Abstract
We report the identification of Cdc7/Dbf4 phosphorylation sites in human MCM2 and the determination of the role of Cdc7/Dbf4 phosphorylation of MCM2 in the initiation of DNA replication. Using immunoblotting, immunofluorescence, and high-speed automated cell-imaging analyses with antibodies specific against MCM2 and Cdc7/Dbf4 phosphorylated MCM2, we show that the chromatin recruitment and phosphorylation of MCM2 are regulated during the cell cycle in HeLa cells. Chromatin-bound MCM2 is phosphorylated by Cdc7/Dbf4 during G1/S, which coincides with the initiation of DNA replication. Moreover, we show that baculovirus-expressed purified MCM2-7 complex and its phosphomimetic MCM2E-7 complex display higher ATPase activity when compared with the nonphosphorylatable MCM2A-7 complex in vitro. Furthermore, suppression of MCM2 expression in HeLa cells by siRNA results in the inhibition of DNA replication. The inhibition can be rescued by the coexpression of wild type MCM2 or MCM2E but not MCM2A. Taken together, these results indicate that Cdc7/Dbf4 phosphorylation of MCM2 is essential for the initiation of DNA replication in mammalian cells.
INTRODUCTION
In all eukaryotic cells, chromosomal DNA replication is a tightly regulated process, which must be strictly coordinated with other cell cycle events, such as cell division, to ensure that the daughter cells maintain the same ploidy as the parental cells. DNA replication occurs at multiple replication origins scattered throughout the genome and each segment of DNA replicates once and once only per cell cycle. It is believed that regulation of DNA replication is exerted primarily at the initiation of DNA synthesis. Perturbation of DNA replication can lead to numerous deleterious events, including mutagenesis and chromosome instability, which can have severe consequences for an organism such as cell death, birth and developmental defects, and cancer (Elledge, 1996).
Great insight into the molecular mechanisms of the initiation of DNA replication in eukaryotic cells has been obtained in the past decades. A large body of genetic and biochemical evidence has now emerged to support a model in which the initiation of DNA replication in eukaryotic cells is controlled by the stepwise establishment of prereplication complexes (pre-RCs) at DNA replication origins in G1 (also known as origin licensing) and the activation of two S phase–promoting kinases, Cdks/cyclins, and Cdc7/Dbf4, in G1/S (for reviews, see Bell and Dutta, 2002; Mendez and Stillman, 2003; Forsburg, 2004). The pre-RCs contain several groups of proteins that are essential for the initiation of DNA replication. These include six subunits of origin recognition complex (ORC), the loading factors Cdc6 and Cdt1 proteins, and the putative DNA replicative helicase MCM2-7 complex (Bell and Stillman, 1992; Chong et al., 1995; Donovan et al., 1997; Maiorano et al., 2000; Nishitani et al., 2000). Origin licensing is sequential, with ORC binding to replication origins, which recruits the Cdc6 and Cdt1 proteins and thereby promotes the loading of MCM proteins (Stillman, 1996; Tye, 1999; Bell and Dutta, 2002; Forsburg, 2004). Although necessary, origin licensing is not sufficient to initiate DNA replication. The initiation of DNA replication requires the activation of two S phase–promoting kinases, Cdks/cyclins and Cdc7/Dbf4 in G1/S (Bell and Dutta, 2002; Henneke et al., 2003; Forsburg, 2004). It is thought that both S phase–promoting kinases phosphorylate critical downstream targets at pre-RCs that trigger the initiation of DNA replication.
The putative DNA replicative helicase, MCM2-7 proteins were originally identified as a group of proteins essential for DNA replication (chromosomal maintenance; Tye, 1999; Forsburg, 2004). They share sequence homology to each other in their nucleotide-binding domains and are a distinct subgroup of the large AAA ATPase family, which has many cellular functions (Lupas and Martin, 2002). Biochemical purification and reconstitution of recombinant proteins indicate that MCM2-7 proteins mainly form a heterohexameric complex with 1:1:1:1:1:1 stoichiometry and likely have a ring-shaped structure that surrounds DNA through its central channel (Lee and Hurwitz, 2000; Prokhorova and Blow, 2000; Sato et al., 2000; Schwacha and Bell, 2001; Davey et al., 2003; Ying and Gautier, 2005). These results strongly suggest that the MCM complex represents the long-sought cellular replicative helicase, similar to that of bacteriophage T7 Gp4, the bacterial DnaB protein, and the large T antigen (T) of SV40 (SV40). Indeed, a single MCM-related protein encoded by archaeon Methanobacterium thermoautotrophicum ΔH (MtMCM), which assembles into two stacked homohexameric rings, has helicase activity (Fletcher et al., 2003). Similarly, purified recombinant Saccharomyces cerevisiae MCM2-7 complex displays ATPase activity and Schizosaccharomyces pombe, mouse, and human MCM subcomplex composed of the dimeric complex of MCM subunits 4, 6, and 7 (MCM4,6,7)2 has an intrinsic ATP-dependent DNA helicase activity (Ishimi, 1997; Lee and Hurwitz, 2001; Schwacha and Bell, 2001; Davey et al., 2003; You et al., 2003). Depletion or neutralization of MCM proteins after initiation in S. cerevisiae or Xenopus cell-free system blocks the progression of replication forks, indicating that MCM proteins are also required for DNA replication fork progression (Labib et al., 2001; Pacek and Walter, 2004). In addition, MCM proteins are downstream targets of Cdks/cyclins and Cdc7/Dbf4, suggesting that the function of MCM proteins may be regulated by S phase–promoting kinases during DNA replication (Masai and Arai, 2002; Forsburg, 2004 and references therein).
Previously, we and others showed that Cdc7/Dbf4 selectively phosphorylates MCM2 subunit of MCM complex (Lei et al., 1997; Brown and Kelly, 1998; Jiang et al., 1999a; Jares and Blow, 2000). However, the molecular mechanism by which Cdc7/Dbf4 phosphorylation of MCM2 regulates the initiation of DNA replication is not clear. In this study, we report the identification of Cdc7/Dbf4 phosphorylation sites in human MCM2 and the determination of the role of Cdc7/Dbf4 phosphorylation of MCM2 in the initiation of DNA replication in mammalian cells.
MATERIALS AND METHODS
Cell Culture, Transfections, and Synchronization
HeLa cells were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS) and penicillin/streptomycin at 37°C. For transient transfection, HeLa cells were transfected with plasmid(s) and/or siRNA using Oligofectamine according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). For cell cycle synchronization, HeLa cells were synchronized at G1/S by a double-thymidine treatment and released into different stages of the cell cycle by addition of fresh medium as previously described (Jiang et al., 1999b). DNA content of synchronous cells were determined by flow cytometry (FACSort, Becton Dickinson, Lincoln Park, NJ) and analyzed by CELLQuest Ver. 3.3 (Becton Dickinson).
Antibodies
Rabbit polyclonal anti-MCM2S1 antibodies (α-MCM2S1) were generated against a synthetic human MCM2 phosphopeptide, MCM2S1 (CRGLLYDpSDEEDE, amino acid residues 133–144), coupled to keyhole limpet-hemocyanin (KLH) via the added N-terminal Cys. α-MCM2S1 was purified using MCM2S1 phosphopeptide-Sepharose 4B affinity column. Rabbit polyclonal anti-MCM2M1 antibodies (α-MCM2M1) were generated against bacterially expressed His-MCM2M1 (amino acids residues 1–296; Figure 1B) and was purified using GST-MCM2M5 fusion protein affinity column. Anti-MCM3, 4, 5, 6, and 7 and xOrc2 antibodies were kindly provided by Drs. John Newport (UCSD, San Diego, CA), Ralf Knippers (Konstanz, Germany), and Ron Laskey (University of Cambridge, United Kingdom). Anti-MCM2 (BM28), RPA, α-tubulin, FLAG M2, and bromodeoxyuridine (BrdU) monoclonal antibodies were purchased from BD Biosciences PharMingen (610701, clone 46; San Diego, CA), Oncogene Research Products (Ab-2; Boston, MA), Sigma (T5168; St. Louis, MO), Sigma (F3165), and Serotec (OBT0030S; Raleigh, NC), respectively. All secondary antibodies used in the study were purchased from Southern Biotechnology (Birmingham, AL) and Jackson ImmunoResearch Laboratories (West Grove, PA).
Figure 1.
