Abstract
Most eukaryotic cells spend most of their life in a quiescent state, poised to respond to specific signals to proliferate. In Saccharomyces cerevisiae, entry into and exit from quiescence are dependent only on the availability of nutrients in the environment. The transition from quiescence to proliferation requires not only drastic metabolic changes but also a complete remodeling of various cellular structures. Here, we describe an actin cytoskeleton organization specific of the yeast quiescent state. When cells cease to divide, actin is reorganized into structures that we named “actin bodies.” We show that actin bodies contain F-actin and several actin-binding proteins such as fimbrin and capping protein. Furthermore, by contrast to actin patches or cables, actin bodies are mostly immobile, and we could not detect any actin filament turnover. Finally, we show that upon cells refeeding, actin bodies rapidly disappear and actin cables and patches can be assembled in the absence of de novo protein synthesis. This led us to propose that actin bodies are a reserve of actin that can be immediately mobilized for actin cables and patches formation upon reentry into a proliferation cycle.
INTRODUCTION
The control of cell growth and proliferation is a key feature of life. Most prokaryotic and eukaryotic cells spend the greatest part of their life span in a nonproliferating state termed quiescence. Astonishingly, whereas an immense body of information on various cellular processes controlling the cell proliferation cycle is available, little is known on the biology of the quiescent cycle. The quiescent cycle is a process in which cells exit from the proliferation cycle; enter and maintain a stable, nonproliferating state; and finally, under specific environmental conditions, return to the cell division cycle (Gray et al., 2004). Deciphering the mechanisms involved in the transitions from proliferation to quiescence is a vast and emergent research field. In multicellular organisms, cell proliferation primarily relies on growth factors and hormones but also probably on parameters involving the whole organism environment. Studying the quiescent cycle in such organisms is therefore very challenging. In contrast, proliferation of unicellular microorganisms such as yeast S. cerevisiae depends “solely” on the availability of nutrients in the environment and is consequently more suitable to study the processes involved in entry into and exit from the quiescent cycle.
On nutrient exhaustion, proliferating yeast cells complete the cell cycle, cease to grow and enter into quiescence as unbudded cells. The correct establishment of the quiescent state clearly involves active processes that are controlled by a complex set of signaling cascades, such as the TOR and RAS pathways. Moreover, yeast cells entering quiescence radically modify their metabolism. They accumulate carbohydrates, polyphosphates, and considerably reduce their rate of protein synthesis (for review, see Herman, 2002; Gray et al., 2004). Recently, DNA microarray analyses have shown that whereas most genes are down-regulated upon entry into quiescence, the transcription of some specific genes is induced and furthermore, that some particular transcripts are enriched in quiescent cells (Gasch et al., 2000; Radonjic et al., 2005). These studies thus revealed a specific reprogramming of the expression profile that occurs upon entry into quiescence. At the cellular level, yeast cells entering quiescence develop a thick cell wall conferring them a higher resistance to temperature, and osmotic or chemical stress. Furthermore, quiescent yeast cells display condensed chromosomes (Pinon, 1978) and punctate mitochondrial network (Gourlay and Ayscough, 2005); yet, little is known on the potential remodeling of the other cellular structures upon entry into quiescence.
The actin cytoskeleton is one of the most dynamic cellular machineries and is crucial for a variety of cellular processes such as cell morphology, migration, polarization, contraction, division, or intracellular traffic. The actin network is highly responsive to a vast number of signaling pathways that coordinate the rapid assembly and disassembly of actin filaments (F-actin) and regulate their incorporation into various structures such as stress fibers or the contractile ring (for review, see Pollard and Borisy, 2003). Actively proliferating yeast cells display three F-actin–containing structures: actin cables, which serve as tracks for polarized transport of vesicles, organelles, and mRNA; actin patches that are required for endocytosis; and an acto-myosin cytokinetic ring (Pruyne et al., 2004). A large set of conserved actin-binding proteins (ABPs) tightly orchestrates the formation and the maintenance of these highly dynamic F-actin–containing structures (Goode and Rodal, 2001; Winder and Ayscough, 2005). Although a number of studies describe the complex network of molecular interactions that regulate the actin cytoskeleton in actively dividing yeast cells, very little is known about its organization in quiescent cells.
Here, we cautiously depict the budding yeast actin cytoskeleton remodeling upon entry into and exit from quiescence. We describe a quiescent cell-specific actin cytoskeleton organization that we named “actin bodies.” We further address the composition, localization, and dynamic of this new structure. Finally, we document the fate of actin bodies when cells exit from quiescence upon refeeding. Based on our data, we propose that actin bodies serve as actin reserves that can be made immediately available for cells reentering a proliferating cycle.
MATERIALS AND METHODS
Strains, Media, and Plasmids
Strains.
All the S. cerevisiae strains used in this study are, unless specified, isogenic to BY4742 or BY4741 (s288c background) available from Euroscarf (Frankfurt, Germany) and are listed in Table 1. Yeast strains carrying green fluorescent protein (GFP) fusions were from Invitrogen (Carlsbad, CA).
Table 1.
Saccharomyces cerevisiae strains
| Strain | Genotype | Reference |
|---|---|---|
| BY4741 (Y1025) | Mata, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 | Invitrogen, Euroscarf |
| BY4742 (Y1026) | Matα his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0 | Invitrogen, Euroscarf |
| BY4743 (Y1568) | Mata/Matα, his3Δ1/his3Δ1, leu2Δ0/leu2Δ, lys2Δ0/LYS2, ura3Δ0/ura3Δ0, met15Δ0/MET15 | Invitrogen, Euroscarf |
| FY23 (Y1441) | Mata, ura3–52, trp1Δ63, leu2Δ1 | Brachmann et al. (1998) |
| FL100 (Y767) | Mata | F. Lacroute |
| Y217 | Mata, his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; IVL3-GFP-HIS3 | Invitrogen, Euroscarf |
| Y250 | Mata, his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; PHO88-GFP-HIS3 | Invitrogen, Euroscarf |
| Y247 | Mata, his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; SCP1-GFP-HIS3 | Invitrogen, Euroscarf |
| Y209 | Mata, his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; SAC6-GFP-HIS3 | Invitrogen, Euroscarf |
| Y244 | Mata, his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; CAP1-GFP-HIS3 | Invitrogen, Euroscarf |
| Y55 | Mata, his3Δ1; leu2Δ0; met15Δ0; ura3Δ0, ABP140–3×GFP-LEU2 | This study |
| Y2754 | Mata, his3Δ1; leu2Δ0; met15Δ0; ura3Δ0, ABP1–3×GFP-URA3 | This study |
| Y115 | Mata, his3Δ1; leu2Δ0; met15Δ0; ura3Δ0, CAP2–3×GFP-URA3 | This study |
| Y1832 | Matα, rvs161::KanR; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0 | Invitrogen, Euroscarf |
| Y2740 | Matα, scp1::KanR; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0 | Invitrogen, Euroscarf |
| Y1354 | Matα, sac6::KanR; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0 | Invitrogen, Euroscarf |
Media.
