Abstract
Mitochondria are the major site of cellular iron utilization for the synthesis of essential cofactors such as iron–sulfur clusters and haem. In the present study, we provide evidence that GTP in the mitochondrial matrix is involved in organellar iron homoeostasis. A mutant of yeast Saccharomyces cerevisiae lacking the mitochondrial GTP/GDP carrier protein (Ggc1p) exhibits decreased levels of matrix GTP and increased levels of matrix GDP [Vozza, Blanco, Palmieri and Palmieri (2004) J. Biol. Chem. 279, 20850–20857]. This mutant (previously called yhm1) also manifests high cellular iron uptake and tremendous iron accumulation within mitochondria [Lesuisse, Lyver, Knight and Dancis (2004) Biochem. J. 378, 599–607]. The reason for these two very different phenotypic defects of the same yeast mutant has so far remained elusive. We show that in vivo targeting of a human nucleoside diphosphate kinase (Nm23-H4), which converts ATP into GTP, to the matrix of ggc1 mutants restores normal iron regulation. Thus the role of Ggc1p in iron metabolism is mediated by effects on GTP/GDP levels in the mitochondrial matrix.
Keywords: carrier protein, GTP, iron homoeostasis, mitochondrion, nucleoside diphosphate kinase, yeast
Abbreviations: CoxIV, cytochrome c oxidase subunit IV; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HA, haemagglutinin; mtDNA, mitochondrial DNA; Mu, mutant; NDPK, nucleoside diphosphate kinase; NTP, nucleoside triphosphate; WT, wild-type
INTRODUCTION
Iron is an essential transition metal for virtually all organisms. However, iron overload is deleterious and has been implicated in various human diseases, including Friedreich's ataxia and sideroblastic anaemia [1,2], sparking an intense interest in identifying the cause of this abnormal cellular iron distribution [3]. Iron entering eukaryotic cells must travel to mitochondria, where it is inserted into haem and Fe-S (iron–sulfur) clusters. These cofactors are then incorporated into a variety of critical proteins that mediate oxidation–reductions, respiration and biosynthetic pathways. Iron in various forms must also be exported from mitochondria for incorporation into extra-mitochondrial proteins. The toxic form of iron associated with various diseases is likely to be mitochondrial iron because of its proximity to nearby sources of free radicals, e.g. leaked intermediates from the respiratory chain. Thus mechanisms must exist (in yeast and human) for maintaining mitochondrial iron levels within a tightly controlled range.
The components of the cellular iron uptake system of Saccharomyces cerevisiae include surface ferric reductases and a permease–multicopper oxidase complex. The expression of the corresponding genes is controlled by Aft1p and Aft2p transcription factors, being induced by iron deprivation and repressed by iron availability [4]. Whereas cellular iron uptake machinery has been extensively studied, the molecular mechanisms underlying mitochondrial iron distribution (import or export) and use (Fe-S cluster biogenesis) are poorly understood [5].
Mitochondrial inner membrane contains a family of 35 carrier proteins that allow exchange of various substrates across this membrane [6,7]. In an effort to identify genes that might be involved in mitochondrial iron distribution, Lesuisse et al. [8] examined a collection of yeast mutants, each deleted for a single member of the mitochondrial carrier family. A strain deleted for YHM1 exhibited increased and misregulated surface ferric reductase and high-affinity ferrous transport activities. The effect of YHM1 deletion on cellular iron uptake was entirely dependent on an intact copy of the iron-sensing regulator AFT1. In the Δyhm1 mutant (YHM1 gene deleted), approx. 30-fold excess iron accumulated in the mitochondrial matrix. Such a phenotype is often linked to a failure to form Fe-S clusters. Indeed, the activity levels of some of the mitochondrial (e.g. aconitase) and cytosolic (e.g. isopropylmalate isomerase) Fe-S proteins were significantly reduced in the Δyhm1 mutant. Isolated Δyhm1 mitochondria also showed decreased rates of haem synthesis [8]. Although Yhm1p belongs to the family of mitochondrial carrier proteins, how this protein may participate in mitochondrial iron metabolism was not clear from these studies.