Identification of Cdc7/Dbf4 phosphorylation sites in human MCM2. (A) Cdc7wt/Dbf4 (wild type) or Cdc7kd/Dbf4 (kinase dead) complex was produced in Sf9 cells using a baculovirus-expression system and purified by anti-FLAG antibody affinity chromatography followed by gel filtration chromatography. Silver staining of purified Cdc7wt/Dbf4 and Cdc7kd/Dbf4 on an SDS/polyacrylamide gel is shown. (B) (a) Schematic representation of bacterially expressed His-MCM2 protein (wt) and its deletion mutants used in in vitro Cdc7/Dbf4 kinase assay. (b) Bacterially expressed purified His-MCM2 and its deletion mutant proteins described in (a) were incubated with baculovirus-expressed purified Cdc7/Dbf4 in the presence of [γ-32P]ATP. Proteins were resolved by SDS/PAGE and visualized by autoradiography (right) or Coomassie blue staining (left). (C) 32P-labeled MCM2 in B and 32P-labeled endogenous MCM2 immunoprecipitated by α-MCM2M1 were eluted from SDS/polyacrylamide gels and digested with trypsin. The resulting phosphopeptides were separated by electrophoresis (pH 1.9 buffer) in the horizontal dimension (anode on the left) and chromatography (phospho-chromatography buffer) in the vertical dimension. Two-dimensional (2-D) tryptic phosphopeptide maps of His-MCM2 (a), His-MCM2N (unpublished data), His-MCM2M1 (unpublished data), and His-MCM2M5 (b) were very similar to that of endogenous MCM2 (c). A schematic map is shown in d. Note: the phosphopeptides (1–6) and the corresponding phosphorylation sites (Ser27, Ser41, and Ser139) are labeled. Multiple phosphopeptides represent partial tryptic digestion products of a single phosphorylation site on the maps. For details, see text, Materials and Methods, and Supplementary Figure S2. (D) HeLa cells were transfected with control luciferase siRNA (siLuc), Cdc7 siRNA (siCdc7), or Cdk2 siRNA (siCdk2) using the Oligofectamine method. Three days after transfection, cells were lysed with RIPA lysis buffer, and cell lysates were subjected to SDS/PAGE, transferred to the PVDF membrane, and immunoblotted with α-Cdc7 (a), α-Cdk2 (b), α-MCM2M1 (c), α-Cyclin A (d), α-Cyclin E (e), or anti-α-tubulin antibody (f). Same cell lysates used in a–f were immunoprecipitated with α-Cdk2, and immunoprecipitates were incubated with 1 μg of histone H1 in the presence of [γ-32P]ATP. After incubation, reactions were resolved by SDS/PAGE and visualized by autoradiography (g). HeLa cells were transfected with siLuc, siCdc7, or siCdk2 as in a–g. Three days after transfection, cells were metabolically labeled with 32P-orthophosphate for 6 h and 32P-labeled endogenous MCM2 from siLuc-, siCdc7-, or siCdk2-transfected cells was immunoprecipitated by α-MCM2M1. After washing, the immunoprecipitates were subjected to SDS/PAGE and visualized by autoradiography (h). 32P-labeled MCM2 proteins in panel h were eluted from SDS/polyacrylamide gel and digested with trypsin. The resulting phosphopeptides were separated by electrophoresis (pH 1.9 buffer) in the horizontal dimension (anode on the left) and chromatography (phospho-chromatography buffer) in the vertical dimension. Shown are 2-D tryptic phosphopeptide maps of 32P-labeled endogenous MCM2 from cells transfected with siLuc (i, 2000 cpm), 32P-labeled endogenous MCM2 from cells transfected siCdc7 (j, 2000 cpm), and 32P-labeled endogenous MCM2 from cells transfected siCdk2 (k, 2000 cpm). Note: the intensities of phosphopeptides 1–6 (corresponding to Ser27, Ser41, and Ser139 phosphorylation in panel j) were greatly diminished when compared with those in i and k. The asterisks represent nonspecific phosphopeptides, which show similar intensities in i–k.
MCM2 Constructs, In Vitro Kinase Assay, In Vivo Metabolic Labeling, Two-dimensional Tryptic Peptide Mapping, and Mass-Spectrometric Analyses
Human MCM2 cDNA and its deletion mutants were generated by PCR and subcloned into EcoRI/SalI sites of pGEXKG, pHis8, or pFLAG plasmid. pHisMCM2M5S27A, pHisMCM2M5S41A, pHis8MCM2M5S139A, pHisMCM2M5S26AS139A, pHisMCM2M5S40AS139A, pHisMCM2M5A, pFLAG-MCM2S139A, pFLAG-MCM2A, and pFLAG-MCM2E, in which Ser-27, Ser-41, Ser-139, Ser26 and Ser139, Ser40 and Ser139 or Ser27, Ser41 and Ser139 were substituted to Ala or Glu were generated by Quick-Change mutagenesis method according to the manufacturer's description (Stratagene, La Jolla, CA). All constructs were fully sequenced.
For in vitro kinase assay, 1–2 μg of purified MCM2 proteins was incubated with 20–50 ng of purified Cdc7/Dbf4 kinase in the presence of [γ-32P]ATP at 30°C for 30 min as previously described (Jiang et al., 1999a). The kinase reactions were terminated by adding an equal volume of 2× sample buffer, and the reaction products were separated by SDS/polyacrylamide gels, stained with Coomassie blue and then dried before autoradiography.
For in vivo metabolic labeling, untransfected HeLa cells or HeLa cells transfected with siRNA for 3 d were washed once with phosphate-free medium and then incubated with the same phosphate-free medium supplemented with 10% dialyzed FBS and 2 mCi/ml [32P]orthophosphate for 6 h. Cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.4, 1% Nonidet P-40, 1% sodium-deoxycholate, 0.1% SDS, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, 1 mM Na3VO4, 1 mM NaF, 10 U/ml aprotinin, and 20 μg/ml leupeptin). After clarification by centrifugation, MCM2 protein was immunoprecipitated by α-MCM2M1. The immunoprecipitates were separated by SDS/polyacrylamide gels, stained with Coomassie blue, and dried before autoradiography.
32P-labeled MCM2 proteins were excised from SDS/polyacrylamide gels and digested with 1 μg of TPCK-treated trypsin in 50 mM NH4HCO3 for 3 h at 37°C and with additional 1 μg trypsin overnight. The supernatant was lyophilized twice, desalted, and dissolved in 1.9 buffer (500∼2000 cpm/μl). Samples were subjected to two-dimensional (2-D) tryptic phosphopeptide mapping analysis as described (Boyle et al., 1991).
For mass spectrometry analysis, MCM2 proteins were resolved by SDS-PAGE and digested in-gel with trypsin (Shevchenko et al., 1996). Tryptic digests were loaded onto a 360-μm OD × 75-μm ID microcapillary precolumn containing 5 cm 5 μm Monitor C18 particles (Column Engineering, Ontario, CA). The precolumn was rinsed with 20 μl of 0.1% acetic acid and connected to an analytical column (360 μm OD × 75 μm; 10 cm 5 μm Monitor C18) with an integrated ESI emitter tip (Ficarro et al., 2005). Peptides were eluted into a linear ion trap mass spectrometer (LTQ; Thermo Electron, San Jose, CA) at a flow rate of ∼100 nl/min with an HPLC gradient of 0–50% B in 30 min (A = 0.1 M acetic acid, B = 0.1 M acetic acid in acetonitrile).
Immunoblotting, Immunofluorescence, and High-Throughput Microscopy Analyses
Standard immunoblotting and immunofluorescence analyses were performed as previously described (Jiang et al., 1999a). Cell images were photographed using a DM IRE2 fluorescence microscopy (Leica, Deerfield, IL). To make a 3-D image of a cell, a series of pictures from the top to the bottom of a cell were taken (every 0.1 μm/picture, 50 pictures/cell) and reconstituted using Volocity 3-D reconstruction imaging software (Improvision, Lexington, MA).
For high-throughput microscopy analysis, asynchronous growing cells stained with α-MCM2S1 or α-MCM2M1 and Hoechst 33258 were sorted and photographed using an EIDAQ 100 high-throughput automated microscopy system (Q3DM, San Diego, CA) and more than 1000 cells per experiment were analyzed. The expression and phosphorylation levels of MCM2 protein and DNA content in cells were determined by intensities of α-MCM2S1, α-MCM2M1, or Hoechst 33258 staining using Cytoshop software (Q3DM).
Purification of Cdc7/Dbf4 and MCM Complexes
The details of mammalian Cdc7/Dbf4 and MCM2-7 baculovirus construction are available on request. To facilitate protein purification and detection, we added an epitope tag at N-terminus of each mammalian Cdc7, Dbf4, and MCM protein (Myc tag for Cdc7 and Cdc7kd; FLAG tag for Dbf4, MCM2, MCM2A, MCM2E, MCM5, and MCM6; and HA tag for MCM3, MCM4, and MCM7). For purification of Cdc7/Dbf4 or MCM complex, 500 ml of Sf9 insect cells (2 × 106 cells/ml) was coinfected with Cdc7/Dbf4, Cdc7kd/Dbf4, MCM2-7, MCM2A-7, or MCM2E-7 baculovirus for 48 h. The infected cells were harvested, washed once with ice-cold PBS, and then resuspended in 10 ml of buffer A (20 mM Tris-HCl, pH 7.5, 0.3 M sodium glutamate, 2 mM magnesium acetate, 0.05% Nonidet P-40, 0.25% Triton X-100, 10% glycerol, 1 mM PMSF, 10 U/ml aprotinin, and 20 μg/ml leupeptin). After incubation on ice for 30 min, the supernatant were collected by centrifugation at 35,000 × g at 4°C for 30 min and then mixed with anti-FLAG M2 Ab-agarose beads (Sigma) equilibrated with buffer A, and incubated at 4°C for 3 h. The beads were collected and washed with three times with 10 bead volumes of buffer B (50 mM Tris-HCl, pH7.5, 150 mM NaCl, 0.1 mM EDTA, 10% glycerol, 1 mM PMSF, 10 U/ml aprotinin, and 20 μg/ml leupeptin). Bound proteins were eluted three times by incubation at 4°C for 30 min with an equal bead volume of buffer B containing 0.2 mg/ml the 3xFLAG peptide (Sigma). The eluted proteins were dialyzed against buffer B, concentrated using Ultrafree-MC (100-kDa cut, Biomax-100; Millipore, Bedford, MA), loaded to a Superose-6 gel filtration column (HR 10/30, Amersham Biosciences, Piscataway, NJ) equilibrated with buffer B, and fractionated through AKTA-FPLC system (Amersham Biosciences). The quality of each eluted fraction was measured by SDS/polyacrylamide gels followed by silver staining, and protein concentrations were determined by the Bradford method (Bio-Rad, Richmond, CA). For Cdc7/Dbf4 purification, peak fractions were pooled, frozen in liquid nitrogen, and stored at −70°C. For MCM purification, fractions 23 and 24, which contained all six MCMs with 1:1:1:1:1:1 stoichiometry and approximately ∼600 kDa, were pooled, frozen in liquid nitrogen, and stored at −70°C.