The YPDA medium was described previously (Martinez et al., 2004). The SD casa medium (Escobar-Henriques and Daignan-Fornier, 2001) was supplemented with 1.8 mM uracil, 0.2 mM tryptophan, and 0.3 mM adenine. The 2% lactate-containing medium was described in (Chateaubodeau et al. 1976), and the N3 glycerol-containing medium was described in (Lefebvre-Legendre et al. 2001) was supplemented with 2% ethanol. To test the viability upon starvation, we used an SD casa WAU medium containing 0.1% glucose and erythrosine as described in (Bonneu et al. 1991). For chronological aging assay, cells were grown for 7 d at 30°C in liquid YPDA medium, and serial dilutions were plated onto YPDA medium in duplicate. Percentages were standardized to 100% of survival for the wild-type strain and are the average of two independent experiments.
Plasmids.
P2893 integrates three tandem copies of GFP at the 3′ end of the ABP140 coding sequence locus. P2932 integrates three tandem copies of GFP at the 3′ end of the CAP2 coding sequence locus. The CAP2–3xGFP fusion protein is functional, i.e., not synthetic lethal with sac6Δ. P2934 integrates three tandem copies of GFP at the 3′ end of the SAC6 coding sequence locus. The SAC6–3xGFP fusion protein is functional (i.e., not synthetic sick with abp1Δ). P3006 integrates three tandem copies of GFP at the 3′ end of the ABP1 coding sequence locus. The ABP1-3xGFP fusion protein is functional (i.e., not synthetic sick with sac6Δ). Details of the constructions and genetic assays are available upon request.
Microscopy
Epifluorescence Microscopy.
Cells were observed in a fully automated Zeiss 200M inverted microscope (Carl Zeiss, Thornwood, NY) equipped with an MS-2000 stage (Applied Scientific Instrumentation, Eugene, OR), a Lambda LS 175 W xenon light source (Sutter Instrument, Novato, CA), a 100 × 1.4 numerical aperture Plan-Apochromat objective, and a five position filter turret. Filter cubes were as follows: for Alexa-phalloidin 568: Cy3 (Ex: HQ535/50–Em: HQ610/75–BS: Q565lp), for live cells GFP: Endow GFP long-pass (Ex: HQ470/40–Em: HQ500lp–BS: Q495lp), and for fixed cells GFP colocalization Narrowband HQ fluorescein isothiocyanate (Ex: HQ487/25–Em: HQ535/40–BS: Q505lp) (Chroma Technology, Rockingham, VT). Images were acquired using a CoolSnap HQ camera (Roper Scientific, Tucson, AZ). The microscope, camera, and shutters (Uniblitz, Rochester, NY) were controlled by SlideBook software (Intelligent Imaging Innovations, Denver, CO).
Fluorescence recovery after photobleaching (FRAP) was performed using the Micropoint wavelength-tunable laser system (Photonics Instruments, St. Charles, IL) attached to the epi-illuminator port of the microscope. This system uses a nitrogen pulse laser (VSL-337ND-S; Laser Science, Franklin, MA) coupled to the microscope by a fiber optic cable. Incoherent light was synchronized and amplified using a coumarin blue chemical chamber, which emits at a wavelength of 440 nm. Simultaneous photobleaching and fluorescence illumination were achieved using a beamsplitter (50% illumination/50% laser) to direct both the fluorescence light and the laser beam into the fluorescence light train of the microscope. The XY targeting of the laser is controlled by two computer driven galvanometer-based steering lenses. Fluorescence intensity of a 12-pixel area was measured on maximal projection of Z-stacks. The fluorescence intensity In was calculated as follows: In = (Iregion of interest − Ibackground)/(Icontrol region − Ibackground). The fluorescence ratio was I0/In. For reference, see (Luedeke et al. 2005).
For GFP imaging, yeast cells were grown at 30°C in SD casa medium appropriately supplemented. For time-lapse live cell microscopy, 2–2.5 μl of the culture was spotted onto a glass slide and immediately imaged at room temperature (∼22°C). Phalloidin staining was done as described previously (Sagot et al., 2002) with Alexa-phalloidin 568 (Invitrogen). All the pictures (unless specified) are maximum projection of Z-stacks acquired with a Z step of 0.3 μM.
Electron Microscopy.
Yeast cells were grown for 3 d in 50 ml of YPDA at 30°C with 220 rpm agitation. Cell pellets were placed on the surface of a copper electron microscopy grid (400 mesh) that had been coated with Formvar. Each loop was very quickly submersed in precooled liquid propane and held at −180°C by liquid nitrogen. The loops were then transferred into a precooled solution of 0.1% glutaraldehyde in dry acetone for 3 d at −82°C. Samples were rinsed with acetone at −20°C and progressively embedded at −20°C in LR Gold resin (Electron Microscopy Sciences, Hatfield, PA). Resin polymerization was carried out at −20°C for 7 d under UV illumination. Ultrathin LR Gold sections were collected on nickel grids coated with Formvar. Sections were first incubated for 5 min with 1 mg/ml glycine and for 5 min with fetal calf serum (1:20). The grids were incubated 45 min at room temperature with mouse anti-actin monoclonal antibody diluted 1/200 (Chemicon International, Temecula, CA), rinsed with Tris-buffered saline:0.1% bovine serum albumin, and incubated for 45 min at room temperature with anti-mouse IgG conjugated to 10-nm gold particles (British Biocell, Cardiff, United Kingdom). The sections were rinsed with distilled water, contrasted 5 min with 2% uranyl acetate in water, followed by 1% lead citrate for 1 min. Specimens were observed with Philips Tecnai 12 Biotwin (120-kV) electron microscope (SERCOMI, Université Victor Ségalen Bordeaux 2, France).
Western Blot
Total protein extracts were prepared using the trichloroacetic acid and glass beads method described in (Reid and Schatz 1982). Antibodies were a generous gift from B. Goode (Rosenstiel Center, Brandeis University, Waltham, MA). Chicken anti-yeast actin antibodies (Aves Labs, Tigard, OR) were used at 1/5000; chicken anti-yeast Abp1p antibodies (Aves Labs) were used at 1/5000; chicken anti-yeast Cap1/2p antibodies (Aves Labs) were used at 1/2500; chicken anti-yeast fimbrin (Sac6p) antibodies (Aves Labs) were used at 1/5000; and mouse anti-Scp1p antibodies (Goodman et al., 2003) were used at 1/1000. HRP-conjugated anti-chicken and anti-mouse secondary antibodies (Pierce Chemical, Rockford, IL) were used at 1/10,000.
Miscellaneous
Glucose concentration was measured using the d-glucose/d-fructose UV test kit (Roche Diagnostics, Mannheim, Germany). Latrunculin-A was a generous gift of B. Goode and was used at the concentration of 200 μM. Cycloheximide (Sigma-Aldrich, St. Louis, MO) was used at the final concentration of 100 μg/ml.
