13235), H2a-614 (accession no. AY158924), H1t gene (accession no. AY158908) and mouse U7 snRNA (accession no. X54165) have been reconstructed by shotgun oligonucleotides ligation. All constructs were verified by DNA sequencing.
In vitro transcription and labelling
In vitro transcription of the 3′-UTRs was performed as previously described (23). Transcripts were separated by denaturing 10% PAGE and electro-eluted from gel slices using a Biotrap apparatus (Schleicher and Schuell). The pure uncapped transcripts were dephosphorylated using bovine alkaline phosphatase (Fermentas) and 5′-radiolabelled with T4 polynucleotide kinase (New England Biolabs). Radioactive transcripts were further purified by denaturing 10% PAGE and eluted overnight at 4°C in buffer A (0.3 M NaCl, 0.5 mM EDTA, 10 mM Tris–HCl pH 7.5). Before use, transcripts were folded in water by incubation at 80°C for 2 min, slow cooling to 35°C and kept on ice.
Enzymatic and chemical RNA probing
Enzymatic and chemical structural probing of RNA were conducted by established procedures (24). Probing mixtures (15 μl) containing radioactive transcripts (50 000 c.p.m) were incubated for 20 min on ice in buffer B (10 mM Tris–HCl pH 7.5, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 10% glycerol) supplemented with 2 mM vanadyl ribonucleoside RNase inhibitor (New England Biolabs), 30 U RNasin (Promega) and 10 pmoles of yeast bulk tRNA. Statistical RNA cleavage was achieved after 10 min incubation at 20°C with 5.9.10−5 U of RNase V1, 1.27 U RNase T1 or 5.6 U of RNase T2. Nucleases V1, T2 and T1 were from Kemotex, Invitrogen and BRL, respectively, and lead acetate from Merck. Reactions were stopped by rapid cooling on ice and addition of 15 μl of stop mix A (0.6 M sodium acetate pH 6, 3 mM EDTA and 0.1 mg/ml total tRNA). Lead probing required a 5 min incubation at 20°C with 10.7 mM final concentration of a freshly prepared lead-acetate solution. Rapid cooling on ice and addition of EDTA (17 mM final concentration) was used for quenching the reactions. After phenol–chloroform extraction, RNA was ethanol precipitated in the presence of glycogen (1 μg) and washed twice with 80% ethanol. Pellets were dried and dissolved in formamide dye. Samples were migrated on 10% denaturing-PAGE in parallel with RNase T1 and alkaline ladders of the corresponding end-labelled RNA for band assignments. RNase T1 ladders were made as previously described (25). Alkaline ladders were obtained by incubation of the labelled transcript with 1 μg of yeast bulk tRNA for 10 min at 80°C in a buffer containing 80 mM Na2CO3/NaHCO3 pH 9. As control, we compared probed RNAs with RNAs treated similarly but without probe.
Enzymatic and chemical footprinting
Footprinting reactions were done as described above in the presence of the SLBP. Recombinant human His-tagged SLBP was obtained as previously described (26). The amount of SLBP used for each footprint was determined by EMSA (see below) in order to obtain at least 80% of the transcript (50 000 c.p.m) shifted. Before use, appropriate amounts of SLBP were dialysed against buffer B and then mixed with the radioactive transcripts (50 000 c.p.m).
Electrophoretic mobility shift assay with U7 snRNA transcripts
For electrophoretic mobility shift assay (EMSA), 50 000 c.p.m of 5′-labelled U7 snRNA transcript were incubated in buffer B on ice for 30 min, with H4-12 transcript (0.01 μM) in the presence or in absence of the SLBP (10 μM). The competitions experiments were conducted by pre-annealing the competitor HDE oligonucleotide (5′-AAGAGCTGTAACACTT-3′) with U7 snRNA transcript or by pre-incubating the SLBP with competitor stem–loop RNA (wild-type stem–loop competitor: 5′-GGAGCUCAACAAAAGGCCCUUUUCAGGGCCACCC-3′; *mutated stem–loop competitor: 5′-GGAGCUCAACAAAACCGGAAAGCCUUCCGGACCC-3′, with an underlined stem–loop sequence completely mutated) on ice for 30 min. The complexes were separated by native 5% PAGE and visualized by phosphorimaging.