Palmieri and co-workers [9] identified Yhm1p as the GTP/GDP carrier of the mitochondrial inner membrane and renamed the protein Ggc1p. They were able to reconstitute bacterially expressed and purified Ggc1p in liposomes and showed that GTP or GDP was transported by this carrier protein in vitro. In vivo, the Δggc1 mutant lacked this exchange activity, and mitochondrial matrix GTP levels were decreased and mitochondrial matrix GDP levels were increased [9]. These results demonstrate that Ggc1p allows GTP to cross the inner membrane barrier in exchange for matrix GDP. Until this report, the mitochondrial inner membrane was considered to be impermeable to GTP, and this might explain why the possible role of matrix GTP in various metabolic processes has not received much attention. GTP in the matrix is utilized for mitochondrial protein synthesis and for incorporation into various RNAs. The observation that yhm1 [8] or ggc1 [9] mutants convert into rho0 is consistent with secondary mtDNA (mitochondrial DNA) damage from iron accumulation and/or nucleotide imbalance. In an earlier study, YHM1 was identified as a high copy suppressor of mtDNA instability associated with an ABF2 deletion [10], but phenotypic defects associated with iron or GTP metabolism were not reported.
In the present study, we show that the main function of Ggc1p is to transport cytosolic GTP across the mitochondrial inner membrane into the matrix as suggested in [9]. More importantly, our results suggest that the role of Ggc1p in iron metabolism is mediated by effects on GTP/GDP levels in the mitochondrial matrix. These findings are completely novel, since a role of GTP in mitochondrial iron metabolism has never been described.
EXPERIMENTAL
Expression of Nm23-H4 constructs
Bacterial expression
A cDNA clone encoding human Nm23-H4 was purchased from Open Biosystems (Huntsville, AL, U.S.A.). Nm23-H4 constructs were cloned into pET21b (Novagen, Madison, WI, U.S.A.), which introduces a His6 tag at the C-terminus of the proteins. Bacterial expression and purification of these proteins were performed as described in [11].
Expression in yeast
Wild-type (WT) and mutant (Mu) forms of CoxIV (cytochrome c oxidase subunit IV)-Nm23 with a C-terminal HA3 (three haemagglutinin) tag were cloned into a LEU2-marked plasmid (pRS425) for expression in yeast from the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) promoter. A haploid BY4741 strain was generated in which the Δggc1::HIS3 allele was covered by GGC1-HA3 on an URA3-marked plasmid. Nm23-H4 constructs were transformed into this GGC1 shuffle strain. Following counter-selection against the URA3 gene on the covering plasmid (GGC1) in the presence of 5-fluoro-orotic acid, the absence of Ggc1p and the expression of Nm23-H4 constructs were monitored by immunoblotting using anti-HA antibodies. Experiments were performed with five strains carrying pRS425, a high-copy-number plasmid requiring selection on leucine-deficient media: (i) parental strain BY4741 with the empty plasmid pRS425, (ii) rho0 derivative of the parent generated by exposure to ethidium bromide also carrying the empty plasmid pRS425, (iii) Δggc1 carrying the empty plasmid pRS425, (iv) Δggc1 carrying pRS425 with GAPDH-WT Nm23 targeted to mitochondria, and (v) Δggc1 carrying pRS425 with GAPDH-Mu Nm23, mutated Nm23-H4 (H151G) targeted to mitochondria.
NDPK (nucleoside diphosphate kinase) activity of Nm23-H4
Autophosphorylation of Nm23-H4 was examined as described in [12]. To determine NDPK activity, samples containing Nm23-H4 were incubated with [γ-32P]ATP (PerkinElmer Life Sciences, Boston, MA, U.S.A.) and unlabelled GDP in a total volume of 50 μl at 4 °C for 10 min. Reactions were stopped by the addition of 5% (w/v) trichloroacetic acid. Then 2% of each reaction mixture (1 μl) was separated on poly(ethyleneimine)–cellulose TLC plates using 0.75 M potassium phosphate (pH 3.4) as a solvent, air-dried and exposed to a film for autoradiography [11,13].
Miscellaneous
The procedure for isolation and purification of mitochondria has been described elsewhere [14]. Import of CoxIV-Nm23 into isolated mitochondria was performed in the presence of ATP (4 mM) and GTP (1 mM) [15]. Cellular iron uptake rate was determined in an 1 h assay as described in [8]. For measurement of mitochondrial iron levels, the strains transformed with pRS425 were grown in small cultures of standard defined media lacking leucine (100 ml volume) spiked with 100 nM radioactive iron 55Fe added as ferric citrate (100 Ci/g; CNL Scientific, San Francisco, CA, U.S.A.). These cultures were diluted to a uniform density of 0.1 D600 unit in larger 1 litre cultures (also containing 100 nM radioactive iron) and grown to a D600 of 2 units. Mitochondria were isolated, and radioactivity and protein levels were determined.