RESULTS
The Actin Cytoskeleton of Quiescent Yeast Cells Is Organized in Bodies
In this study, we chose to follow the operational definition of quiescent cells proposed by (Gray et al. 2004) and previously used for transcriptome studies (Radonjic et al., 2005), i.e., quiescent cells obtained in a stationary phase culture in glucose-rich liquid medium at 30°C.
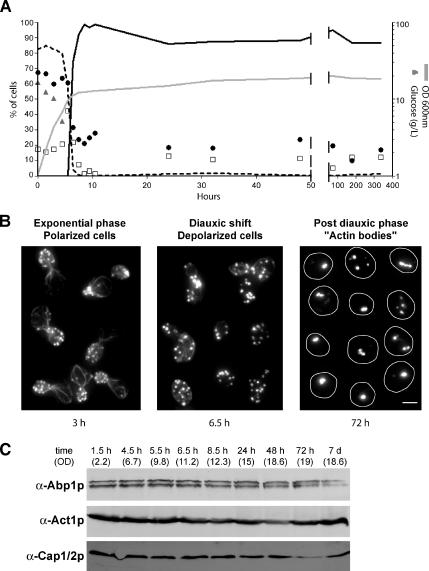
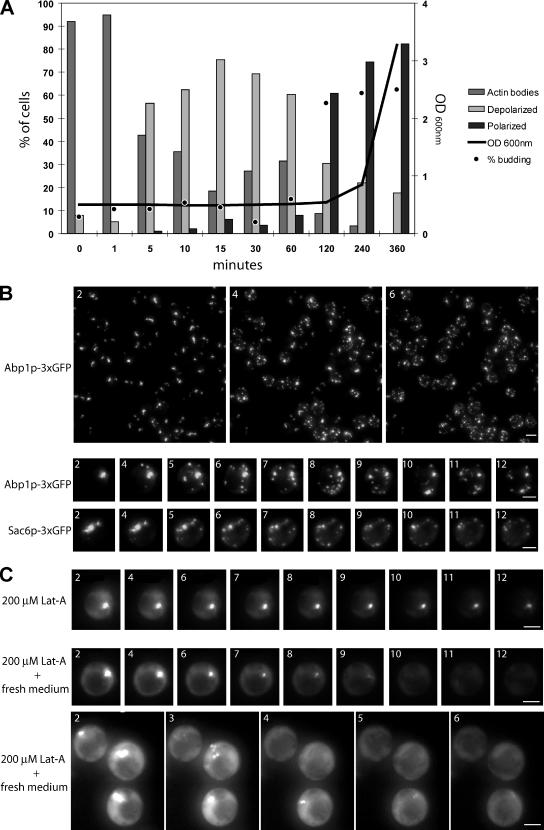
We followed the reorganization of the yeast actin cytoskeleton during the different stages of the growth of wild-type yeast cells cultured in glucose-rich medium. As extensively described previously, actively proliferating yeast cells displayed an actin cytoskeleton composed of three F-actin–containing structures: actin cables, actin patches, and an actin cytokinetic ring (for review, see Pruyne et al., 2004). Cables and patches were polarized to the active growth sites (the bud tip and the bud neck upon cytokinesis; Figure 1B, left). At the diauxic shift, corresponding to glucose exhaustion, cables became disorganized and patches depolarized within the cell. Then, cables disappeared and structures resembling actin patches remained depolarized (Figure 1B, middle). Finally, patches disappeared, while a new actin organization that we have called actin bodies appeared (Figure 1B, right). Within a few hours, actin bodies were found in the majority of the cell population (Figure 1A, black curve). Because they could be stained with phalloidin, actin bodies contained bona fide F-actin. Actin bodies did not have a specific shape; they could either be spherical or elongated. When elongated, actin bodies displayed variable thickness and curvature. Furthermore, each cell could bear one or more actin bodies. Typical examples are shown in Figure 1B (right). The actin cytoskeleton remained organized in actin bodies during the entire stationary phase, and we were able to observe actin bodies in cells grown for as long as 2 months (our unpublished data). Based on these observations, it was clear that a notable amount of F-actin was still present in quiescent cells. Consistently, the steady-state level of total cellular actin was not affected in stationary phase (Figure 1C). Of note, the amount of the ABPs Abp1p and Cap1/2 did not significantly change all along the different steps of the actin cytoskeleton reorganization (Figure 1C).
Figure 1.
Actin cytoskeleton reorganization upon entry into quiescence. (A) Wild-type yeast cells were grown in YPDA at 30°C. Cell density was monitored by measuring OD600 nm (gray line). At various stage of growth, samples were taken, glucose concentration in the medium was measured (gray triangles), and cells were stained with Alexa-phalloidin. For each time point, cells with polarized actin cytoskeleton (black dashed curve), depolarized actin cytoskeleton (open squares), or actin bodies (black curve) were counted (n > 200 for each time point). The budding index (percentage of budding cells) is shown as black circles (n > 200 for each time point). (B) Left, examples of polarized cells in exponential phase of growth (time point 3 h in A). Middle, examples of depolarized cells upon diauxic shift (time point 6.5 h in A). Right, examples of quiescent cells bearing actin bodies (time point 72 h in A). Images are two-dimensional (2D) maximal projection of three-dimensional (3D) image stacks. Bar, 2 μm. (C) Western blot analysis of the steady-state levels of Abp1p, Act1p, and Cap1/2p upon entry into quiescence. Time points and OD correspond to time points in A.
As shown in Figure 1A, actin bodies occurred after the diauxic shift, when glucose was exhausted. We thus asked whether this particular actin cytoskeleton organization could be observed in cells grown on nonfermentable carbon sources. As shown in Table 2, actin bodies were found in cells grown for 7 d in lactate or glycerol/ethanol-containing medium. Finally, actin bodies were observed in both haploid and diploid cells and in various strain backgrounds (Table 3). Therefore, actin bodies define a new actin cytoskeleton organization specifically found in nonproliferating yeast cells.
Table 2.
Actin bodies in cells grown in different culture media
| Rich medium carbon source | Cells with actin bodies (%) | Budding index (%) |
|---|---|---|
| Glucose | 91 ± 8 | 20 ± 3 |
| Lactate | 88 ± 1 | 11 ± 4 |
| Glycerol/ethanol | 93 ± 7 | 22 ± 3 |
Strain BY4741 grown for 7 d at 30°C and 220 rpm (n > 200; 2 experiments).
Table 3.
Actin bodies in different strain backgrounds
| Strain | Cells with actin bodies (%) | Budding index (%) |
|---|---|---|
| BY4741 | 91 ± 7 | 20 ± 3 |
| FY23 | 95 ± 5 | 5 ± 2 |
| FL100 | 94 ± 6 | 18 ± 11 |
| BY4743 | 94 ± 6 | 18 ± 3 |
Strains grown for 7 d in YPDA at 30°C and 220 rpm (n > 200; 2 experiments).