RESULTS
Methodological considerations
Three transcripts corresponding to the 3′-UTR of histone pre-mRNAs H4-12, H1t and H2a-614 (94, 85 and 113 nt, respectively) have been synthesized by T7 run-off transcription. These sequences start downstream of the stop codons and end after the HDE sequences. They were chosen according to previous studies, which showed that they were active during processing (12,16) (Figure 1). For better T7 transcription yield, two G residues have been inserted at the 5′ end of the sequence. For run-off transcription, C residues were added at the 3′ ends in order to create a SmaI restriction site (Figure 1). The structure of the three RNA has been probed by RNases T1, T2, V1 and by lead. RNase T2 and lead cut preferentially after unpaired residues and RNase T1 cuts after unpaired G residues. RNase V1 cuts in double-stranded sequences or higher order structures. The background hydrolysis by water or traces of contaminating metal cations were discriminated from probe induced-cuts by control experiments performed without probes. As an additional control, we checked by chemical and enzymatic probing that the folding of the 3′-UTR was identical in full-length pre-mRNAs and that the structure of the 3′-UTR was not influenced by sequences located into the open reading frame. For this, the probed uncapped pre-mRNAs were extended by reverse transcription using a primer annealed to the 15 last residues at the 3′ end of the transcript (data not shown).
The H4-12 3′-UTR contains four structured domains
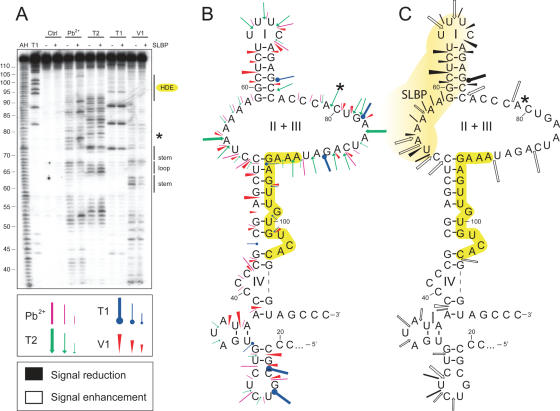
The 94 nt-long T7 transcript from the H4-12 3′-UTR was synthesized and 5′-end labelled by T4 polynucleotide kinase. The 3′-UTR was probed by lead and RNase T2, T1, V1 (see above). As the probes exhibit different specificities, a map of the single stranded and double stranded regions of the RNA molecule could be drawn. These results were used as constraints for the M-fold software (27). The resulting 2D-model of the 3′-UTR of H4-12 precursor mRNA is shown in Figure 2. It contains a cloverleaf structure comprising four domains named I, II, III and IV. The presence of the highly conserved hairpin structure (domain I), which is bound by the SLBP, is clearly visible. This hairpin is highly stable since no T1 cuts are detected after the five G residues of stem I (G37, G38, G48, G49 and G50) even under denaturing conditions (Figure 2A, lane T1). More strikingly, the HDE is embedded in two hairpin structures named domains III and IV. The 5′ part of the HDE is involved in domain III, a 6 bp-stem and 4 nt-loop. The stem from domain III is hardly cut by RNase V1 and T1. V1 cuts are only detected on one side of the helix. On the other side of the helix, three G residues from the HDE (G71, G72 and G74) are not cleaved at all by T1, even under denaturing conditions, indicating inaccessibility of domain III-stem to RNase hydrolysis and high stability in denaturing conditions. Such behaviour suggests that the stem of domain III may be buried in the tertiary structure of the H4 UTR. In contrast G77 which lies in the 3′ part of HDE, in the stem of domain IV, is readily cut by T1 under denaturing conditions (Figure 2A, lane T1) suggesting that stem IV can unfold more easily than stem III. The processing site clearly lies in a single-stranded sequence as indicated by the presence of lead, T1 and T2 cuts and the absence of V1 cuts.
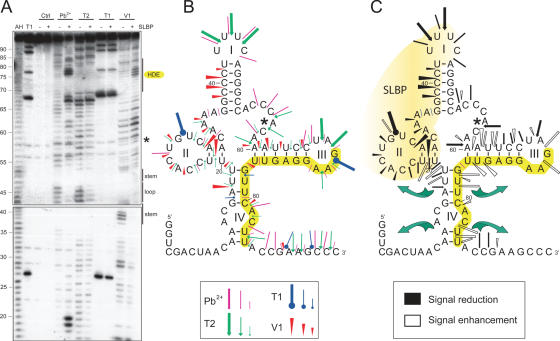
Figure 2.