RESULTS AND DISCUSSION
The gene name YHM1 has been changed to GGC1 for GTP/GDP carrier. This change was made because of the demonstrated nucleotide exchange activity of the purified protein [9]. A yeast mutant lacking Ggc1p exhibits decreased levels of matrix GTP and increased levels of matrix GDP. On the other hand, this mutant also manifests tremendous iron accumulation within mitochondria [8]. These findings were quite surprising, considering that a role of GTP in mitochondrial iron metabolism has never been described. These reports therefore prompted us to investigate whether Ggc1p is a bifunctional protein involved in GTP/GDP transport and also in mitochondrial iron distribution, or whether the role of Ggc1p in iron metabolism is mediated by effects on GTP/GDP levels in the mitochondrial matrix.
In mammalian cells, GTP is synthesized in the mitochondrial matrix and cytosol. In the mammalian tricarboxylic acid cycle, succinyl-CoA ligase converts succinyl-CoA into succinate with the generation of GTP. In yeast, the enzyme produces ATP instead of GTP [16,17], and thus yeast mitochondria are dependent on cytosolic GTP supply. Our strategy was therefore to target a GTP-synthesizing enzyme to the mitochondrial matrix of the Δggc1 mutant in vivo and determine if iron-related phenotypic defects are corrected even in the absence of Ggc1p. For this purpose we chose a human GTP-synthesizing enzyme (an NDPK) that is normally present in human mitochondrial matrix, but the corresponding yeast homologue is absent from yeast mitochondrial matrix (see below).
NDPKs catalyse the transfer of a γ phosphate from NTPs (nucleoside triphosphates) to NDPs [18]. NDPK transiently phosphorylates itself on a conserved histidine residue at the catalytic site and then transfers the high-energy phosphate to an NDP [19]. The high-energy phosphate is usually supplied by ATP, and the enzyme regulates the crucial balance between ATP and GTP or other NTPs. In yeast, NDPK is encoded by a single nuclear gene YNK1 [20], and the corresponding protein (Ynk1p) and activity are present in the cytosol and mitochondrial intermembrane space [12]. Ynk1p mediates the final stage of a major route of GTP synthesis (ATP+GDP→ADP+GTP) [20,21] and may serve as a local GTP supplier [12,15].
Unlike the case for yeast, the human Nm23/NDPK family consists of eight related genes and widely expressed proteins termed Nm23-H1 to Nm23-H8. One of these isoforms, Nm23-H4, contains a cleavable mitochondrial targeting signal and is exclusively localized to the matrix [22]. We therefore chose Nm23-H4 for our experiments. This isoform, however, is so far less characterized compared with other isoforms such as Nm23-H1 and Nm23-H2.
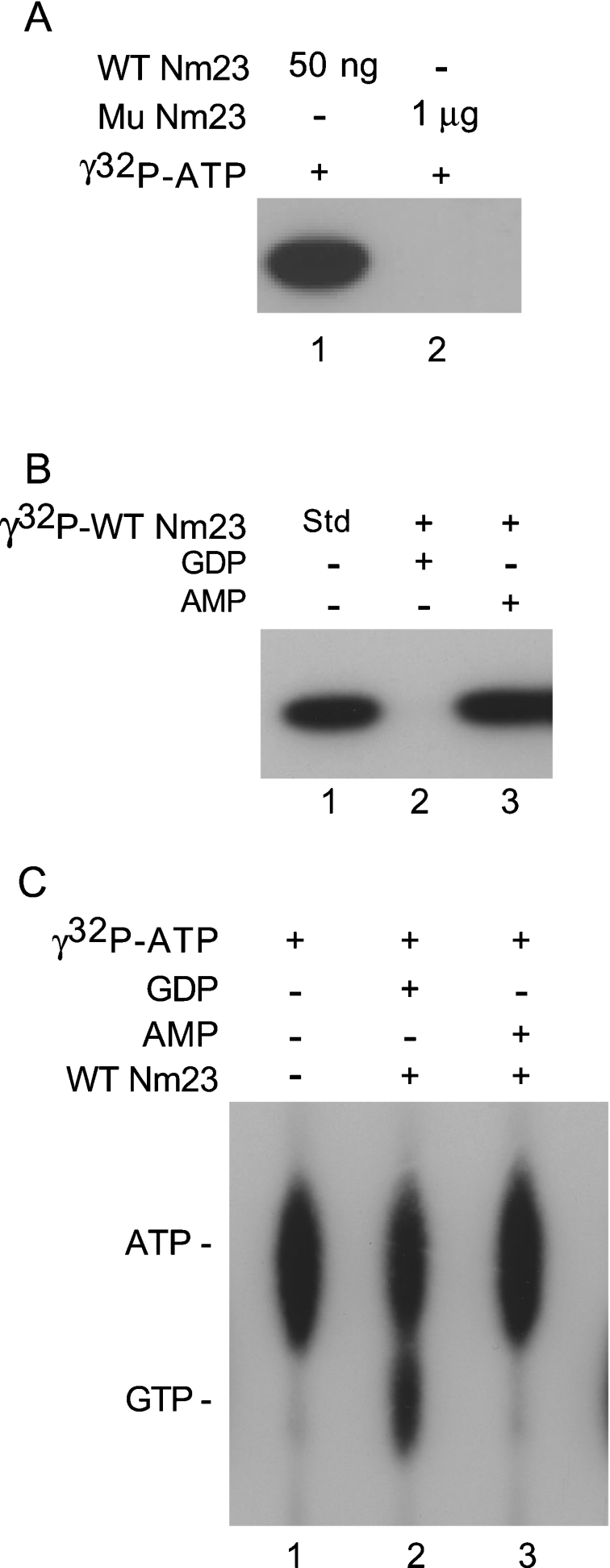