ATPase Assay
For the ATPase assay, purified MCM2-7, MCM2A-7, or MCM2E-7 complex was incubated in ATPase buffer (20 mM Tris-HCl, pH 7.5, 0.1 mg/ml bovine serum albumin, 0.5 mM DTT, 10 mM MgCl2, and 25 μM cold ATP) in the presence of 1 μCi of [γ-32P]ATP at 30°C for 30 min. After incubation, 0.5 μl of the reaction was spotted on a poly(ethyleneimine)-cellulose TLC plate (10078; Selecto Scientific, Suwanee, GA) and developed in 0.8 M LiCl and 0.8 M glacial acetic acid as described (Herbig et al., 1999).
Pre-RC Formation Assay Using the Xenopus laevis System
Xenopus interphase egg extracts and demembraned sperm nuclei were prepared as described (Kubota et al., 1997; Tada et al., 2001). Immunodepletion of MCM2 (three rounds, 45 min each) were carried out at 4°C with protein A-Sepharose beads prebound with α-MCM2M1. To analyze the proteins bound to chromatin during DNA replication, demembraned sperm nuclei were mixed with egg extracts (3000 sperm nuclei/μl extracts) and an ATP-regenerating system and incubated at 22°C for 30 min. Baculovirus-expressed purified human MCM2–7, MCM2A-7, and MCM2E-7 (100 nM) or bacterially purified GST-geminin (100 nM) was added in egg extracts before the incubation with sperm nuclei. After the incubation, the mixture was diluted in ELB buffer (10 mM HEPES-KOH, pH 7.7, 2.5 mM MgCl2, 50 mM KCl, 250 mM sucrose, and 1 mM DTT) containing 0.1% Triton X-100 and was centrifuged at 6000 × g for 2 min at 4°C onto ELB buffer containing 500 mM sucrose to collect chromatin. After washing with ELB buffer containing 0.1% Triton X-100 for three times, the pellet was resuspended with sample buffer, boiled for 3 min, and subjected to SDS-PAGE followed by immunoblotting.
RNA Interference
A small interference RNA duplex (siRNA) mixture that targets a 5′-untranslated region of human MCM2 (AAUCAUCGGAAUCCTTCACCA) and 3′-untranslated region of human MCM2 (AAGGAUUCCUUGGGAUUCUGG), and siRNA that targets the coding region of human Cdc7 (AAGCUGUACCACAGCUUAGUA or AAGCAGTCAAAGACTGTGGAT) were purchased from Dharmacon Research (Boulder, CO). Escherichia coli RNase III–prepared Cdc2 esiRNA, Cdk2 esiRNA, and control luciferase esiRNA were generated as described (Zhu and Jiang, 2005).
For the DNA replication assay, cells were transfected with were 100 nM of MCM2 siRNA mixture or control siRNA using Oligofectamine (Invitrogen). Twenty-four hours after transfection, cells were transfected again with 100 nM of MCM2 siRNA mixture or control siRNA, along with pFLAG-MCM2, pFLAG-MCM2A, or pFLAG-MCM2E using Lipofectamine 2000 (Invitrogen). Twenty-four hours after the second transfection, cells were trypsinized and replated on the coverslips in fresh medium containing 10 μM BrdU. Sixteen hours after incubation, cells were fixed and stained with mouse anti-FLAG antibody and rat anti-BrdU antibody and secondary antibodies and then mounted as previously described (Jiang et al., 1999a). BrdU-positive cells in FLAG positive cells (>500) were scored.
RESULTS
Identification of Cdc7/Dbf4 Phosphorylation Sites in Human MCM2
To identify Cdc7/Dbf4 phosphorylation sites in MCM2, bacterially expressed purified human MCM2 and its deletion mutants were incubated with baculovirus-expressed purified human Cdc7/Dbf4 or the kinase dead mutant Cdc7kd/Dbf4 in the presence of [γ-32P]ATP (Figure 1, A and B, and Supplementary Figure S1A). The MCM2 protein and its deletion mutants were mainly phosphorylated by Cdc7/Dbf4, but not Cdc7kd/Dbf4, at MCM2 N-terminus (amino acid residues 1–178) in vitro (Figure 1B and Supplementary Figure S1A). We performed 2-D tryptic phosphopeptide mapping analysis of MCM2 and its mutants phosphorylated by Cdc7/Dbf4 in vitro and demonstrated that the N-terminal region of MCM2 (residues 1–178) was phosphorylated by Cdc7/Dbf4 at multiple sites (spots, Figure 1C). Mass-spectrometric analysis of these trypsin-digested peptides indicated that MCM2 was mainly phosphorylated by Cdc7/Dbf4 at Ser27, Ser41, and Ser139 (described in Materials and Methods and Supplementary Figure S1B). To confirm mass-spectrometry results, we generated several point mutant MCM2M5 proteins, in which Ser27, Ser41, Ser139, all three, as well as Ser26 and Ser139 or Ser40 and Ser139 were substituted by Ala (S27A, S41A, S139A, 3A, S26AS139A, or S40AS139A). 2-D tryptic phosphopeptide mapping analysis of point mutant MCM2M5 proteins revealed that only peptides bearing S27A, S41A, or S139A could not be phosphorylated by Cdc7/Dbf4 (see Supplementary Figure S2D). Together, these results demonstrated that Ser27, Ser41, and Ser139 in MCM2 are in vitro Cdc7/Dbf4 phosphorylation sites.
To determine the in vivo Cdc7/Dbf4 phosphorylation sites in MCM2, HeLa cells were metabolically labeled with 32P-orthophosphate, and endogenous 32P-labeled MCM2 protein was immunoprecipitated by anti-MCM2–specific antibodies (α-MCM2M1, see below). The immunoprecipitates were subjected to SDS/PAGE and ∼120-kDa MCM2 was identified by autoradiography. 2-D tryptic phosphopeptide mapping analyses of in vivo 32P-labeled MCM2 were performed and then compared with those of bacterially expressed purified human MCM2 phosphorylated by Cdc7/Dbf4 in vitro. Multiple phosphopeptides (spots) in the 2-D map of in vivo 32P-labeled MCM2 observed were similar to the phosphopeptides observed in the 2-D maps of MCM2 phosphorylated by Cdc7/Dbf4 in vitro (Figure 1C and Jiang et al., 1999a). In addition, mass-spectrometric analysis on the trypsin-digested peptides of in vivo 32P-labeled MCM2 indicated that MCM2 was phosphorylated mainly at Ser27, Ser41, and Ser139 and, to a lesser extent, at Ser53 and Ser108 (representative data showed in Supplementary Figure S2, A–C). Phosphorylation of Ser108 on MCM2 by ATR was previous described (Cortez et al., 2004; Yoo et al., 2004). Because MCM2 was phosphorylated in vivo at Ser27, Ser41, and Ser139, which were phosphorylated by Cdc7/Dbf4 in vitro, the results suggested that Ser27, Ser41, and Ser139 are in vivo Cdc7/Dbf4 phosphorylation sites in MCM2.
Analysis of the surrounding sequences of Ser27, Ser41, and Ser139 suggested that the Ser27 and Ser41 sites were also similar to the Cdk phosphorylation consensus site (X-S/T-P-X-K/R). Because baculovirus-expressed purified Cdk2/cyclin E could also phosphorylate Ser27 and Ser41 but not Ser139 of MCM2 in vitro (T. Tsuji and W. Jiang, unpublished observation), we wanted to ascertain that phosphorylation of Ser27, Ser41, and Ser139 in MCM2 in vivo was Cdc7/Dbf4 dependent. We first treated HeLa cells with two commonly used Cdk inhibitors, roscovitine and olomoucine. To our surprise, administration of these two Cdk inhibitors in cells resulted in strongly inhibition of Cdc7/Dbf4 kinase activity by reduction the expression levels of Cdc7 protein (unpublished data). Therefore, we performed knockdown experiments in which we reduced Cdc7 or Cdk2 expression by siRNA treatment and monitored phosphorylation of MCM2 in these cells by 32P-orthophosphate labeling. Immunoblotting analysis indicated that transfection of Cdc7 siRNA or Cdk2 siRNA, but not control (Luciferase) siRNA, effectively reduced expression of Cdc7 (>70%) or Cdk2 (>95%) in HeLa cells (Figure 1D, a and b). No reduction in MCM2, cyclin A, or α-tubulin expression was detected in Cdc7, Cdk2, or control siRNA-treated cells (Figure 1D, c, d, and f). However, depletion of Cdc7 expression by Cdc7 siRNA, but not depletion of Cdk2 expression by Cdk2 siRNA, resulted in an accumulation of the slower-migrating band of MCM2 that represents an underphosphorylated form of MCM2 (Figure 1Dc and see below). In addition, depletion of Cdc7 expression by Cdc7 siRNA increased the expression level of cyclin E and the Cdk2 associated histone H1 kinase activity, presumably due to block of cell cycle progression at G1/S (Figure 1D, e and g).