Actin Bodies Do Not Localize to a Particular Region of the Cell
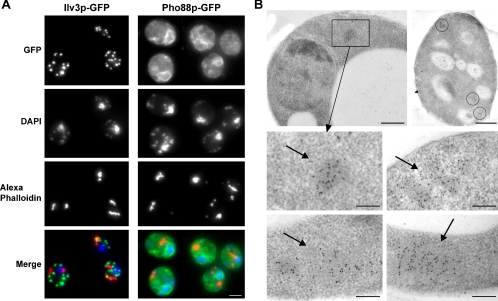
We then addressed the possibility that actin bodies could be located in a particular subcellular region. We thus compared the localization of actin bodies with mitochondria, nucleus, and endoplasmic reticulum by using Ilv3p-GFP, 4,6-diamidino-2-phenylindole (DAPI), and Pho88p-GFP, respectively (Huh et al., 2003). As shown in Figure 2A, actin bodies did not colocalize with mitochondria or nucleus and were not associated with any particular region of the endoplasmic reticulum. These results were confirmed by electron microscopy experiments based on immunogold staining with anti-actin antibodies on cells grown in rich medium for 3 d. As shown in Figure 2B, anti-actin antibodies clustered on slightly electron-dense structures. Importantly, such clusters of actin antibodies were not detected in exponentially growing cells (our unpublished data). Based on their similarity of size, shape, and localization, we could reasonably conclude that the structures on which the anti-actin antibodies were clustering were the actin bodies observed by fluorescence microscopy by using Alexa-phalloidin staining. This experiment confirmed that even at the ultrastructural level, actin bodies do not localize to a particular region of the cytoplasm. Furthermore, no tight association between actin bodies and cellular membrane could be observed.
Figure 2.
Localization of actin bodies. (A) Localization of actin bodies within cells. Wild-type yeast cells expressing either Ilv3p-GFP (left) or Pho88p-GFP (right) staining the mitochondria or the endoplasmic reticulum, respectively, were grown for 7 d at 30°C in SD casa medium, fixed, and stained with DAPI and Alexa-phalloidin. In the merged bottom images, green is GFP-, blue is DAPI-, and red is Alexa-phalloidin–stained F-actin. Images are 2D maximal projection of 3D image stacks. Bar, 2 μm. (B) Immunogold localization of anti-actin antibodies linked to 10-nm gold particles on wild-type yeast cells grown for 3 d at 30°C in YPDA medium. Arrows point to clusters of gold particles. In the right top image, three clusters of gold particles within a cell are circled. Bar, 500 nm (top); 200 nm (bottom).
Several Actin-binding Proteins Colocalize with Actin Bodies
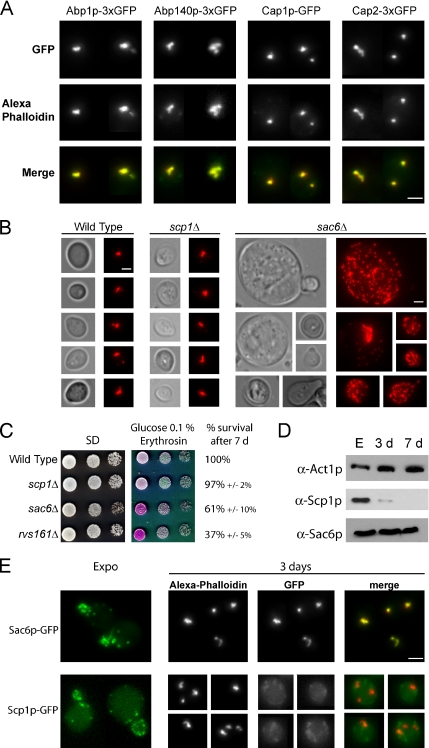
In actively proliferating cells, a vast number of ABPs colocalize with F-actin–containing structures. We thus wondered whether some ABPs were localized in the actin bodies in quiescent cells. Twenty-nine GFP-fused ABPs were tested for localization in quiescent cells (Supplemental Table 1). Among them, six showed a partial colocalization with actin bodies (Arc15p, Arc18p, Arc35p, Arp2p, Crn1p, and Srv2p), and five others (Abp1p, Abp140p, Cap1p, Cap2p, and Sac6p) strictly colocalized with actin bodies. Abp1p and Cap1p/Cap2p are ABPs detected only in actin patches (Drubin et al., 1988; Amatruda and Cooper, 1992; Doyle and Botstein, 1996; Waddle et al., 1996), Abp140p is a protein preferentially found in actin cables (Asakura et al., 1998; Yang and Pon, 2002), and Sac6p has been detected in both actin cables and patches (Drubin et al., 1988; Doyle and Botstein, 1996). To confirm those results and increase when necessary the GFP signal in quiescent cells, 3xGFP fusion proteins were constructed and shown to be functional (see Materials and Methods). As shown in Figure 3A, Abp1p-3xGFP, Abp140p-3xGFP, Cap1p-GFP, and Cap2p-3xGFP colocalized with actin bodies. Similar results were obtained with Sac6p (Figure 3E). Thus, proteins found in actin cables and/or patches colocalized with actin bodies in nonproliferating cells.
Figure 3.
(A) Colocalization of actin bodies with various actin-binding proteins. Wild-type yeast cells expressing different ABPs fused to three tandem GFP copies at a native level were grown for 2 d at 30°C in SD casa medium. Cells were fixed and stained with Alexa-phalloidin. In the merged bottom panel, GFP is shown in green, and Alexa-phalloidin–stained F-actin is shown in red. Images are 2D maximal projection of 3D image stacks. (B) Actin bodies formation in two actin-bundling protein mutants: sac6Δ and scp1Δ. Wild-type (left), scp1Δ (middle), and sac6Δ (right) cells were grown for 3 d in YPDA medium, fixed, and stained with Alexa-phalloidin. Bar, 2 μm. (C) Actin-bundling mutants survival upon starvation and in stationary phase. Serial dilutions were spotted onto regular SD casa WAU medium or synthetic medium containing casa, WUA, 0.1% glucose, and erythrosine, a dye that stains dead cells pink (Bonneu et al., 1991). Viability of the strains after 7 d of growth at 30°C in YPDA medium is indicated. Viability was assessed as described in Materials and Methods. (D) Steady-state levels of Sac6p and Scp1p at different growth phases. Samples of wild-type cells were taken in exponential phase, after 3 d (3 d) or 7 d (7 d) of culture at 30°C in YPDA medium. (E) Localization of Sac6p and Scp1p in stationary phase. Yeast cells expressing either Sac6p-GFP or Scp1p-GFP were grown in SD casa medium at 30°C. Left, GFP fluorescence is shown in living exponentially growing cells. In the following panels, cells grown for 3 d at 30°C were fixed and stained with Alexa-phalloidin. In the merged panel, GFP is shown in green, and Alexa-phalloidin–stained F-actin is shown in red. Images are 2D maximal projection of 3D image stacks. Bar, 2 μm.