To characterize human Nm23-H4, we expressed the protein with a C-terminal His6 tag in bacteria and purified it to homogeneity (results not shown). When purified Nm23-H4 was supplemented with [γ-32P]ATP, it was able to autophosphorylate, as judged by SDS/PAGE and autoradiography (Figure 1A, lane 1). A mutant, Nm23 (H151G), in which the critical histidine residue at the predicted catalytic site of the enzyme was replaced by glycine, did not show any activity (lane 2). The high-energy radiolabelled phosphate from preformed autophosphorylated Nm23-H4 was specifically removed by NDPs (e.g. GDP), but not by nucleoside monophosphates such as AMP (Figure 1B).
Figure 1. Purified human Nm23-H4 can convert ATP into GTP.
(A) Bacterially expressed and purified wild-type Nm23-H4 (50 ng) or mutant Nm23-H4 (H151G) (1 μg) was incubated with [γ-32P]ATP (1 μCi; 3000 Ci/mmol) for 10 min on ice, and analysed by SDS/PAGE and autoradiography. (B) Wild-type Nm23-H4 (50 ng) was pre-incubated with [γ-32P]ATP (1 μCi) to form autophosphorylated enzyme intermediate. Samples were then supplemented with GDP (0.2 mM) or AMP (0.2 mM) and incubated for 10 min at 4 °C, and analysed by SDS/PAGE and autoradiography. (C) Wild-type Nm23-H4 (50 ng) was incubated with [γ-32P]ATP (1 μCi) plus unlabelled ATP (1 mM), in the absence or presence of unlabelled GDP (1 mM) or AMP (1 mM), for 10 min at 4 °C. Samples were analysed by TLC and autoradiography.
To demonstrate directly that the radiolabelled phosphate removed in the presence of NDPs was used to generate corresponding NTPs, Nm23-H4 was incubated with [γ-32P]ATP in the absence (Figure 1C, lane 1) or presence (lane 2) of unlabelled GDP. Samples were analysed by TLC followed by autoradiography [11]. Nm23-H4 was able to convert [γ-32P]ATP and unlabelled GDP into [γ-32P]GTP. Unlabelled AMP (lane 3) served as a negative control.
The precursor form of human Nm23-H4 has a cleavable 33-amino-acid-long N-terminal mitochondrial targeting sequence [22,23]. To achieve an efficient import into yeast mitochondria, we replaced the N-terminal 33 amino acids of Nm23-H4 with the cleavable targeting signal of an authentic yeast mitochondrial protein, cytochrome c oxidase subunit IV. The resulting chimaeric protein is called CoxIV-Nm23. When incubated with isolated yeast mitochondria, a significant portion of the 35S-labelled CoxIV-Nm23 precursor (Figure 2A, lane 1, ‘p’, no mitochondria) was found to be imported (lane 2, plus mitochondria), and the mature form (‘m’) thus generated in the matrix remained quantitatively protected from externally added trypsin (lane 3). Import efficiency (∼40%) was comparable with that observed with authentic yeast mitochondrial precursor proteins [15,24]. As control, no import was detected when the membrane potential across the inner membrane was dissipated by valinomycin (Val, lane 4).