Similar results were also obtained used another Cdc7 siRNA (Montagnoli et al., 2004 and unpublished data). Analysis of in vivo 32P-labeled MCM2 protein immunoprecipitated by α-MCM2M1 revealed that phosphorylation of MCM2 in Cdc7 siRNA-treated cells was markedly reduced when compared with that in Cdk2 or control siRNA-treated cells (Figure 1Dh). 2-D tryptic phosphopeptide mapping analysis indicated that reduction of phosphorylation of MCM2 in Cdc7 siRNA-treated cells was mainly due to inhibition of phosphorylation of MCM2 at Ser27, Ser41, and Ser139 sites (Figure 1D, i–k). Taken together, these results strongly suggest that MCM2 is an in vivo Cdc7, but not Cdk2, substrate and Ser27, Ser41, and Ser139 in MCM2 are in vivo Cdc7/Dbf4 phosphorylation sites.
Chromatin-associated MCM2 Is Phosphorylated by Cdc7/Dbf4 in G1/S, which Coincides with the Initiation of DNA Replication
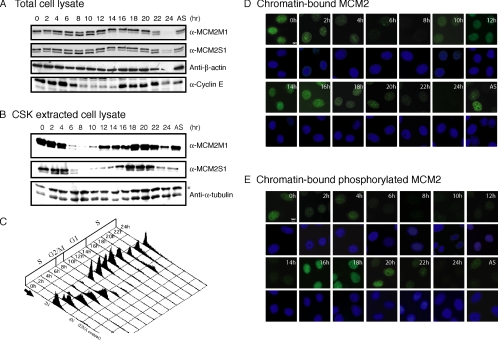
To determine the functional role of Cdc7/Dbf4 phosphorylation of MCM2 in regulation of DNA replication, we generated rabbit polyclonal antibodies against bacterially expressed purified human MCM2 N-terminal fragment M1 (α-MCM2M1) and a chemically synthesized MCM2 phosphoserine-139 peptide (α-MCM2S1; Supplementary Figure S3). Immunoblotting analysis indicated that α-MCM2S1 specifically recognizes MCM2 protein that is phosphorylated at Ser139, whereas α-MCM2M1 recognizes both phosphorylated and unphosphorylated MCM2 proteins (Supplementary Figure S3). Using these antibodies, we examined the expression, chromatin association, and Cdc7/Dbf4 phosphorylation of MCM2 during the cell cycle. HeLa cells were synchronized at G1/S by a double-thymidine block and then released into different stages of the cell cycle by addition of fresh medium. The cell cycle distributions of the synchronous cells were determined by FACS analysis (Figure 2C). The expression, chromatin association, and Cdc7/Dbf4 phosphorylation of MCM2 were determined by immunoblotting and immunofluorescence analyses using α-MCM2M1 and α-MCM2S1. Consistent with previous reports (Todorov et al., 1995; Jiang et al., 1999a), immunoblotting analysis showed that the expression levels of MCM2 were constant during the cell cycle and the phosphorylation levels of MCM2 were slightly regulated during the cell cycle (Figure 2A). In contrast, immunoblotting and immunofluorescence analyses revealed that chromatin association of MCM2 and Cdc7/Dbf4 phosphorylation of chromatin-bound MCM2 were clearly regulated during the cell cycle. As shown in Figure 2, B, D, and E, chromatin-bound MCM2 and Cdc7/Dbf4 phosphorylation of chromatin-bound MCM2 were detected in early S phase cells and gradually decreased in late S and G2/M phase cells. As cells entered the next round of the cell cycle, association of MCM2 with chromatin was detected in cells at G1 phase (Figure 2, B and D, 12–14 h). However, Cdc7/Dbf4 phosphorylation of chromatin-bound MCM2 was not detected in cells at this stage of the cell cycle. In contrast, the levels of Cdc7/Dbf4 phosphorylation of chromatin-bound MCM2 increased dramatically and reached maximum in G1/S and early S phase (Figure 2, B and E, 16–18 h). These results indicated that chromatin-bound MCM2 and Cdc7/Dbf4 phosphorylation of chromatin-bound MCM2 were regulated during the cell cycle. MCM2 was not phosphorylated by Cdc7/Dbf4 in early G1 and unphosphorylated MCM2 accumulated on chromatin. Chromatin-bound MCM2 was phosphorylated by Cdc7/Dbf4 during G1/S phase, and phosphorylated MCM2 detached from chromatin in late S and G2/M.
Figure 2.
Immunoblotting and immunofluorescence analyses of chromatin-association and phosphorylation of MCM2 during the cell cycle in synchronous cells. HeLa cells were synchronized at the G1/S transition by a double-thymidine block and then released into different stages of the cell cycle by addition of fresh medium. (A) Cells were lysed in 1% NP-40 buffer. Cell lysates were separated by SDS/PAGE and immunoblotted with either α-MCM2M1, α-MCM2S1, anti-β-actin antibody (loading control), or α-Cyclin E (G1/S marker). (B) Cells were extracted with CSK/0.5% Triton X-100 buffer. After extraction, the insoluble fraction of cells was lysed into sample buffer. After sonication, cell lysates were separated by SDS/PAGE and immunoblotted with either α-MCM2M1, α-MCM2S1, or anti-α-tubulin antibody (loading control, nonspecific band was marked by the asterisk). (C) Shown are the cell cycle parameters of synchronous cells determined by FACS analysis. (D and E) Cell grown in coverslips were extracted with CSK/0.5% Triton X-100 buffer as in B before fixation. Cells were then stained with α-MCM2M1 (green) and Hoechst 33258 (blue; D) or α-MCM2S1 (green) and Hoechst 33258 (blue; E).
We also examined the phosphorylation and association of MCM2 with chromatin during the cell cycle in asynchronous HeLa cells by immunofluorescence analysis with α-MCM2M1 and α-MCM2S1 using high-throughput automated microscopy (see Materials and Methods). Asynchronous HeLa cells grown on coverslips were extracted with or without CSK buffer (remove cytosolic and nucleosolic proteins but retain the chromatin-bound proteins) before fixation. The expression or phosphorylation levels of MCM2 during the cell cycle were determined by intensities of α-MCM2M1 or α-MCM2S1 staining. To verify DNA content, cells were also costained with Hoechst 33258. At least a minimum of 1000 cells was analyzed per experiment. As shown in Figure 3, typical cell cycle parameters of asynchronous cell cultures (histograms of G1-S-G2/M, bottom panels) were obtained based on the intensities of Hoechst 33258 staining in cells. Consistent with the results obtained from the immunoblotting analysis (Figure 2A), the intensities of α-MCM2M1 staining in CSK unextracted cells were constant at different phases of the cell cycle (Figure 3A, middle panel), indicating that the expression levels of MCM2 protein were not regulated during the cell cycle. The intensities of α-MCM2S1 staining in CSK unextracted cells at different phases of the cell cycle fluctuated slightly, indicating that phosphorylation levels of MCM2 by Cdc7/Dbf4 were regulated during the cell cycle. The levels of Cdc7/Dbf4 phosphorylation of MCM2 were low in G1 cells, increased in G1/S-to-S phase cells, and reached maximum in G2/M cells (Figure 3B, middle panel).
Figure 3.
Determination of expression, phosphorylation, and chromatin association of MCM2 during the cell cycle in asynchronous cells using high-throughput automated microscopy. (A) HeLa cells were plated on coverslips and stained with α-MCM2M1 and Hoechst 33258. DNA content of the culture and expression levels of MCM2 in cells were determined by Hoechst staining (bottom) and α-MCM2M1 staining (middle). Cells that had 2 N DNA content were gated as G1 cells. Cells that had a little bit of more than 2 N DNA content were gated as G1/S-early S cells. Cells that had >2 N but <4 N DNA content were gated as S phase cells. Cells that had nearly 4 N DNA content were gated as late S/G2 cells. Cells that had 4 N DNA content were gated as G2/M cells. Quantitative data (middle) and representative photos (top) of expression levels of MCM2 in cells at different stages of the cell cycle (G1, G1S-early S, S late S/G2, and G2/M) are shown. (B) HeLa cells were plated on coverslips and stained with α-MCM2S1 and Hoechst 33258. DNA content of the culture and phosphorylation levels of MCM2 in cells were determined by Hoechst staining (bottom) and α-MCM2S1 staining (middle). Cells at different stages of the cell cycle were gated as in A. Quantitative data (middle) and representative photos (top) of phosphorylation levels of MCM2 in cells at different stages of the cell cycle (G1, G1S-early S, S late S/G2, and G2/M) are shown. (C) HeLa cells were plated on coverslips, extracted with CSK buffer, fixed, and stained with α-MCM2M1 and Hoechst 33258 as in A. DNA content of the culture and chromatin associated MCM2 in cells were determined by Hoechst staining (bottom) and α-MCM2M1 staining (middle). Cells at different stages of the cell cycle were gated as in A. Quantitative data (middle) and representative photos (top) of chromatin-associated MCM2 in cells at different stages of the cell cycle (G1, G1S-early S, S late S/G2, and G2/M) are shown. (D) HeLa cells were plated on coverslips, extracted with CSK buffer, fixed, and stained with α-MCM2S1 and Hoechst 33258 as in B. DNA content of the culture and phosphorylation levels of chromatin associated MCM2 in cells were determined by Hoechst staining (bottom) and α-MCM2S1 staining (middle). Cells at different stages of the cell cycle were gated as in A. Quantitative data (middle) and representative photos (top) of phosphorylation levels of chromatin associated MCM2 in cells at different stages of the cell cycle (G1, G1S-early S, S late S/G2, and G2/M) are shown.