Establishment of an experiment-based 2D model of the 3′-UTR of histone H4-12 pre-mRNAs in free state and effect of SLBP binding. (A) Chemical and enzymatic structural probing. Probing was done in the presence (+) or absence (−) of recombinant SLBP. AH and T1 are ladders of the RNA under denaturing condition; Ctrl is the control without any probe. Probes were Pb2+, RNase T2, RNase T1 and RNase V1. Domains of the stem–loop structure, the processing site (*) and the spacer element are indicated on the right part of the gel. (B) 2D model of the 3′-UTR of histone H4-12 pre-mRNAs with probing data. The colour code for the probes is indicated in the figure. Three intensities of cuts/modifications for each probe are shown (strong, medium, moderate). The spacer element is highlighted in yellow. (C) Changes in the probing signals on SLBP binding shown on the RNA folding. Protection by SLBP is shown in black (signal reduction) and cut/modification appearance and enhancement (signal enhancement) are shown in white; symbols for each probe are the same as in (B). The presumed SLBP binding domain, deduced from cleavage protections, is coloured in light yellow. Green arrows symbolize opening of stem IV on SLBP binding.
The SLBP footprint experiments reveal structural rearrangements in H4-12 HDE
Using recombinant SLBP produced in baculovirus, we performed footprinting of the SLBP–3′-UTR complex (Figure 2). As expected, the SLBP binding prevents the V1 cleavages that occur in the 5′ part of the stem I and the single-strand specific cleavages in loop I (lead and T2). More surprisingly, the SLBP also protects residues 20–36 in domain II upstream of its target domain I indicating that SLBP also interacts with domain II (Figure 2C, shown in black).
The binding of SLBP to the 3′-UTR not only leads to protection but also induces the appearance of new cleavages or enhancement of existing cuts. This shows that binding of the SLBP induces structural rearrangements of the 3′-UTR precursor (Figure 2C, shown in white). All these SLBP-enhanced cuts are exclusively located in domains III and IV containing the HDE. In the well-defined stem–loop III, V1 and lead reactivity increase is clearly visible. This variation in the accessibility to the probes suggests that the global conformation of the UTR has changed. On the contrary, the structure of domain IV is significantly altered by the binding of the SLBP. The strong lead cleavages that appear on both sides of stem IV indicate that the stem is progressively melted on SLBP binding. Altogether, these data suggest that the reactivity changes observed after SLBP binding might result from significant conformational changes, which lead to a more accessible 3′ end of the HDE.
The SLBP enables U7 snRNA anchoring on H4-12 3′-UTR
One obvious consequence of the conformation changes observed on SLBP binding would be to facilitate U7 snRNP anchoring. To validate this hypothesis, we performed EMSAs with the 3′-UTR of H4-12 pre-mRNAs transcript and [32P]-labelled U7 snRNA transcript. The H4-12 3′-UTR and U7 snRNA were mixed in the presence or absence of the SLBP and complexes separated by native PAGE. Figure 3 shows that incubation of the H4-12 3′-UTR and U7 snRNA with the SLBP improves the anchoring of the U7 snRNA (lane 5) since more U7 is shifted in the SLBP/H4-12/U7 ternary complex in the presence of the SLBP. The ternary complex formation was prevented using a RNA stem–loop corresponding to the SLBP binding site (lane 9) and by a DNA-oligonucleotide corresponding to the HDE sequence (lane 7). This provided unambiguous ternary complex U7/3′-UTR/SLBP identification. In addition, a mutated RNA stem–loop was unable to displace the SLBP from the complex (lane 11).
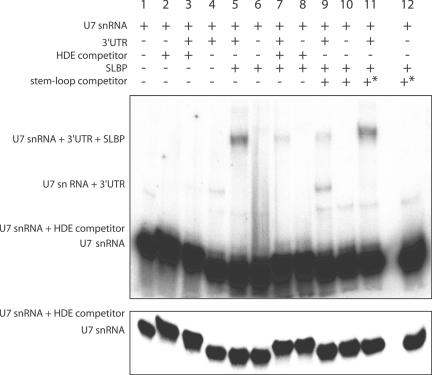
Figure 3.