Figure 2. In vitro and in vivo targeting of human Nm23-H4 to the yeast mitochondrial matrix.
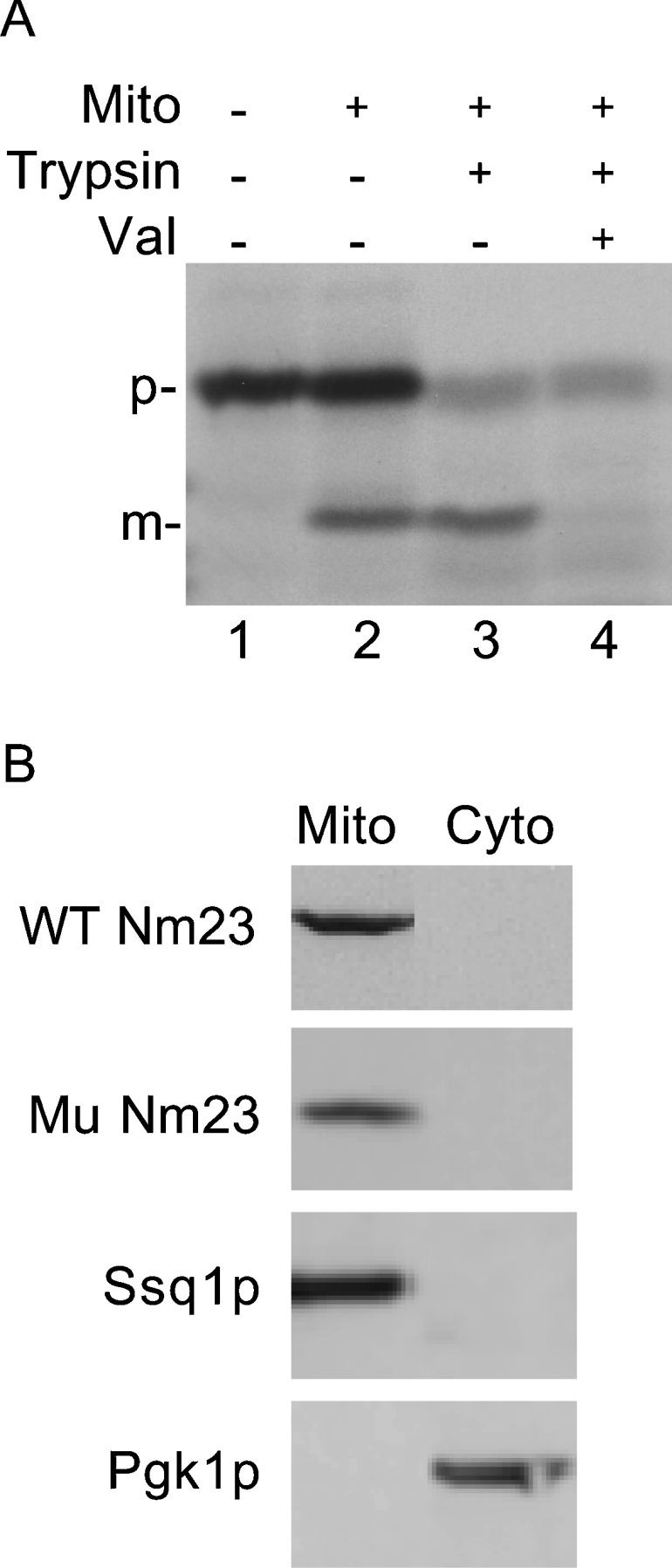
(A) 35S-labelled CoxIV-Nm23 was synthesized in reticulocyte lysate and incubated with isolated yeast mitochondria in the presence of ATP (4 mM) and GTP (1 mM) for 10 min at 30 °C. Valinomycin (5 μg/ml) was included as indicated (Val). Following import, samples were treated with trypsin (0.2 mg/ml) where indicated and analysed by SDS/PAGE and autoradiography. (B) Wild-type and mutant forms of Nm23-H4 with a C-terminal HA3 tag were expressed in yeast. Mitochondria and cytosolic fractions were analysed by anti-HA, anti-Ssq1p and anti-Pgk1p antibodies. Ssq1p and Pgk1p served as markers for the mitochondria and cytosol respectively.