Actin bodies occur as tight F-actin–containing structures and are therefore likely to be maintained by actin-bundling proteins. Scp1p and Sac6p are two yeast actin-bundling proteins that act together to regulate the stability and the organization of the actin cytoskeleton in proliferating cells (Goodman et al., 2003, Winder et al., 2003). We thus asked whether these two proteins were involved in the actin cytoskeleton organization in quiescent cells. Actively proliferating scp1Δ cells displayed a wild-type actin cytoskeleton organization (Goodman et al., 2003; Winder et al., 2003) and 92 ± 2% of scp1Δ quiescent cells displayed actin bodies after 3 d of culture (Figure 3B). In contrast, sac6Δ cells presented a strongly altered actin cytoskeleton organization in both exponential and stationary phase (Adams et al., 1991; Figure 3B). Importantly, sac6Δ cells displayed a reduced viability upon starvation and stationary phase (Figure 3C; Gourlay et al., 2004). Of note, deletion of SCP1 did not significantly exacerbate the sac6Δ phenotype (see Supplemental Figure 1). Thus, it is tempting to speculate that the actin-bundling activity of fimbrin/Sac6p is required to maintain actin bodies in stationary phase, whereas the activity of the calponin homologue Scp1p is not. Consistently, the steady-state level of Sac6p remained stable in stationary phase, whereas Scp1p steady-state level strongly decreased upon starvation (Figure 3D). Finally, whereas both Scp1-GFP and Sac6p-GFP were detected in exponentially growing cells, in nonproliferating cells, Sac6p-GFP colocalized with actin bodies but no Scp1-GFP signal could be detected, as expected according to the steady-state level of this protein (Figure 3E).
Actin Bodies Are Stable Structures
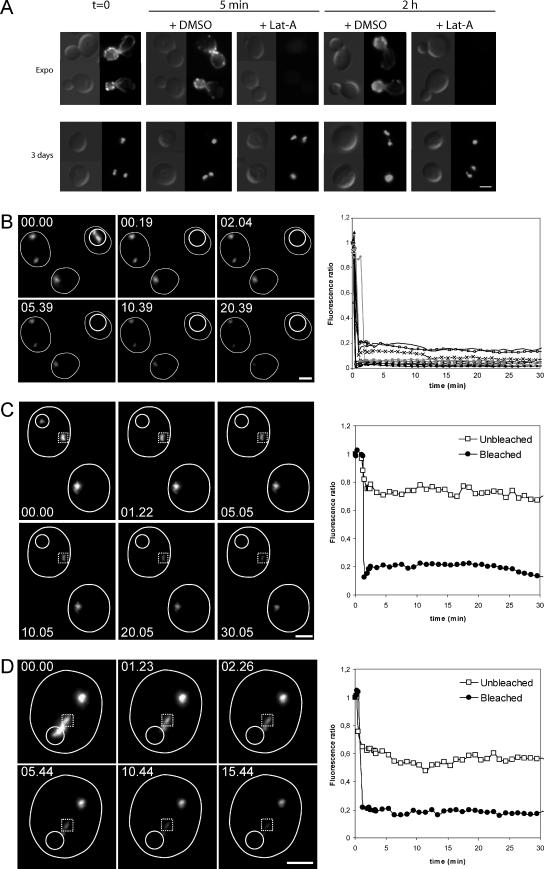
In yeast, the F-actin–containing structures observed in proliferating cells are highly motile (Doyle and Botstein, 1996; Yang and Pon, 2002). Within these structures, protein exchange is very dynamic (Yang and Pon, 2002; Kaksonen et al., 2003), and the actin turnover in the filaments that form actin cables and patches is extremely rapid. Indeed, actin patches are very short-lived structures, the half-life of which is estimated to be 20–40 s (Waddle et al., 1996). Consistently, in living cells, actin filaments rapidly disappear upon treatment with latruculin A (Lat-A), a drug that prevents G-actin polymerization (Ayscough et al., 1997). We therefore used Lat-A to estimate the turnover of actin filaments in the bodies. As expected, in actively proliferating cells, a 200 μM Lat-A treatment caused the disappearance of all F-actin containing structures (actin cables, patches, and cytokinetic rings) in <5 min (Figure 4A, top). By contrast, for cells grown for 3 or 7 d, the same treatment did not affect actin bodies, even after prolonged incubation in the presence of the drug (Figure 4A, top, and Supplemental Figure 2). Thus, actin bodies are more resistant to Lat-A treatment than any other known yeast F-actin–containing structure.
Figure 4.
Actin bodies contain stable actin filaments. (A) Actin bodies are resistant to latrunculin A (A). Wild-type cells were grown at 30°C in YPDA medium. During exponential growth (top) or after 3 d of culture (bottom), cells were incubated 5 min or 2 h with either 200 μM Lat-A or dimethyl sulfoxide as a control. Cells were then fixed and stained with Alexa-phalloidin. (B) Fluorescence recovery after laser ablation of the Abp1p-3xGFP signal of an actin body in a cell with a single actin body. Wild-type cells expressing Abp1p-3xGFP were grown for 3 d at 30°C in SD casa medium. Images on the left are 2D maximal projection of 3D image stacks time-lapse series. Time is indicated in the top left corner. The small white circle indicates the bleached region. The graph on the right shows the kinetics of fluorescence course obtained for 10 different cells. The same kind of curves was obtained for 32 cells. (C) Fluorescence recovery after laser ablation of the Abp1p-3xGFP signal of an actin body in a cell displaying two actin bodies. Wild-type cells expressing Abp1p-3xGFP were grown for 3 d at 30°C in SD casa medium. Images on the left are 2D maximal projection of 3D image stacks time-lapse series. Time is indicated in the top left corner. The small white circle indicates the bleached region; the dotted line square indicates the unbleached actin body in the same cell. The graph on the right shows the kinetics of fluorescence course measured for the bleached (black dots) and the unbleached (open squares) regions. The same kind of curve was obtained for 12 different cells. (D) Fluorescence recovery after laser ablation of the Abp1p-3xGFP signal of one-half of an actin body. Wild-type cells expressing Abp1p-3xGFP were grown for 3 d at 30°C in SD casa medium. Images on the left are 2D maximal projection of 3D image stacks time-lapse series. Time is indicated in the top left corner. The small white circle indicates the bleached region; the dotted line square indicated the unbleached region of the same actin body. The graph on the right shows the kinetics of fluorescence course measured for the bleached (black dots) and the unbleached (open squares) regions. The same kind of curve was obtained for 14 different actin bodies. Bar, 2 μm.