Electrophoretic Mobility Shift Assay using U7 snRNA, H4-12 pre-mRNAs transcripts, SLBP protein and competitors. The 5′-radiolabelled U7 snRNA was incubated with 0.01 μM H4-12 pre-mRNAs in the absence (lane 4) or presence (lane 5) of recombinant SLBP (10 μM). In order to assess the specificity of hybridization between U7 snRNA and H4-12 3′-UTR, competition experiments were performed using a DNA–oligonucleotide corresponding to HDE sequence both in the absence (lane 3) or presence (lane 7) of SLBP. The (U7: DNA–oligonucleotide) complex is clearly separated from U7 RNA (see the less exposed autoradiogram in the lower part of the figure). To demonstrate that the observed effects are a direct consequence of SLBP binding to the 3′-UTR, competition experiments were done by adding RNA-competitor stem–loops. Whereas the wild-type competitor stem–loop decreased the level of U7 transcripts annealed to the 3′-UTR (lane 9), the asterisk-mutated competitor stem–loop (*) that exhibits no homology with wild-type stem–loop has no effect (lane 11 versus lane 5, see experimental procedures for the sequences of the stem–loops).
The HDE of H1t and H2a-614 3′-UTR are also embedded in hairpins
Next, we performed similar experiments on two other histone precursor 3′-UTR, namely H1t and H2a-614. These histone genes have been selected for this study because they show different behaviour with respect to the influence of the SLBP as shown by previous in vitro processing experiments (16). The in vitro 3′ end processing of histone H1t pre-mRNAs fully requires the SLBP whereas the processing of H2a-614 does not need the presence of the SLBP.
The probing of the H1t 3′-UTR pre-mRNAs has been performed using the same probes (Figure 4) and the 2D model also contains four domains that have been named I to IV in analogy to H4-12. Domain I, the stem–loop structure bound by the SLBP, is highly stable as shown by the absence of reactivity to G residues of the stem under denaturing conditions and by strong stacking at positions 44–48 in the alkaline ladder (Figure 4A). Stem I is accessible to V1 on both sides in contrast to the half-accessibility observed in the case of H4-12. This suggests that the 3′ part of stem I in H4-12 might be masked by another RNA domain or might adopt a specific conformation inaccessible to RNases. Domains II of H4-12 and H1t are very similar whereas domain III is completely unpaired in H1t. The HDE is almost completely embedded in a structured domain IV.
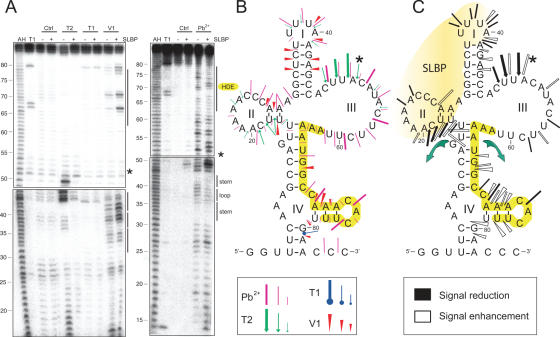
Figure 4.
2D model of the 3′-UTR of histone H1t pre-mRNAs in free state and effect of SLBP binding. (A) Chemical and enzymatic probing data. (B) 2D model of the RNA. (C) Changes in the probing profile induced by SLBP binding are shown on the RNA 2D model. Legend is the same as for Figure 2.
Like for H4-12, the SLBP footprint on the H1t 3′-UTR shows protection in domain I and II but only the 5′ half of stem I is protected (Figure 4C). This is in good agreement with previous experiments which showed that the SLBP interacts only with the 5′ part of domain I to allow binding of another protein, the 3′ hExo, on the second part (28). Binding of the SLBP led to appearance of lead cleavages in stem II and adjacent residues from the upper part of stem IV, suggesting opening of these double stranded regions. Similarly, an enhancement of accessibility to the probes in the 3′ part of domain IV was observed. Like for H4-12, these results suggest that SLBP induces conformational rearrangements resulting in increases of the probe-reactivity of residues from the HDE. However, in the case of the H1t, it is the 5′ part of the HDE that opens on SLBP binding whereas the 3′ part of the HDE is opened in the case of H4-12. To conclude, one can say that the overall 2D-structure of the H1t 3′ UTR is more accessible than H4-12 but the SLBP also induces structural rearrangements in both domains IV.