For expression in yeast, we used two constructs: wild-type CoxIV-Nm23 (WT Nm23), and CoxIV-Nm23 with H151G mutation (Mu Nm23). These constructs with a C-terminal HA3 tag were expressed in Δggc1 using a shuffle strategy. As expected, both wild-type and mutant forms of Nm23-H4 were found in mitochondria and not in the cytosol (Figure 2B). The proteins were present inside the organelle, as they remained protected from external trypsin (results not shown).
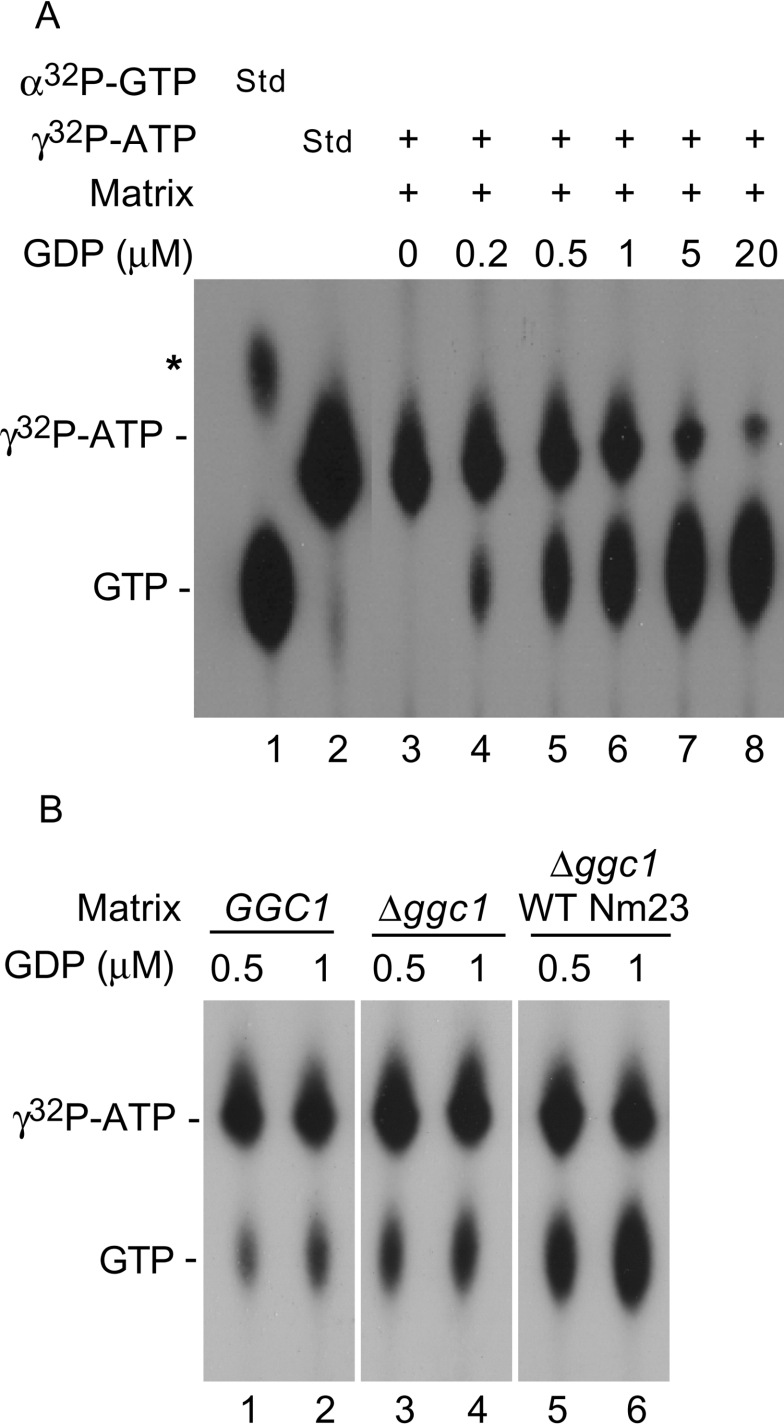
Mitochondrial matrix contains many ATPases and GTPases. To minimize/obviate the effect of these NTPases, we performed NDPK activity assays at 4 °C. Under these conditions, however, purified human Nm23-H4 is active (Figure 1). A soluble matrix fraction isolated from Δggc1 mitochondria with WT Nm23 (targeted in vivo) was incubated with [γ-32P]ATP and increasing concentrations of unlabelled GDP, and samples were analysed by TLC and autoradiography. The formation of [γ-32P]GTP was observed in a GDP dose-dependent manner, and the process was highly efficient (Figure 3A, lanes 3–8). In contrast, only a small amount of [γ-32P]GTP was formed in matrix fractions isolated from wild-type or Δggc1 mitochondria (Figure 3B), likely due to the contaminating NDPK activity of Ynk1p in the intermembrane space. Thus Nm23-H4 targeted in vivo to the matrix of Δggc1 mitochondria is active, and can convert ATP into GTP.