However, unlike in CSK untreated cells, the intensities of α-MCM2M1 staining in CSK-extracted cells fluctuated during the cell cycle (Figure 3C). The α-MCM2M1 staining was detected in G1 phase of CSK-extracted cells and staining intensities increased in G1/S and early S cells. In late S-to-G2/M phases of CSK-extracted cells, the staining of α-MCM2M1 gradually decreased (Figure 3C). These results indicated that the levels of chromatin-bound MCM2 protein were regulated during cell cycle. Intensities of α-MCM2S1 staining in CSK-extracted cells were also regulated during the cell cycle. The intensities of α-MCM2S1 staining were very low in G1 phase of CSK-extracted cells even though chromatin-bound MCM2 were readily detected in cells at this stage of the cell cycle by α-MCM2M1 staining (compare Figure 3, C and D). The intensities of α-MCM2S1 staining increased dramatically in G1/S and early S of CSK-extracted cells, coincident with the activation of Cdc7/Dbf4 as previous reported (Figure 3, C and D; Jiang et al., 1999a). The intensities of α-MCM2S1 staining were markedly decreased in late S phase of CSK-extracted cells and hardly detected in G2/M phase of CSK-extracted cells (Figure 3D). Similar results were also obtained by FACS analysis (Supplementary Figure S4). Taken together, these results demonstrated that chromatin-associated MCM2 is phosphorylated by Cdc7/Dbf4 in G1/S, which coincides with the initiation of DNA replication.
Previous studies showed that high levels of chromatin-bound MCM2 were detected in the nuclei of G1 and G1/S cells. However, after cells entered S phase, a significant amount of MCM2 was not colocalized with replication foci as measured by 5′-BrdU incorporation (Todorov et al., 1994; Dimitrova et al., 1999; Dimitrova and Berezney, 2002). These results suggested that either MCM2 was not preferentially colocalized with sites of DNA replication or that MCM2 could not be detected at replication forks because of its conformational change (i.e., phosphorylation). We examined subcellular localization of Cdc7/Dbf4-phosphorylated MCM2 in BrdU-labeled synchronous G1/S, early S, middle S, and late S cells using 3-D immunofluorescence imaging analysis. HeLa cells were synchronized at G1/S, early S, middle S, and late S phases of the cell cycle and labeled with BrdU for 2 min. After CSK buffer extraction, cells were fixed and costained with α-MCM2S1 and anti-BrdU antibody. A series of pictures from the top to the bottom of cells were taken, and 3-D images were reconstructed using Volocity 3-D reconstruction imaging software (Improvision; see Materials and Methods). Consistent with the results obtained from Figure 2B, high levels of Cdc7/Dbf4 phosphorylated chromatin-bound MCM2 were detected in G1/S and early S phase cells when DNA replication was initiated. In middle S and late S phase cells, low levels of Cdc7/Dbf4 phosphorylated chromatin-bound MCM2 were detected (Figure 4). However, Cdc7/Dbf4-phosphorylated chromatin-bound MCM2 was not colocalized with BrdU staining in all G1/S and S phase cells (Figure 4, A–D). In contrast, RPA, an essential DNA replication protein that binds to single-stranded DNA at replication forks, colocalized with sites of DNA replication throughout S phase (Figure 4E and unpublished data). Thus, our results, together with previous reports, indicated that chromatin bound MCM2 was not preferentially colocalized with sites of DNA replication.
Figure 4.
Cdc7/Dbf4 phosphorylated MCM2 does not colocalize with sites of DNA replication. HeLa cells were synchronized by a double-thymidine block and released into different stages of the cell cycle as in Figure 2. Cells in G1/S (A), early S (B and E), middle S (C), and late S (D) were labeled with BrdU for 2 min, extracted with CSK buffer, fixed, and stained with anti-BrdU antibodies (red) and α-MCM2S1 (green, A–D) or anti-RPA antibodies (green, E). A series of pictures taken from the top to the bottom of cells were taken and 3-D images were reconstructed by Velocity 3-D imaging software (Improvision).
Cdc7/Dbf4 Phosphorylation of MCM2 Affects the ATPase Activity of MCM2-7 Complex In Vitro
Previous studies showed that the purified yeast and Xenopus recombinant MCM2-7 heterohexamer complex displayed ATPase activity in vitro (Schwacha and Bell, 2001; Davey et al., 2003; You et al., 2003; Ying and Gautier, 2005). To determine if Cdc7/Dbf4 phosphorylation of MCM2 affects the complex formation and/or ATPase activity of MCM2-7 proteins, we generated baculoviruses expressing N-terminal epitope-tagged versions of all six mammalian MCM proteins, MCM2-7. We also generated baculoviruses expressing N-terminal epitope tagged MCM2 mutants, MCM2A and MCM2E, where Cdc7/Dbf4 phosphorylation sites (Ser27, Ser41, and Ser139) were substituted by Ala or Glu, respectively. Previous studies indicated that addition of epitope tags at N-terminal of yeast MCM proteins did not affect their functions in vivo and the ATPase activity of MCM complex in vitro (Schwacha and Bell, 2001; Davey et al., 2003). The recombinant MCM proteins, MCM2–7, MCM2A-7, or MCM2E-7, were coexpressed in insect Sf9 cells by coinfection with the corresponding baculoviruses, and the MCM heteromeric complexes were copurified using anti-FLAG tag antibody affinity chromatography, followed by gel filtration (see Materials and Methods). The purity and stoichiometry of MCM complexes were determined by silver staining of SDS/PAGE gel.
Figure 5A and Supplementary Figure S5 show purified ∼600-kDa MCM2-7, MCM2A-7, or MCM2E-7 complex on SDS/PAGE gels by silver staining. Multiple bands migrating around 90–155 kDa were detected. Immunoblotting analysis using anti-tag or anti-individual MCM-specific antibodies demonstrated that they were MCM2-7 proteins (unpublished data). These results indicated that purified MCM2-7, MCM2A-7, or MCM2E-7 complex was a heterohexamer complex. Thus, copurification of MCM2 and its Cdc7/Dbf4 phosphorylation mutants with other MCM proteins indicates that Cdc7/Dbf4 phosphorylation of MCM2 did not significantly affect MCM2-7 complex formation. Previously, we and others showed that differential phosphorylated forms of mammalian MCM2 proteins could be resolved on SDS/PAGE (Todorov et al., 1995; Jiang et al., 1999a). The slower migrating band of MCM2 represents the underphosphorylated form of MCM2, whereas the several faster-migrating bands of MCM2 reflect the hyperphosphorylated forms of MCM2. The MCM2 protein in purified MCM2–7 complex migrated as several bands around 130–155 kDa on the gel. These results suggested that MCM2 might be phosphorylated in Sf9 cells by endogenous insect Cdc7/Dbf4 (or other nonspecific kinases). In contrast, the MCM2A in purified MCM2A-7 complex only migrated as a major band of 155 kDa and a minor band of 145 kDa on the gel. The MCM2E in purified MCM2E-7 complex migrated as two major bands of 155 and 145 kDa and a minor band of 130 kDa on the gel.
Figure 5.
Purification and ATPase analysis of baculovirus-expressed purified MCM2-7 and its Cdc7/Dbf4 phosphorylation mutants, MCM2A-7 and MCM2E-7, complexes. (A) MCM2-7, MCM2A-7, or MCM2E-7 complex was produced in Sf9 cells using a baculovirus-expression system and purified using anti-FLAG antibody affinity chromatography followed by gel filtration chromatography. Silver staining of MCM2-7 complexes on SDS/SDS/polyacrylamide gels was shown. (B) Baculovirus-expressed purified MCM2-7, MCM2A-7, or MCM2E-7 complex as in A was incubated in ATPase buffer in the presence of 1 μCi of [γ-32P]ATP at 30°C for 30 min. Released 32P was determined using TLC, and ATPase activity of the protein was calculated. The position of released Pi, ATP, and origin are indicated.
We measured the ATPase activity of MCM2–7, MCM2A-7 or MCM2E-7 complex by TLC using [γ-32P]ATP as the substrate. Like purified yeast MCM2–7 complex (Schwacha and Bell, 2001; Davey et al., 2003), purified mammalian MCM2-7 complex clearly displayed detectable ATPase activity (0.7 pmol ATP/min/pmol protein) as shown by the appearance of 32Pi hydrolyzed from [γ-32P]ATP (Figure 5B). Similar ATPase activity (0.7 pmol ATP/min/pmol protein) was also detected in the Cdc7/Dbf4 phosphomimetic MCM2E-7 complex (Figure 5B). By contrast, the Cdc7/Dbf4 nonphosphorylatable MCM2A-7 complex showed very poor ATPase activity (0.2 pmol ATP/min/pmol protein). These results indicated that phosphorylation of MCM2 by Cdc7/Dbf4 is critical for the ATPase activity of MCM2–7 complex in vitro.
Cdc7/Dbf4 Phosphorylation of MCM2 Is Essential for the Initiation of DNA Replication
We examined the consequences of Cdc7/Dbf4 phosphorylation of MCM2 in regulation of the initiation of DNA replication in human cells. To eliminate the contribution of endogenous MCM2 protein, we repressed endogenous MCM2 protein expression using RNA interference (RNAi). Because MCM proteins are very abundant in all eukaryotic cells, we transfected HeLa cells with control siRNA or MCM2 siRNA (a mixture corresponding to a 21-nucleotide [nt] sequence unique to human MCM2 5′UTR and a 21-nt sequence unique to human MCM2 3′UTR) twice within 24 h to achieve more effective knockdown of MCM2 expression. As shown in Figure 6A, the expression levels of MCM2 were dramatically inhibited in cells transfected with MCM2 siRNA but not in cells transfected with control siRNA. No reduction in expression levels of α-tubulin was detected in either MCM2 siRNA or control siRNA treated cells.