We then wondered whether the pool of ABPs localized in actin bodies was dynamic. For this purpose, we used FRAP to study the turnover of Abp1p-3xGFP in actin bodies. Because the recovery of Abp1-3xGFP reflects the exchange of Abp1p-3xGFP within actin bodies or with the Abp1p-3xGFP cytoplasmic pool, it gives indications on actin bodies dynamic. Actin bodies of cells grown for 3 d were photobleached and even after 30 min, no significant recovery of Abp1-3xGFP was detected. This was true when an actin body was entirely bleached in cells displaying one or two separated actin bodies (0/32 and 0/12 cells, respectively; Figure 4, B and C) but also when an actin body was partially bleached (0/14; Figure 4D). In conclusion, no Abp1-3xGFP exchange could be detected, strongly suggesting that there was no significant Abp1-3xGFP cytoplasmic pool and/or that Abp1p-3xGFP was not moving within an actin body. Similar results were obtained using Sac6p-3xGFP (our unpublished data). Furthermore, as it could be observed in Figure 4, B–D, actin bodies were generally nonmobile within the cells. These data indicate that actin bodies are stable and nonmotile structures in which the turnover of actin filaments and ABPs is extremely slow.
Actin Bodies Rapidly Disappear upon Cells Refeeding
In nonproliferating cells, actin bodies behaved as stable structures. We thus wondered how actin bodies reorganize when cells exit from quiescence and reenter the proliferative cell cycle. To address this question, a wild-type yeast culture was grown to stationary phase (7 d) in rich medium, cells were then diluted into fresh medium, and actin cytoskeleton reorganization was followed by actin phalloidin staining. As shown in Figure 5A, <5 min after cells refeeding, >50% of the actin bodies disappeared and depolarized actin cables and patches reappeared. Full polarization of these structures and bud emergence occurred 2 h after cells refeeding. To follow more precisely actin body disappearance, we used wild-type yeast cells expressing Abp1p-3xGFP, which were grown for 2 d, diluted into fresh medium after a brief centrifugation, and immediately imaged. As shown in Figure 5B and in Supplemental Movies S1 and S2, 4 min after refeeding, highly motile small Abp1-3xGFP dots occurred simultaneously in most of the cells, whereas actin bodies concomitantly disappeared. Similar results were obtained for cells expressing Sac6p-3xGFP (Figure 5B, bottom, and Supplemental Movie S3), and no changes in actin bodies organization could be observed when cells were transferred into water instead of fresh medium (our unpublished data). When cells were treated with 200 μM Lat-A before refeeding, most actin bodies disappeared within 15 min. In contrast with the untreated cells, no patches reappeared concomitantly (Figure 5C, Supplemental Movie S4, and Supplemental Table 3). We concluded that actin bodies cannot directly give rise to actin patches without any actin filament turnover.
Figure 5.
Reorganization of the actin cytoskeleton upon stationary phase exit. (A) Wild-type cells were grown for 7 d at 30°C in YPDA medium. Cells were then washed and resuspended in fresh YPDA medium and grown at 30°C. Cell density was monitored by measuring OD600 nm (gray line). For each time points, cells were stained with Alexa-phalloidin. Cells with polarized actin cytoskeleton (dark gray bar), depolarized actin cytoskeleton (light gray bar), or actin bodies (gray bar) were counted (n > 200; SD <5%). The budding index (percentage of budding cells) is shown as black dots (n > 200; SD <5%). (B) Wild-type yeast cells expressing Abp1p-3xGFP (top) or Sac6–3xGFP were grown 2 d at 30°C in SD casa medium. After a brief centrifugation, the cell pellet was resuspended in fresh medium and cells were immediately imaged. Images are 2D maximal projection of 3D image stacks time-lapse series. Time in minutes after medium renewal is indicated in the left corner. Bar, 2 μm. (C) Wild-type yeast cells expressing Abp1p-3x GFP were grown for 3 d at 30°C in SD casa medium. The cell culture was concentrated and Lat-A was added at the final concentration of 200 μM. This concentrated culture was incubated for 30 min at 30°C. After this incubation, 20 μl of the cell suspension was mixed either to 20 μl of water containing 200 μM Lat-A (top) or to 20 μl of 2X fresh SD casa containing 200 μM Lat-A, and immediately imaged (two bottom panels). Images are 2D maximal projection of 3D image stacks time-lapse series. Time in minutes after addition of Lat-A–containing water or fresh medium is indicated in the left corner. Bar, 2 μm.
Actin Bodies Disappearance and Actin Cables and Patches Formation Can Occur in the Absence of Protein Synthesis
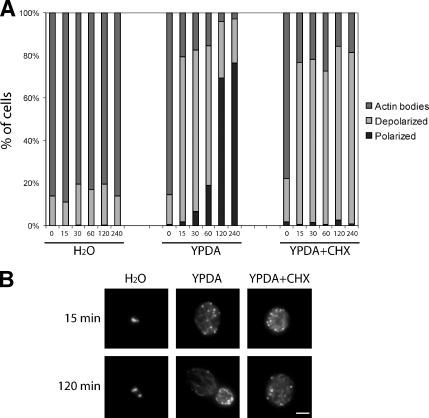
To gain insight into the mechanism of actin patches and cables reappearance upon actin bodies disassembly, we followed actin bodies reorganization upon cells refeeding in the presence of cycloheximide, a drug preventing protein synthesis. Cells were grown for 7 d and then transferred into either water, YPDA, or YDPA containing 100 μg/ml cycloheximide. This concentration of cycloheximide inhibits protein synthesis even in cells grown to stationary phase (Brengues et al., 2005). However, to further ensure cycloheximide efficiency, treated cells were preincubated for 1 h with the drug before refeeding in cycloheximide-containing YPDA. As shown in Figure 6A, whereas actin bodies were not affected by transferring the cells into water, when the cells were transferred into YPDA, containing cycloheximide or not, actin bodies rapidly disappeared and depolarized actin cables and patches appeared (for examples, see Figure 6B). However, polarization of these structures was totally inhibited by cycloheximide (Figure 6, A and B). In conclusion, actin bodies reorganization into actin cables and patches could occur in the absence of de novo protein synthesis.
Figure 6.
Actin cytoskeleton depolarization upon exit from quiescence can happen in the absence of protein synthesis. (A) Wild-type cells were grown for 7 d at 30°C in YPDA medium. Cells were then washed and resuspended in either water (left) or fresh YPDA medium (middle), and grown at 30°C. For cycloheximide (CHX) treatment, wild-type cells were grown for 7 d at 30°C in YPDA medium and then preincubated with 100 μg/ml cycloheximide by addition of the drug directly into the growth medium. After 1 h of pretreatment at 30°C, cells were washed and resuspended in fresh YPDA medium containing 100 μg/ml cycloheximide. Cells were then fixed and stained with Alexa-phalloidin. For each time point, cells with polarized actin cytoskeleton (dark gray bar), depolarized actin cytoskeleton (light gray bar), or actin bodies (gray bar) were counted (n > 200, SD <5%). (B) Images are 2D maximal projection of 3D image stacks of examples of cells observed for the experiment in A. Bar, 2 μm.