Figure 3. Human Nm23-H4, targeted in vivo to the yeast mitochondrial matrix, can convert ATP into GTP.
(A) A matrix fraction isolated from Δggc1 mitochondria with WT Nm23 targeted in vivo was incubated with [γ-32P]ATP and increasing concentrations of unlabelled GDP (lanes 3–8), and samples were analysed by TLC and autoradiography. Lanes 1 and 2 show migration pattern of [α-32P]GTP and [γ-32P]ATP respectively (Std). A minor spot indicated by an asterisk (*) in lane 1 represents contaminating [α-32P]GDP. (B) Matrix fractions isolated from GGC1, Δggc1 and Δggc1 mitochondria with WT Nm23 were incubated with [γ-32P]ATP and two different concentrations of unlabelled GDP as indicated, and samples were analysed by TLC and autoradiography. Data are from the same autoradiogram.
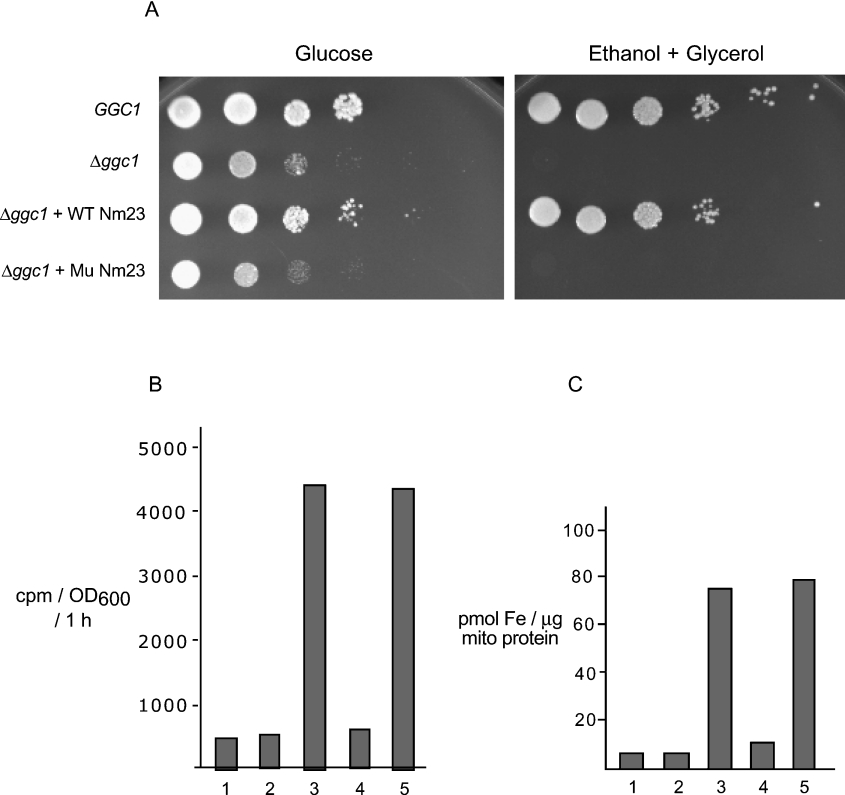
Cells lacking functional mtDNA (rho0), such as Δggc1, cannot grow on non-fermentable media, which requires respiration for energy production. Δggc1 cells expressing WT Nm23, but not Mu Nm23, grew on non-fermentable media and the growth was comparable with that of wild-type (GGC1) cells (Figure 4A, right panel). As controls, all strains grew on fermentable media (left panel). Thus the presence of active Nm23-H4 in the mito-chondrial matrix is critical for correcting the growth defect of Δggc1 cells; it bypasses the need for GTP to be supplied to the mitochondrial matrix from the cytosolic pool. Most likely, Nm23-H4 utilizes matrix ATP and GDP to restore matrix GTP levels in vivo, thereby complementing growth defects of the Δggc1 mutant.