Figure 6.
Cdc7/Dbf4 phosphorylation of MCM2 is essential for the initiation of DNA replication. (A) HeLa cells were transfected with control or MCM2 siRNA. Twenty-four hours after transfection, cells were transfected with control or MCM2 siRNA again together with the indicated mammalian expression vectors. Two days after the second transfection, cells were lysed and lysates were subjected to SDS/PAGE, transferred to the PVDF membrane, and immunoblotted with α-MCM2M1, anti-FLAG antibody, or anti-α-tubulin antibody. (B) HeLa cells transfected with indicated mammalian expression vectors were extracted with CSK buffer, fixed, and costained with α-FLAG (green) and α-MCM6 (red). (C) Xenopus sperm chromatin was incubated with Xenopus mock-depleted EC (egg cytosol, lane 1), EC with geminin (inhibitor of chromatin association of MCMs, lane 2), MCM2-depleted EC (EC/ΔMCM2, lane 3), or MCM2-depleted EC with baculovirus-expressed purified MCM2–7 (lane 4), MCM2A-7 (lane 5), MCM2E-7 (lane 6), or MCM2–7 with geminin (lane7). After a 30-min incubation, sperm chromatin was isolated by centrifugation, subjected to SDS/PAGE, transferred to PVDF membrane, and immunoblotted with α-MCM2M1, α-xMCM7, or α-xOrc2. Note: xMCM7 was not completely codepleted in MCM2-depleted EC and could load onto chromatin with purified MCM complex. (D) Cells were transfected the indicated mammalian expression vectors and siRNA as in A. Twenty-four hours after the second transfection, cells were labeled with BrdU for an additional of 16 h. After fixation, cells were stained with anti-FLAG antibody, anti-BrdU antibody, and Hoechst 33258. BrdU-positive cells in FLAG positive cells (>500) were scored (%).
We cotransfected HeLa cells with MCM2 siRNA and mammalian expression vectors expressing FLAG-tagged MCM2 or its Cdc7/Dbf4 phosphorylation mutants, MCM2A and MCM2E, using the corresponding cDNA that only contains the MCM2 coding sequence but not the native 5′ and 3′ UTR. Immunoblotting analysis indicated that the expression levels of endogenous MCM2 protein were greatly reduced in these cells (Figure 6A). However, the expression levels of exogenous FLAG-MCM2 proteins were not affected (Figure 6A). Thus, these results indicated that MCM2 siRNA could only effectively ablate the expression of endogenous MCM2 but not the expression of the exogenous FLAG-MCM2 proteins in HeLa cells.
To determine if the exogenous FLAG-MCM2 proteins could assemble into pre-RC as normal, we examined chromatin association of MCM proteins in HeLa cells expressing exogenous FLAG-MCM2, MCM2A, or MCM2E by immunofluorescence analysis. As shown in Figure 6B, exogenous FLAG-MCM2, MCM2A, or MCM2E was colocalized with endogenous MCM6 proteins in the nuclei of HeLa cells extracted with CSK buffer, indicating that expression of exogenous FLAG-MCM2 proteins did not affect association of MCM proteins with chromatin. We also examined if phosphorylation of MCM2 affects pre-RC formation using X. laevis cell-free assay. Consistent with previous reports (Kubota et al., 1997; Tada et al., 2001), endogenous Xenopus MCM complex could load onto sperm chromatin within 30 min after the sperm chromatin was added into egg extracts (Figure 6C). The chromatin loading of MCM complex was blocked either by addition of geminin, a DNA replication inhibitor, into egg extracts or by immunodepletion of MCM complex with MCM2 antibodies from egg extract. Addition of baculovirus-expressed purified MCM2-7, MCM2A-7, or MCM2E-7 (Figure 5A) to MCM2-immunodepleted egg extracts promoted loading of the recombinant mammalian MCM complex onto the chromatin. There was no difference among MCM2-7, MCM2A-7, and MCM2E-7 for their loading on the chromatin and the chromatin loading of baculovirus-expressed purified MCM complex was also geminin sensitive. Thus, these results, together with the fact that Cdc7/Dbf4 phosphorylation of MCM2 does not perturb formation of MCM heterohexameric complex in vitro and MCM proteins association with chromatin in vivo (Figures 5A and 6B), indicated that Cdc7/Dbf4 phosphorylation of MCM2 does not affect pre-RC formation.
We determined the effects of Cdc7/Dbf4 phosphorylation of MCM2 on the initiation of DNA replication. HeLa cells were cotransfected with MCM2 siRNA and FLAG (control), FLAG-MCM2, FLAG-MCM2A, or FLAG-MCM2E plasmid and then cultured in medium in the presence of BrdU for additional 16 h. After fixation, cells were stained anti-BrdU and anti-FLAG antibodies to identify cells that had replicated their DNA and expressed exogenous MCM2 proteins. As shown in Figure 6D, when compared with cells transfected control siRNA and control plasmid, cells transfected with MCM2 siRNA and control plasmid showed strong inhibition of BrdU incorporation, indicating that MCM2 is required for the initiation of DNA replication. Similar inhibition of BrdU incorporation was also observed in cells transfected with MCM2 siRNA and FLAG-MCM2A plasmid. However, BrdU incorporation was not inhibited in cells transfected with MCM2 siRNA and either FLAG-MCM2 plasmid or FLAG-MCM2E plasmid. The results indicated that inhibition of DNA replication in cells treated with MCM2 siRNA could be rescued by coexpression of MCM2 or MCM2E but not MCM2A. These results, together with that Cdc7/Dbf4 phosphorylation of MCM2 is important for the ATPase activity of MCM complex in vitro, demonstrated that Cdc7/Dbf4 phosphorylation of MCM2 is essential for the initiation of DNA replication in human cells.
DISCUSSION
In the present study, we report the identification of Cdc7/Dbf4 phosphorylation sites on MCM2 and determine the functional role of Cdc7/Dbf4 phosphorylation of MCM2 in the initiation of DNA replication in human cells. Previous studies demonstrated that the S phase–promoting kinase Cdc7/Dbf4 and the putative DNA replicative helicase MCM2-7 play essential roles in the initiation of DNA replication in all eukaryotes. We and others showed that Cdc7/Dbf4 selectively phosphorylates the MCM2 subunit of the MCM complex, suggesting that Cdc7/Dbf4 may be directly involved in the initiation of DNA replication by targeting MCM2 (Lei et al., 1997; Brown and Kelly, 1998; Jiang et al., 1999a; Jares and Blow, 2000). To determine the importance of Cdc7/Dbf4 phosphorylation of MCM2 in the initiation of DNA replication, we mapped Cdc7/Dbf4 phosphorylation sites in human MCM2. Our results showed that human MCM2 is phosphorylated in vivo at three major sites (Ser27, Ser41, and Ser139) and two minor sites (Ser53 and Ser108). Because Ser27, Ser41, and Ser139 in MCM2 are also specifically phosphorylated by purified Cdc7/Dbf4 in vitro and their in vivo phosphorylation was greatly reduced in Cdc7 siRNA treated cells, it strongly suggests that Ser27, Ser41, and Ser139 are in vivo Cdc7/Dbf4 phosphorylation sites in MCM2.
Sequence alignments show that Ser27, Ser41, and Ser139 of human MCM2 are highly conserved in higher eukaryotes (from Drosophila to mammals). However, the N-terminal sequences of MCM2 between human and lower eukaryotes, such as S. cerevisiae and S. pombe, are highly divergent. Therefore, we could not define analogous phosphorylation sites in MCM2 from these species. Nonetheless, previous studies have demonstrated that yeast MCM2 is a critical downstream target of Cdc7/Dbf4 kinase (Lei et al., 1997; Brown and Kelly, 1998). Careful analysis of the surrounding sequences of Ser27, Ser41, and Ser139 in human MCM2 did not reveal consensus sites for Cdc7/Dbf4 phosphorylation. The Ser27 and Ser41 sites are similar to the Cdk phosphorylation consensus site (X-S/T-P-X-K/R), whereas the Ser139 site is similar to the casein kinase II (CKII) phosphorylation consensus site (X-S/T-X-X-D/E). Previous studies suggested that MCM2 might be a target of Cdk and Ddk (Masai et al., 2000; Montagnoli et al., 2006). Because phosphorylation of MCM2 was not affected in HeLa cells treated with Cdk2 siRNA (or Cdc2 siRNA, T. Tsuji and W. Jiang, unpublished observation), our results indicated that MCM2 was not directly targeted by Cdks in vivo (Figure 1D). It was shown that Cdc7 belongs a subfamily of protein kinase closely related to the Cdk subfamily and the CKII subfamily (Manning et al., 2002). These results together with ours presented in this study indicate that Cdc7 phosphorylates similar primary sequences of substrates as Cdk does. However, recognition of a ternary structure of a substrate by Cdc7 and/or Dbf4 or subcellular colocalization of a substrate with Cdc7/Dbf4 might be crucial for determining the Cdc7/Dbf4 kinase-substrate specificity. Consistent with this idea, unlike the purified MCM2 protein, a MCM2 Ser139 peptide (MRRGLLYDSDEEDEERPA) fused with GST protein could not be phosphorylated by purified Cdc7/Dbf4 in vitro (unpublished data).