Probing of the H2a-614 reveals a rather different organization (Figure 5). Only domain I displays clear V1 cuts on both sides of stem I and single strand-specific cuts in loop I. Domains II and III are completely unpaired while domain IV, which contains the HDE, is composed of 9 bp interrupted by two bulges. The 5′ and 3′ ends of the HDE are unpaired. Similar to H4-12 and H1t, binding of the SLBP induces protections exclusively on domains I and II; however no significant changes are observed in domain III and IV, except five lead cleavages at positions 53, 78, 86, 89 and 105 flanking stem IV (Figure 5C). In contrast to the previous histone 3′-UTRs no changes in the HDE are detected on SLBP binding to the H2a-614 3′-UTR. Altogether, these data suggests that the 3′-UTR of H2a-614 is less structured than H4-12 and H1t and that binding of the SLBP has no effect on the structure of the HDE.
Figure 5.
2D model of the 3′-UTR of histone H2a-614 pre-mRNAs in free state and effect of SLBP binding. (A) Chemical and enzymatic probing data. (B) 2D model of the RNA. (C) Changes in the probing profile induced by SLBP binding are shown on the RNA 2D model. Legend is the same as for Figure 2.
SLBP-binding does not improve the U7 snRNA anchoring on H1t and H2a-614 3′-UTR
We have previously shown that the conformation changes observed on SLBP binding on the H4-12 3′-UTR facilitates U7 snRNP anchoring. We repeated the same experiment with H1t and H2a-614 transcripts. Despite the fact that we observed conformation changes in the H1t 3′-UTR on SLBP binding, we did not observe any improvement of the U7 snRNA hybridization (data not shown). The same absence of stimulation was observed with H2a-614 3′-UTR, which did not display any conformation change on SLBP binding (data not shown).
These data, together with footprint results suggest the following hierarchy of SLBP-dependency for U7 snRNA anchoring. The SLBP induces significant conformation changes in H4-12 3′-UTR and consequently favours the anchoring of the U7 snRNA to H4-12. With H1t, significant conformational changes are observed on SLBP binding but no improvement of the U7 snRNA could be detected. With H2a-614, changes could be detected neither on the 2D structure nor on U7 snRNA binding. These results are in good agreement with previous experiments showing that H4-12 and H1t are SLBP-dependent for efficient processing whereas H2a-614 does not require SLBP for in vitro processing (6,16).
DISCUSSION
The 3′ end processing of histone pre-mRNAs is an intricate process because it requires at least three proteins (SLBP, ZFP100, CPSF-73) and the minor U7 snRNP. Numerous RNA–RNA and RNA–protein interactions enable assembly of the processing machinery. In addition, the unique U7 snRNP triggers 3′ end processing of many different histone pre-mRNAs by annealing to distinct HDEs. In order to decipher the function of each component, we investigated the influence of the SLBP on the anchoring of the U7 snRNA by chemical and enzymatic probing.
The structural probing led to three 2D models in which resemblances are found. In the three cases, the stem–loop dedicated to the SLBP binding (domain I) is clearly present, followed by a single stranded region carrying the cleavage site. Remarkably, nucleotides from the three HDE are embedded in duplex structures, which is unexpected for a sequence dedicated to U7 snRNA hybridization. Duplexes harbouring the HDE nucleotides are formed with proximal sequences, as in stem III of H4-12 pre-mRNAs and by long-distance interactions with sequences located upstream of the SLBP hairpin-binding site (as in stem IV of the three pre-mRNAs).
Footprint experiments revealed that significant probe-reactivity changes occur in H4-12 and H1t 5′-UTR on SLBP binding. First, RNA protections were unambiguously detected in domain I and in adjacent domain II of the three 3′-UTR. In fact, the SLBP interacts mainly with the 5′ half of the hairpin I and with the upstream region of domain II. This is in good agreement with previous experiments showing that the 3′ hExo can bind to the already formed SLBP–hairpin complex and form a ternary complex (28). This implies that the 3′ hExo can start trimming before or even without the release of the SLBP. More surprising was the protection induced by the SLBP on the upstream domain II. It was already known that the four residues preceding stem I are essential for SLBP binding (29,30). We show here that SLBP can protect more residues, from 15 residues for H4-12 to only 6 for H2a-614. This variation also suggests some plasticity in the RNA-protein binding mode, according to the sequence diversity observed in the pre-mRNAs upstream of the canonical stem–loop I.