DISCUSSION
Actin cytoskeleton reorganization upon specific signals is at the basis of crucial aspects of cellular life, such as polarized intracellular transport, cell migration, or cell division. Here, we have described a new actin cytoskeleton organization, the actin bodies, that specifically arise when yeast cells enter the quiescent cycle. These structures are maintained during all quiescence and are rapidly disassembled upon reentry of cells into the proliferation cycle. Actin bodies have to be distinguished from the actin bar described in some yeast mutant strains that display actin cytoskeleton organization defects (Johnston et al., 1991; Holtzman et al., 1993). Actin bars can only be detected using anti-actin antibodies. In contrast, actin bodies can be stained with fluorescently labeled phalloidin, demonstrating that these structures contain bona fide F-actin filaments. Besides, in a previous study, an actin “chunks” phenotype was mentioned for quiescent yeast cells cultured for 28 d (Gourlay et al., 2004) and mutants, such as prs3Δ, whi2Δ, end3Δ, sla1Δ have been reported to display actin “clumps” in early stationary phase (Binley et al., 1999; Care et al., 2004; Gourlay and Ayscough, 2005). Notwithstanding, these structures have not been further studied and to date, no precise description (composition, kinetics, and so on) of the yeast actin cytoskeleton reorganization upon entry into quiescence has been published. Here, we show that actin bodies occur massively when wild-type cells cease to proliferate, whereas actin cables and patches concomitantly disappear. The F-actin filaments embedded into actin bodies can be either “old” actin filaments “recycled” from actin cables, and/or patches or nucleated de novo by an actin nucleator. This nucleator could be a specific and yet not identified nucleator of the actin bodies filaments. Alternatively, it could be a previously characterized yeast actin nucleator, such as the Arp2/3 complex or the formins, that nucleate actin filaments assembled into patches or cables, respectively (Winter et al., 1999; Evangelista et al., 2002; Sagot et al., 2002). Because the inactivation of such nucleators prevents yeast proliferation, addressing the involvement of these molecular machines in the formation of actin bodies is not trivial.
By contrast to actin cables and patches, actin bodies filament turnover seems to be very slow (Figure 4A). What causes this observed lack of dynamics? The corollary to this question is how are actin bodies maintained? Two nonexclusive possibilities can be envisioned: either the filament turnover is reduced by modification of actin itself (or its associated nucleotide), or the filaments are tightly maintained by ABPs. These ABPs could either be specific of the actin bodies or be classic ABPs that would specifically be modified in stationary phase to prevent F-actin turnover. We have shown here that in quiescent cells, several actin-binding proteins colocalize with the actin bodies. Among these ABPs, the capping protein has been shown to block actin filaments barbed ends and to decrease the rate of filament disassembly. Similarly, the actin filament-bundling protein Sac6p (the fimbrin orthologue) also colocalized with actin bodies. Furthermore, Sac6p seemed to be required for the formation and/or maintenance of actin bodies (Figure 3B). Thus, at least two proteins that diminish the actin filament turnover could potentially be implicated in the formation and/or the maintenance of actin bodies. The fact that actin filaments turnover is specifically reduced in stationary phase could be the consequence of active formation of actin bodies. In contrast, we can also envisage that the reduction of filament turnover specifically induced upon stationary phase entry passively leads to the formation of actin bodies. Indeed, it has previously been reported that act1-157 cells (an actin allele with increased actin dynamics) display less actin chunks than the WT counterpart, when grown to early stationary phase (Gourlay et al., 2004). At the molecular level, the deciphering of the actin bodies formation and/or maintenance in stationary phase awaits the finding of mutants specifically impaired in these processes. Our attempt to identify such mutants revealed that among 23 viable ABP deletion mutants, only arc18Δ and sac6Δ showed a clear alteration of actin cytoskeleton in stationary phase (Supplemental Table 2). However, the fact that these mutants already display an altered actin cytoskeleton organization in actively proliferating cells makes interpretation of their specific effect on actin cytoskeleton reorganization upon entry into quiescence rather delicate. Furthermore, the entry into stationary phase is a slow process and to date, despite the great lengths we went to, we did not find any other experimental way to induce the actin bodies formation. This highly complicates the decoding of the signaling cascade that leads to this actin cytoskeleton reorganization upon entry into quiescence. Because actin bodies rapidly and synchronously disappear upon cells refeeding (Figure 5), studying the signals leading to the actin bodies disassembly may be a better route to understand the pathways involved in the maintenance of the actin bodies.
Actin bodies are promptly disassembled upon cells refeeding. Indeed, our movies revealed the concomitant apparition of highly mobile smaller particles. The shape and speed of these particles are very similar to those of actin patches; however, because they move mostly from the original actin bodies toward the cell periphery (see Supplemental Movies), they could not strictly be regarded as bona fide actin patches that form upon endocytosis. After this transitional state, cells rapidly display depolarized actin cables and cell periphery associated actin patches. Two hours after refeeding, most cells are polarized and a new bud has emerged. Because actin cables and patches can be assembled in the absence of de novo protein synthesis (Figure 6), we speculate that actin bodies could be an actin- and ABPs-containing “reservoir” immediately available for cables and patches assembly upon cells refeeding. Whether actin contained in the actin storage bodies is mobilized as monomeric actin, as filaments or as more organized F-actin–containing structures remains to be clarified. However, upon exit from quiescence, we never observed any transitory state in which no F-actin could be detected. Why would cells need such a reserve of actin (and ABPs)? Actin could be required for the cell viability during quiescence. Alternatively, an actin reserve could be crucial to resume cell growth upon exit from stationary phase. At the molecular level, is it critical for the cell to stock actin as a structured actin storage body, or could cells cope with a reserve of unorganized F-actin or even G-actin? Are actin bodies the only way for cells to store actin in stationary phase? The finding of mutants that either would not display actin storage bodies while quiescent or that would be unable to disassemble them upon reentry into the proliferation cycle would help address these questions.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Goodman and J. D'Agostino for technical assistance. We thank C. Plaisant for the 3xGFP fusion proteins construction and characterization. We are especially grateful to B. Goode for providing reagents and support. We express our gratitude to D. Pellman for comments on the manuscript. This work was supported by a young investigator grant (ANR JC05_42065) from the French National Agency for Research (to I.S. and B.P.) and by Grant ARC3719 from the Association pour la Recherche sur le Cancer. The microscope used in this study was financed by The Region Aquitaine and Centre National de la Recherche Scientifique/Victor Ségalen Bordeaux II University, Unité Mixte de Recherche 5095.
Abbreviations used:
- ABP
actin-binding protein
- Lat-A
lantrunculin A
Footnotes

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-04-0282) on August 16, 2006.