While this manuscript was being prepared for submission, Montagnoli et al. (2006) reported identification of MCM2 phosphorylation sites by S phase–regulating kinases. They found that MCM2 was phosphorylated in vitro by Cdks at Ser13, Ser27, and Ser41, by Ddk at Ser40, Ser53, and Ser108, and by CKII at Ser139. The basis for the discrepancies between their results and ours is currently unclear. One possibility is that the purified Ddk used in their and our in vitro phosphorylation studies might have different specific activity. In our study, highly purified Cdc7/Dbf4 strongly phosphorylated MCM2 in vitro, whereas kinase-dead Cdc7kd/Dbf4 did not (Supplementary Figure S1). These results ruled out the possibility that a contaminating kinase(s) copurified with the Cdc7/Dbf4. We showed that Ser27, Ser41, and Ser139 of MCM2 were specifically phosphorylated by Cdc7/Dbf4 but not by Cdc7kd/Dbf4 in vitro and that phosphorylation of these sites was also detected in vivo. In contrast, phosphorylation of Ser26 and Ser40 in MCM2 by Cdc7/Dbf4 was not detected in vitro and phosphorylation of these sites was not detected in vivo either (Supplementary Figure S2). Although the amino acid sequences around Ser26 and Ser40 are the same (LTSSPGR). Montagnoli et al. (2006) could only detect Cdc7/Dbf4 phosphorylation of Ser40, but not Ser26. In addition, Montagnoli et al. showed that MCM2 was phosphorylated by CKII but not Cdc7/Dbf4 in vitro. However, they also showed that phosphorylation of Ser139 in vivo was affected by serum starvation, suggesting that phosphorylation of Ser139 was regulated in vivo. Because the activity of CKII is not regulated during the cell cycle, they were not sure if CKII is responsible for Ser139 phosphorylation in vivo (see discussion section in Montagnoli et al., 2006). In contrast, we clearly showed that MCM2 Ser139 was phosphorylated by Cdc7/Dbf4 in vitro and phosphorylation of this site in vivo is required for chromatin recruitment of MCM complex and the initiation of DNA replication during the cell cycle (Figures 2, 3, and 6). Taken together, our results strongly indicated that Ser27, Ser41, and Ser139 are Cdc7/Dbf4 phosphorylation sites.
We examined chromatin recruitment and phosphorylation of chromatin-bound MCM2 by Cdc7/Dbf4 using immunoblotting, immunofluorescence, and high-speed automated cell-imaging analyses with antibodies specific against MCM2 and Cdc7/Dbf4 phosphorylated MCM2. We found that MCM2 was not phosphorylated by Cdc7/Dbf4 in G1 and unphosphorylated MCM2 accumulated on chromatin. Chromatin-bound MCM2 was phosphorylated by Cdc7/Dbf4 during G1/S and early S phase, which coincided with the initiation of DNA replication. In late S and G2/M, phosphorylated MCM2 gradually disassociated from chromatin. Previous studies showed that, unlike other essential DNA replication proteins such as RPA and PCNA, MCM proteins fail to colocalize with sites of DNA replication through S phase (Todorov et al., 1994; Romanowski et al., 1996; Dimitrova et al., 1999; Laskey and Madine, 2003). The results suggest that either MCM complex does not preferentially colocalize with sites of DNA replication or MCM complex could not be detected at replication sites by certain antibodies due to conformational changes induced by post-translational modification, such as phosphorylation. We show that, like MCM2, chromatin-bound Cdc7/Dbf4 phosphorylated MCM2 does not colocalize with replication foci during G1/S and S phase, indicating that failure of detection of MCM2 at replication foci is not due to the use of antibodies that cannot recognize phosphorylated MCM2. Thus, our results, together with previous studies, indicate that MCM complex does not preferentially colocalize with sites of DNA replication (Todorov et al., 1994; Romanowski et al., 1996; Dimitrova et al., 1999; Laskey and Madine, 2003). How could the DNA replicative MCM helicase fail to colocalize with sites of DNA replication? Recently, Laskey and Madine (2003) proposed a rotary pumping model to explain the discrepancy. They suggested that MCM helicase could work at a distance from replication forks as a rotary motor that pumps DNA along its helical axis by simple rotation, such that the movement resembles that of a threaded bolt through a nut.
All six MCM proteins (MCM2-7) are members of the diverse AAA ATPase family, and it is known that the purified recombinant yeast and Xenopus MCM2-7 heterohexameric complex has ATPase activity (Tye, 1999; Schwacha and Bell, 2001; Davey et al., 2003; You et al., 2003). We examined if Cdc7/Dbf4 phosphorylation of MCM2 affects the formation, conformation, and/or ATPase activity of MCM2-7 complex in vitro. We show that MCM2, its nonphosphorylatable mutant MCM2A or its phosphomimetic mutant MCM2E could copurify with MCM3-7 as a heterohexameric complex from insect Sf9 cells using a baculovirus expression system. These results indicate that phosphorylation of MCM2 does not affect MCM complex formation. However, we show that the MCM2-7 complex or the MCM2E-7 complex displays much higher ATPase activity in vitro than the MCM2A-7 complex. Previous studies showed that the purified yeast globular heterohexamer MCM2-7 complex did not yield any detectable enzymatic or nucleotide binding activities (Adachi et al., 1997). In contrast, the purified ring-shaped (MCM4,6,7)2 subcomplex displayed ATP-dependent DNA helicase activity (Sato et al., 2000). Therefore, it was proposed that activation of helicase activity of heterohexamer MCM 2-7 complex in vivo at replication origins requires a series of post-translational modifications, such as phosphorylation, of MCM proteins that convert the globular inactive heterohexamer MCM2-7 complex conformation to the ring-shaped active heterohexamer MCM 2-7 complex conformation (Tye and Sawyer, 2000). Our unpublished results suggested Cdc7/Dbf4 phosphorylation of MCM2 might induce a conformational change of MCM2-7 complex as it was assayed by a protease partial digestion analysis (T. Tsuji and W. Jiang, unpublished observation and Herbig et al., 1999; Mizushima et al., 2000). Thus, alteration of the confirmation of MCM2-7 complex by Cdc7/Dbf4 phosphorylation of MCM2 could be one of the critical steps for activation of the ATPase-coupled helicase activity of MCM2-7 complex. Further studies, such as electron microscopic observation and the determination of the structure of MCM2–7 complex (Sato et al., 2000; Fletcher et al., 2003), are needed to define how Cdc7/Dbf4 phosphorylation of MCM2 regulates the conformational change and the enzymatic activity of MCM2-7 complex in the future.
We provide direct evidence that Cdc7/Dbf4 phosphorylation of MCM2 is required for the initiation of DNA replication in human cells. Consistent with previous biochemical and genetic studies in various organisms (Todorov et al., 1994; Forsburg et al., 1997; Kubota et al., 1997; Lei et al., 1997), we show that suppression of MCM2 expression by MCM2 siRNA inhibits DNA replication in HeLa cells, indicating that MCM2 is essential for DNA replication in human cells. The inhibition of DNA replication in MCM2 siRNA-treated cells can be rescued by coexpression of MCM2 or its Cdc7/Dbf4 phosphomimetic MCM2E mutant but not its Cdc7/Dbf4 nonphosphorylatable MCM2A mutant. These results, together with the result that Cdc7/Dbf4 phosphorylation of MCM2 is important for the ATPase activity of MCM2–7 complex in vitro, demonstrate that Cdc7/Dbf4 phosphorylation of MCM2 is essential for the initiation of DNA replication in human.
On the basis of our results, together with the current model for the initiation of DNA replication (Bell and Dutta, 2002; Mendez and Stillman, 2003; Forsburg, 2004), we propose a model for regulation of the initiation of DNA replication by Cdc7/Dbf4 phosphorylation of MCM2 in human cells. In G1, when Cdc7/Dbf4 activity is at a minimum, MCM2 is not phosphorylated by Cdc7/Dbf4. Unphosphorylated MCM2 together with other MCMs (MCM2-7 complex) is recruited to chromatin, presumably to the replication origins by the DNA replication loading factors Cdc6 and Cdt1 to establish pre-RCs. During G1/S and early S, activation of Cdc7/Dbf4 results in phosphorylation of chromatin-bound MCM2. Phosphorylation of MCM2 in the MCM2-7 complex by Cdc7/Dbf4 induces the conformational change of the complex and activates its helicase activity, which is essential for DNA replication. MCM2-7 helicase works at a distance from the replicative forks, pumping DNA along its helical axis by ATPase-coupled rotation. In late S and G2/M, MCM2-7 complex dissociates from chromatin, presumably by additional post-translational modifications to prevent DNA rereplication.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Xinyu Wang for participating the initial work of this study; Drs. John Newport (UCSD, San Diego), Ralf Knippers (Konstanz, Germany), and Ron Laskey (University of Cambridge, United Kingdom) for antibodies; Drs. Zeev Ronai, Gary Chiang, Robert Abraham, and Nanxin Li for critical reading of the manuscript; Jill Meisenhelder for helping 2-D tryptic phosphopeptide mapping analysis; and Ningning Sai for laboratory support. This work was supported by grants from the National Science Foundation (NSF-0233997) and National Institutes of Health (GM67859) to W.J.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-03-0241) on August 9, 2006.