In addition to RNA protection, the binding of the SLBP- induces increased reactivity that can be interpreted as a progressive opening of the duplexes containing H4-12 and H1t HDE. The reactivity changes in H4-12 affect stem–loop III, which becomes more accessible to double-strand specific probes and to lead. The adjacent stem IV is characterized by new lead cuts which suggests melting of the stem. In the case of H1t, reactivity changes are less pronounced but a clear increase of the accessibility of stem IV containing the HDE is observed. Stem IV becomes reactive to single-strand specific probes, suggesting melting of the upper part of the stem, whereas the lower part becomes more accessible to V1 cuts. Thus, in both cases, the reactivity changes observed on SLBP binding reveal significant structural changes and dynamic rearrangements. One obvious consequence of the HDE melting would be to favour U7 snRNA hybridization and we showed that these rearrangements actually facilitate the anchoring of the U7 snRNA in vitro. However, improvement of the U7 snRNA binding is only observed with H4-12 and not with H1t although it displays also the HDE melting. On the other hand, the H2a-614 3′-UTR is much more static and shows no significant conformation change on SLBP binding. These results are in good agreement with previous investigations that showed that the 3′ end processing of H1t is more dependent than H2a on SLBP (16). Altogether, the results observed with the three different UTRs suggest that distinct mechanisms govern the first step of the processing reaction. However, we cannot exclude that the presence of other trans-acting factors like ZFP100 or Sm/Lsm proteins in U7 snRNP might improve the efficiency of U7 hybridization with H1t and H2a-614 and reach a level comparable to that of H4-12.
The current data clearly show that H4-12 3′-UTR binds the SLBP and undergoes dynamic structural rearrangements that favour U7 snRNA hybridization. Such reaction is reminiscent of U6 snRNA which displays a very compact structure in its naked form, whereas in presence of Prp24p and Lsm proteins the RNA structure is much more opened, more accessible to V1 cuts, which would facilitate pairing with U4 RNA (31). A role of RNA chaperones was proposed for Prp24p and Lsm proteins, acting in order to open the RNA structure and stabilize the active U6 snRNA structure, which would not be sufficiently stable by its own. Binding of the SLBP on H4-12 and H1t 3′-UTR induces comparable effects as Prp24p and Lsm proteins on the U6 snRNA. The histone RNA structures become more accessible to the lead, RNases, U7 snRNA and also water molecules. Indeed, spontaneous hydrolysis of the 3′-UTR at the cleavage site was stimulated after SLBP binding, suggesting that the cleavage process mediated by CPSF-73 might be water-mediated.
An interesting consequence of the SLBP role in promoting U7 binding is that 3′ end processing reaction of histone pre-mRNAs is an ordered process. The SLBP binds first, facilitates anchoring of U7 snRNP and finally the whole complex is locked by the ZFP100, which bridges the SLBP-hairpin complex to the U7 snRNP. This model implies that SLBP-binding might initiate and control pre-mRNAs processing. In the absence of the SLBP, the processing machinery would be unable to promote processing and to accumulate processing intermediate complexes (histone pre-mRNAs/U7snRNP/ZFP100). To avoid intermediate-products accumulation, it makes sense to think that SLBP is the first trans-acting factor to bind newly synthesized histone pre-mRNAs. The fact that the HDE of histone precursor mRNAs is ‘hidden’ in the RNA secondary structure from U7 snRNA anchoring might be looked at as a safety mechanism to prevent processing and therefore histone biosynthesis in non-S phase especially at the G1-S transition phase when histone transcription is increasing and SLBP is not yet available (32,33). SLBP is the cornerstone of histone expression, its binding on histone pre-mRNAs ‘gives a go’ to the whole process.
To conclude, the SLBP is necessary for 3′ end processing of histone pre-mRNAs. In addition, the SLBP is essential for cell-cycle regulation of histone expression. The SLBP is one of the three actually known cell cycle regulated factors that are involved in the 3′ end processing reaction of the histone pre-mRNAs (32,33). Together with Symplekin and CstF-64, two components of the cleavage/polyadenylation machinery (11,34,35), the SLBP regulates histone expression at the 3′-end processing level.