REFERENCES
- Adams A. E., Botstein D., Drubin D. G. Requirement of yeast fimbrin for actin organization and morphogenesis in vivo. Nature. 1991;354:404–408. doi: 10.1038/354404a0. [DOI] [PubMed] [Google Scholar]
- Amatruda J. F., Cooper J. A. Purification, characterization, and immunofluorescence localization of Saccharomyces cerevisiae capping protein. J. Cell Biol. 1992;117:1067–1076. doi: 10.1083/jcb.117.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura T. Isolation and characterization of a novel actin filament-binding protein from Saccharomyces cerevisiae. Oncogene. 1998;16:121–130. doi: 10.1038/sj.onc.1201487. [DOI] [PubMed] [Google Scholar]
- Ayscough K. R., Stryker J., Pokala N., Sanders M., Crews P., Drubin D. G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley K. M., Radcliffe P. A., Trevethick J., Duffy K. A., Sudbery P. E. The yeast PRS3 gene is required for cell integrity, cell cycle arrest upon nutrient deprivation, ion homeostasis and the proper organization of the actin cytoskeleton. Yeast. 1999;15:1459–1469. doi: 10.1002/(SICI)1097-0061(199910)15:14<1459::AID-YEA472>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Bonneu M., Crouzet M., Urdaci M., Aigle M. Direct detection of yeast mutants with reduced viability on plates by erythrosine B staining. Anal. Biochem. 1991;193:225–230. doi: 10.1016/0003-2697(91)90013-j. [DOI] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiaeS288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Care A., Vousden K. A., Binley K. M., Radcliffe P., Trevethick J., Mannazzu I., Sudbery P. E. A synthetic lethal screen identifies a role for the cortical actin patch/endocytosis complex in the response to nutrient deprivation in Saccharomyces cerevisiae. Genetics. 2004;166:707–719. doi: 10.1534/genetics.166.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateaubodeau G. A., Guerin M., Guerin B. Permeability of yeast mitochondrial internal membrane: structure-activity relationship. Biochimie. 1976;58:601–610. doi: 10.1016/s0300-9084(76)80230-1. [DOI] [PubMed] [Google Scholar]
- Doyle T., Botstein D. Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc. Natl. Acad. Sci. USA. 1996;93:3886–3891. doi: 10.1073/pnas.93.9.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin D. G., Miller K. G., Botstein D. Yeast actin-binding proteins: evidence for a role in morphogenesis. J. Cell Biol. 1988;107:2551–2561. doi: 10.1083/jcb.107.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Henriques M., Daignan-Fornier B. Transcriptional regulation of the yeast GMP synthesis pathway by its end products. J. Biol. Chem. 2001;276:1523–1530. doi: 10.1074/jbc.M007926200. [DOI] [PubMed] [Google Scholar]
- Evangelista M., Pruyne D., Amberg D. C., Boone C., Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 2002;4:260–269. doi: 10.1038/ncb770. [DOI] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., Brown P. O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode B. L., Rodal A. A. Modular complexes that regulate actin assembly in budding yeast. Curr. Opin. Microbiol. 2001;4:703–712. doi: 10.1016/s1369-5274(01)00272-7. [DOI] [PubMed] [Google Scholar]
- Goodman A., Goode B. L., Matsudaira P., Fink G. R. The Saccharomyces cerevisiae calponin/transgelin homolog Scp1 functions with fimbrin to regulate stability and organization of the actin cytoskeleton. Mol. Biol. Cell. 2003;14:2617–2629. doi: 10.1091/mbc.E03-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay C. W., Ayscough K. R. Identification of an upstream regulatory pathway controlling actin-mediated apoptosis in yeast. J. Cell Sci. 2005;118:2119–2132. doi: 10.1242/jcs.02337. [DOI] [PubMed] [Google Scholar]
- Gourlay C. W., Carpp L. N., Timpson P., Winder S. J., Ayscough K. R. A role for the actin cytoskeleton in cell death and aging in yeast. J. Cell Biol. 2004;164:803–809. doi: 10.1083/jcb.200310148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. V., Petsko G. A., Johnston G. C., Ringe D., Singer R. A., Werner-Washburne M. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman P. K. Stationary phase in yeast. Curr. Opin. Microbiol. 2002;5:602–607. doi: 10.1016/s1369-5274(02)00377-6. [DOI] [PubMed] [Google Scholar]
- Holtzman D. A., Yang S., Drubin D. G. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J. Cell Biol. 1993;122:635–644. doi: 10.1083/jcb.122.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Johnston G. C., Prendergast J. A., Singer R. A. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J. Cell Biol. 1991;113:539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M., Sun Y., Drubin D. G. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Lefebvre-Legendre L., Vaillier J., Benabdelhak H., Velours J., Slonimski P. P., di Rago J. P. Identification of a nuclear gene (FMC1) required for the assembly/stability of yeast mitochondrial F(1)-ATPase in heat stress conditions. J. Biol. Chem. 2001;276:6789–6796. doi: 10.1074/jbc.M009557200. [DOI] [PubMed] [Google Scholar]
- Luedeke C., Frei S. B., Sbalzarini I., Schwarz H., Spang A., Barral Y. Septin-dependent compartmentalization of the endoplasmic reticulum during yeast polarized growth. J. Cell Biol. 2005;169:897–908. doi: 10.1083/jcb.200412143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M. J., Roy S., Archuletta A. B., Wentzell P. D., Anna-Arriola S. S., Rodriguez A. L., Aragon A. D., Quinones G. A., Allen C., Werner-Washburne M. Genomic analysis of stationary-phase and exit in Saccharomyces cerevisiae: gene expression and identification of novel essential genes. Mol. Biol. Cell. 2004;15:5295–5305. doi: 10.1091/mbc.E03-11-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon R. Folded chromosomes in non-cycling yeast cells: evidence for a characteristic go form. Chromosoma. 1978;67:263–274. doi: 10.1007/BF02569039. [DOI] [PubMed] [Google Scholar]
- Pollard T. D., Borisy G. G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Legesse-Miller A., Gao L., Dong Y., Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- Radonjic M., Andrau J. C., Lijnzaad P., Kemmeren P., Kockelkorn T. T., van Leenen D., van Berkum N. L., Holstege F. C. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol. Cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Reid G. A., Schatz G. Import of proteins into mitochondria Yeast cells grown in the presence of carbonyl cyanide m-chlorophenylhydrazone accumulate massive amounts of some mitochondrial precursor polypeptides. J. Biol. Chem. 1982;257:13056–13061. [PubMed] [Google Scholar]
- Sagot I., Klee S. K., Pellman D. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 2002;4:42–50. doi: 10.1038/ncb719. [DOI] [PubMed] [Google Scholar]
- Waddle J. A., Karpova T. S., Waterston R. H., Cooper J. A. Movement of cortical actin patches in yeast. J. Cell Biol. 1996;132:861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder S. J., Ayscough K. R. Actin-binding proteins. J. Cell Sci. 2005;118:651–654. doi: 10.1242/jcs.01670. [DOI] [PubMed] [Google Scholar]
- Winder S. J., Jess T., Ayscough K. R. SCP1 encodesan actin-bundling protein in yeast. Biochem. J. 2003;375:287–295. doi: 10.1042/BJ20030796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D. C., Choe E. Y., Li R. Genetic dissection of the budding yeast Arp2/3 complex: a comparison of the in vivo and structural roles of individual subunits. Proc. Natl. Acad. Sci. USA. 1999;96:7288–7293. doi: 10.1073/pnas.96.13.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. C., Pon L. A. Actin cable dynamics in budding yeast. Proc. Natl. Acad. Sci. USA. 2002;99:751–756. doi: 10.1073/pnas.022462899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.