REFERENCES
- Adachi Y., Usukura J., Yanagida M. A globular complex formation by Nda1 and the other five members of the MCM protein family in fission yeast. Genes Cells. 1997;2:467–479. doi: 10.1046/j.1365-2443.1997.1350333.x. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex [see comments] Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Boyle W. J., van der Geer P., Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- Brown G. W., Kelly T. J. Purification of Hsk1, a minichromosome maintenance protein kinase from fission yeast. J. Biol. Chem. 1998;273:22083–22090. doi: 10.1074/jbc.273.34.22083. [DOI] [PubMed] [Google Scholar]
- Chong J. P., Mahbubani H. M., Khoo C. Y., Blow J. J. Purification of an MCM-containing complex as a component of the DNA replication licensing system [see comments] Nature. 1995;375:418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- Cortez D., Glick G., Elledge S. J. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. USA. 2004;101:10078–10083. doi: 10.1073/pnas.0403410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M. J., Indiani C., O'Donnell M. Reconstitution of the Mcm2–7p heterohexamer, subunit arrangement, and ATP site architecture. J. Biol. Chem. 2003;278:4491–4499. doi: 10.1074/jbc.M210511200. [DOI] [PubMed] [Google Scholar]
- Dimitrova D. S., Berezney R. The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J. Cell Sci. 2002;115:4037–4051. doi: 10.1242/jcs.00087. [DOI] [PubMed] [Google Scholar]
- Dimitrova D. S., Todorov I. T., Melendy T., Gilbert D. M. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S., Harwood J., Drury L. S., Diffley J. F. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Ficarro S. B., Salomon A. R., Brill L. M., Mason D. E., Stettler-Gill M., Brock A., Peters E. C. Automated immobilized metal affinity chromatography/nano-liquid chromatography/electrospray ionization mass spectrometry platform for profiling protein phosphorylation sites. Rapid Commun. Mass Spectrom. 2005;19:57–71. doi: 10.1002/rcm.1746. [DOI] [PubMed] [Google Scholar]
- Fletcher R. J., Bishop B. E., Leon R. P., Sclafani R. A., Ogata C. M., Chen X. S. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat. Struct. Biol. 2003;10:160–167. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- Forsburg S. L. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S. L., Sherman D. A., Ottilie S., Yasuda J. R., Hodson J. A. Mutational analysis of Cdc19p, a Schizosaccharomyces pombe MCM protein. Genetics. 1997;147:1025–1041. doi: 10.1093/genetics/147.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneke G., Koundrioukoff S., Hubscher U. Multiple roles for kinases in DNA replication. EMBO Rep. 2003;4:252–256. doi: 10.1038/sj.embor.embor774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig U., Marlar C. A., Fanning E. The Cdc6 nucleotide-binding site regulates its activity in DNA replication in human cells. Mol. Biol. Cell. 1999;10:2631–2645. doi: 10.1091/mbc.10.8.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex [published erratum appears in J. Biol. Chem. 1998 Sep 4;273(36):23616] J. Biol. Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- Jares P., Blow J. J. Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev. 2000;14:1528–1540. [PMC free article] [PubMed] [Google Scholar]
- Jiang W., McDonald D., Hope T. J., Hunter T. Mammalian Cdc7-Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J. 1999a;18:5703–5713. doi: 10.1093/emboj/18.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Wells N. J., Hunter T. Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc. Natl. Acad. Sci. USA. 1999b;96:6193–6198. doi: 10.1073/pnas.96.11.6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y., Mimura S., Nishimoto S., Masuda T., Nojima H., Takisawa H. Licensing of DNA replication by a multi-protein complex of MCM/P1 proteins in Xenopus eggs. EMBO J. 1997;16:3320–3331. doi: 10.1093/emboj/16.11.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K., Kearsey S. E., Diffley J. F. MCM2-7 proteins are essential components of prereplicative complexes that accumulate cooperatively in the nucleus during G1-phase and are required to establish, but not maintain, the S-phase checkpoint. Mol. Biol. Cell. 2001;12:3658–3667. doi: 10.1091/mbc.12.11.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey R. A., Madine M. A. A rotary pumping model for helicase function of MCM proteins at a distance from replication forks. EMBO Rep. 2003;4:26–30. doi: 10.1038/sj.embor.embor706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. K., Hurwitz J. Isolation and characterization of various complexes of the minichromosome maintenance proteins of Schizosaccharomyces pombe. J. Biol. Chem. 2000;275:18871–18878. doi: 10.1074/jbc.M001118200. [DOI] [PubMed] [Google Scholar]
- Lee J. K., Hurwitz J. Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc. Natl. Acad. Sci. USA. 2001;98:54–59. doi: 10.1073/pnas.98.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M., Kawasaki Y., Young M. R., Kihara M., Sugino A., Tye B. K. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A. N., Martin J. AAA proteins. Curr. Opin. Struct. Biol. 2002;12:746–753. doi: 10.1016/s0959-440x(02)00388-3. [DOI] [PubMed] [Google Scholar]
- Maiorano D., Moreau J., Mechali M. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature. 2000;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Masai H., Arai K. Cdc7 kinase complex: a key regulator in the initiation of DNA replication. J. Cell. Physiol. 2002;190:287–296. doi: 10.1002/jcp.10070. [DOI] [PubMed] [Google Scholar]
- Masai H., Matsui E., You Z., Ishimi Y., Tamai K., Arai K. Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a critical threonine residue of Cdc7 by Cdks. J. Biol. Chem. 2000;275:29042–29052. doi: 10.1074/jbc.M002713200. [DOI] [PubMed] [Google Scholar]
- Mendez J., Stillman B. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays. 2003;25:1158–1167. doi: 10.1002/bies.10370. [DOI] [PubMed] [Google Scholar]
- Mizushima T., Takahashi N., Stillman B. Cdc6p modulates the structure and DNA binding activity of the origin recognition complex in vitro. Genes Dev. 2000;14:1631–1641. [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A., Tenca P., Sola F., Carpani D., Brotherton D., Albanese C., Santocanale C. Cdc7 inhibition reveals a p53-dependent replication checkpoint that is defective in cancer cells. Cancer Res. 2004;64:7110–7116. doi: 10.1158/0008-5472.CAN-04-1547. [DOI] [PubMed] [Google Scholar]
- Montagnoli A., Valsasina B., Brotherton D., Troiani S., Rainoldi S., Tenca P., Molinari A., Santocanale C. Identification of Mcm2 phosphorylation sites by S-phase regulating kinases. J. Biol. Chem. 2006;281:10281–10290. doi: 10.1074/jbc.M512921200. [DOI] [PubMed] [Google Scholar]
- Nishitani H., Lygerou Z., Nishimoto T., Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- Pacek M., Walter J. C. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 2004;23:3667–3676. doi: 10.1038/sj.emboj.7600369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhorova T. A., Blow J. J. Sequential MCM/P1 subcomplex assembly is required to form a heterohexamer with replication licensing activity. J. Biol. Chem. 2000;275:2491–2498. doi: 10.1074/jbc.275.4.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski P., Madine M. A., Laskey R. A. XMCM7, a novel member of the Xenopus MCM family, interacts with XMCM3 and colocalizes with it throughout replication [see comments] Proc. Natl. Acad. Sci. USA. 1996;93:10189–10194. doi: 10.1073/pnas.93.19.10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Gotow T., You Z., Komamura-Kohno Y., Uchiyama Y., Yabuta N., Nojima H., Ishimi Y. Electron microscopic observation and single-stranded DNA binding activity of the Mcm4,6,7 complex. J. Mol. Biol. 2000;300:421–431. doi: 10.1006/jmbi.2000.3865. [DOI] [PubMed] [Google Scholar]
- Schwacha A., Bell S. P. Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol. Cell. 2001;8:1093–1104. doi: 10.1016/s1097-2765(01)00389-6. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Tada S., Li A., Maiorano D., Mechali M., Blow J. J. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov I. T., Attaran A., Kearsey S. E. BM28, a human member of the MCM2–3-5 family, is displaced from chromatin during DNA replication. J. Cell Biol. 1995;129:1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov I. T., Pepperkok R., Philipova R. N., Kearsey S. E., Ansorge W., Werner D. A human nuclear protein with sequence homology to a family of early S phase proteins is required for entry into S phase and for cell division. J. Cell Sci. 1994;107:253–265. doi: 10.1242/jcs.107.1.253. [DOI] [PubMed] [Google Scholar]
- Tye B. K. Mcm proteins in DNA replication. Annu. Rev. Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- Tye B. K., Sawyer S. The hexameric eukaryotic MCM helicase: building symmetry from nonidentical parts. J. Biol. Chem. 2000;275:34833–34836. doi: 10.1074/jbc.R000018200. [DOI] [PubMed] [Google Scholar]
- Ying C. Y., Gautier J. The ATPase activity of MCM2–7 is dispensable for pre-RC assembly but is required for DNA unwinding. EMBO J. 2005;24:4334–4344. doi: 10.1038/sj.emboj.7600892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. Y., Shevchenko A., Dunphy W. G. Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. J. Biol. Chem. 2004;279:53353–53364. doi: 10.1074/jbc.M408026200. [DOI] [PubMed] [Google Scholar]
- You Z., Ishimi Y., Mizuno T., Sugasawa K., Hanaoka F., Masai H. Thymine-rich single-stranded DNA activates Mcm4/6/7 helicase on Y-fork and bubble-like substrates. EMBO J. 2003;22:6148–6160. doi: 10.1093/emboj/cdg576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Jiang W. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc. Natl. Acad. Sci. USA. 2005;102:343–348. doi: 10.1073/pnas.0408438102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.