Acknowledgments
We are grateful to A. Clénet for skilful technical assistance, to S. Barends for critical reading of the manuscript, P. Romby, H. Moine and J.-C. Paillart for helpful discussions. This work was supported by grants from the Centre National de la Recherche Scientifique (CNRS), Ministère de l'Education Nationale, de la Recherche et de la Technologie (MENRT), Université Louis Pasteur and Association pour Recherche sur le Cancer (ARC# 3814). Funding to pay the Open Access publication charges for this article was provided by Association pour la Recherche sur le Cancer (ARC# 3814).
Conflict of interest statement. None declared.
REFERENCES
- 1.DeBry R.W., Marzluff W.F. Selection on silent sites in the rodent H3 histone gene family. Genetics. 1994;138:191–202. doi: 10.1093/genetics/138.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huynen M., Konings D., Hogeweg P. Equal G and C contents in histone genes indicate selection pressures on mRNA secondary structure. J. Mol. Evol. 1992;34:280–291. doi: 10.1007/BF00160235. [DOI] [PubMed] [Google Scholar]
- 3.Marzluff W.F. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr. Opin. Cell Biol. 2005;17:274–280. doi: 10.1016/j.ceb.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Jaeger S., Barends S., Giegé R., Eriani G., Martin F. Expression of metazoan replication-dependent histone genes. Biochimie. 2005;87:827–834. doi: 10.1016/j.biochi.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z.F., Whitfield M.L., Ingledue T.C., 3rd, Dominski Z., Marzluff W.F. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNAs processing. Genes Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 6.Martin F., Schaller A., Eglite S., Schümperli D., Müller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pillai R.S., Will C.L., Lührmann R., Schümperli D., Müller B. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 2001;20:5470–5479. doi: 10.1093/emboj/20.19.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominski Z., Erkmann J.A., Yang X., Sanchez R., Marzluff W.F. A novel zinc finger protein is associated with U7 snRNP and interacts with the stem–loop binding protein in the histone pre-mRNP to stimulate 3′-end processing. Genes Dev. 2002;16:58–71. doi: 10.1101/gad.932302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azzouz T., Gruber A., Schümperli D. U7 snRNP-specific Lsm11 protein: dual binding contacts with the 100 kDa zinc finger processing factor (ZFP100) and a ZFP100-independent function in histone RNA 3′ end processing. Nucleic Acids Res. 2005;33:2106–2117. doi: 10.1093/nar/gki516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominski Z., Yang X.C., Marzluff W.F. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNAs processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Kolev N., Steitz J.A. Symplekin and multiple other polyadenylation factors participate in 3′ end maturation of histone mRNAs. Genes Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Streit A., Koning T.W., Soldati D., Melin L., Schümperli D. Variable effects of the conserved RNA hairpin element upon 3′ end processing of histone pre-mRNAs in vitro. Nucleic Acids Res. 1993;21:1569–1575. doi: 10.1093/nar/21.7.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho D.C., Scharl E.C., Steitz J.A. Decreasing the distance between the two conserved sequence elements of histone pre-messenger RNA interferes with 3′ processing in vitro. RNA. 1995;1:905–914. [PMC free article] [PubMed] [Google Scholar]
- 14.Scharl E.C., Steitz J.A. The site of 3′ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J. 1994;13:2432–2440. doi: 10.1002/j.1460-2075.1994.tb06528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scharl E.C., Steitz J.A. Length suppression in histone messenger RNA 3′-end maturation: processing defects of insertion mutant premessenger RNAs can be compensated by insertions into the U7 small nuclear RNA. Proc. Natl Acad. Sci. USA. 1996;93:14659–14664. doi: 10.1073/pnas.93.25.14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominski Z., Zheng L.X., Sanchez R., Marzluff W.F. Stem–loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNAs. Mol. Cell Biol. 1999;19:3561–3570. doi: 10.1128/mcb.19.5.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pandey N.B., Williams A.S., Sun J.H., Brown V.D., Bond U., Marzluff W.F. Point mutations in the stem–loop at the 3′ end of mouse histone mRNA reduce expression by reducing the efficiency of 3′ end formation. Mol. Cell Biol. 1994;14:1709–1720. doi: 10.1128/mcb.14.3.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pettitt J., Crombie C., Schümperli D., Müller B. The Caenorhabditis elegans histone hairpin-binding protein is required for core histone gene expression and is essential for embryonic and postembryonic cell division. J. Cell Sci. 2002;115:857–866. doi: 10.1242/jcs.115.4.857. [DOI] [PubMed] [Google Scholar]
- 19.Kodama Y., Rothman J.H., Sugimoto A., Yamamoto M. The stem–loop binding protein CDL-1 is required for chromosome condensation, progression of cell death and morphogenesis in Caenorhabditis elegans. Development. 2002;129:187–196. doi: 10.1242/dev.129.1.187. [DOI] [PubMed] [Google Scholar]
- 20.Zhao X., McKillop-Smith S., Müller B. The human histone gene expression regulator HBP/SLBP is required for histone and DNA synthesis, cell cycle progression and cell proliferation in mitotic cells. J. Cell Sci. 2004;1:6043–6051. doi: 10.1242/jcs.01523. [DOI] [PubMed] [Google Scholar]
- 21.Wagner E.J., Berkow A., Marzluff W.F. Expression of an RNAi-resistant SLBP restores proper S-phase progression. Biochem. Soc. Trans. 2005;33:471–473. doi: 10.1042/BST0330471. [DOI] [PubMed] [Google Scholar]
- 22.Godfrey A.C., Kupsco J.M., Burch B.D., Zimmerman R.M., Dominski Z., Marzluff W.F., Duronio R.J. U7 snRNA mutations in Drosophila block histone pre-mRNAs processing and disrupt oogenesis. RNA. 2006;12:396–409. doi: 10.1261/rna.2270406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perret V., Garcia A., Grosjean H., Ebel J.P., Florentz C., Giegé R. Relaxation of a transfer RNA specificity by removal of modified nucleotides. Nature. 1990;344:787–789. doi: 10.1038/344787a0. [DOI] [PubMed] [Google Scholar]
- 24.Giegé R., Helm M., Florentz C. Classical and novel chemical tools for RNA structure probing. In: Söll D., Nishimura S., Moore P.B., editors. RNA. Amsterdam: Pergamon/Elsevier; 2001. pp. 79–89. [Google Scholar]
- 25.Peattie D., Gilbert W. Chemical probes for higher-order structure in RNA. Proc. Natl Acad. Sci. USA. 1980;77:4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin F., Michel F., Zenklusen D., Müller B., Schümperli D. Positive and negative mutant selection in the human histone hairpin-binding protein using the yeast three-hybrid system. Nucleic Acids Res. 2000;28:1594–1603. doi: 10.1093/nar/28.7.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuker M., Stiegler P. Optimal computer folding of larger RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dominski Z., Yang X.C., Kaygun H., Dadlez M., Marzluff W.F. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell. 2003;12:295–305. doi: 10.1016/s1097-2765(03)00278-8. [DOI] [PubMed] [Google Scholar]
- 29.Williams A.S., Marzluff W.F. The sequence of the stem and flanking sequences at the 3′ end of histone mRNA are critical determinants for the binding of the stem–loop binding protein. Nucleic Acids Res. 1995;23:654–662. doi: 10.1093/nar/23.4.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Battle D.J., Doudna J.A. The stem–loop binding protein forms a highly stable and specific complex with the 3′ stem–loop of histone mRNAs. RNA. 2001;7:123–132. doi: 10.1017/s1355838201001820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karaduman R., Fabrizio P., Hartmuth K., Urlaub H., Luhrmann R. RNA structure and RNA–protein interactions in purified yeast U6 snRNPs. J. Mol. Biol. 2006;356:1248–1262. doi: 10.1016/j.jmb.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 32.Whitfield M.L., Zheng L.X., Baldwin A., Ohta T., Hurt M.M., Marzluff W.F. Stem–loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng L., Dominski Z., Yang X.C., Elms P., Raska C.S., Borchers C.H., Marzluff W.F. Phosphorylation of stem–loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol. Cell Biol. 2003;23:1590–1601. doi: 10.1128/MCB.23.5.1590-1601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lüscher B., Schümperli D. RNA 3′ processing regulates histone mRNA levels in a mammalian cell cycle mutant. A processing factor becomes limiting in G1-arrested cells. EMBO J. 1987;6:1721–1726. doi: 10.1002/j.1460-2075.1987.tb02423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martincic K., Campbell R., Edwalds-Gilbert G., Souan L., Lotze M.T., Milcarek C. Increase in the 64-kDa subunit of the polyadenylation/cleavage stimulatory factor during the G0 to S phase transition. Proc. Natl Acad. Sci. USA. 1998;95:11095–11100. doi: 10.1073/pnas.95.19.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]