Figure 4. Matrix-targeted wild-type Nm23-H4 complements Δggc1 growth on non-fermentable media and also restores normal iron regulation.
(A) Cells were grown to saturation in glucose media lacking leucine to maintain selection for pRS425-based plasmids. Cells were harvested and washed with water. Equal numbers of cells were diluted 10-fold sequentially and plated on fermentable (2% glucose) or non-fermentable (2% ethanol and 3% glycerol) minus leucine medium and grown at 30 °C for 2 days. (B) Cells were incubated with 1 μM 55Fe radionuclide for 1 h at 30 °C, and iron uptake was calculated from cell-associated radioactivity. (C) Cells were grown in 5 μM 55Fe radionuclide for 16 h at 30 °C, and iron content of isolated mitochondria was determined. In (B) and (C), lanes 1–5 correspond to GGC1, rho0, Δggc1, Δggc1+WT Nm23 and Δggc1+Mu Nm23 respectively.
Δggc1 cells show high cellular iron uptake and accumulate excess iron in mitochondria [8]. These phenotypes were almost completely corrected by the presence of wild-type Nm23-H4 in the mitochondrial matrix (Figures 4B and 4C, respectively). Mu Nm23 failed to complement abnormal iron regulation. A chemically induced rho0 strain did not exhibit abnormal iron regulation and served as control. These results suggest that the localization of active Nm23-H4 to the mitochondrial matrix is crucial for rescuing iron-related phenotypic defects in Δggc1.
In summary, the results presented here strongly suggest that GTP in the mitochondrial matrix plays an important role in organellar iron homoeostasis. We have shown that the role of the mitochondrial GTP/GDP carrier, Ggc1p, in iron metabolism is mediated by effects on GTP/GDP levels in the mitochondrial matrix. Lower mitochondrial matrix GTP is associated with mitochondrial iron accumulation. Targeting of human Nm23-H4 to the mitochondrial matrix of Δggc1 reverts this phenotype and returns mitochondrial iron levels to normal. The Nm23-H4 enzyme exhibits activity capable of converting ATP and GDP into GTP in the mitochondrial matrix, and it is this activity that is responsible for rescuing the iron phenotypic defects of the mutant. This also implies that there may exist a Ggc1p-independent pathway that delivers sufficient levels of GDP, but not GTP, to the matrix of Δggc1 mitochondria. GDP transport into mitochondria (in the absence of Ggc1p) could be mediated by one of the other mitochondrial carrier proteins. This large protein family includes exchangers of nucleotides, oxo acids and amino acids, which often lack perfect substrate specificity. In vitro transport studies have shown that several of these proteins are capable of transporting more than one molecule: the specific metabolite with high efficiency, and also relatively unrelated molecules with lower efficiency [7,25,26]. How GDP enters the matrix of Δggc1 mitochondria in vivo remains to be determined.
The compartmentalization of GTP synthesis is different in yeast and human cells, and a human homologue of the yeast GTP/GDP carrier Ggc1p has not been identified based on amino acid sequence homology. However, in mammalian cells, other members of the carrier family could carry out the transport of GTP or GDP. For example, isolated rat heart mitochondria have been reported to transport guanine nucleotides, and two distinct transport mechanisms have been identified [7,27,28]. The targeting of a human NDPK (Nm23-H4) to the mitochondrial matrix of Δggc1 rescues the iron phenotypic defects of the yeast mutant. This result may point to a role of GTP in mitochondrial iron metabolism in human mitochondria. Basic mitochondrial functions in yeast are often highly conserved with mammalian cells. These include mitochondrial protein import, mitochondrial dynamics, and mitochondrial iron and nucleotide metabolism [29–33]. Links between nucleotides and iron homoeostasis are also likely to be conserved, such that conclusions from yeast studies may be extrapolated to human physiology.
How might GTP in the mitochondrial matrix regulate iron metabolism? A possibility is that the effect is mediated via a GTPase. The major function of GTPases in the mitochondrial matrix is to participate in organellar protein synthesis. A speculative scenario is as follows. Starvation or other cellular stresses might convey a signal to mitochondria mediated by lowered GTP levels. The lowered mitochondrial GTP levels in turn might (via target matrix GTPases) trigger disassembly of mitochondrial ribosomes and arrest translation of mitochondria-encoded proteins. These GTPases might also simultaneously signal (directly or indirectly) a freeze of iron use and distribution leading to accumulation in mitochondria. Iron stored in mitochondria in response to this cellular stress signal would nonetheless be bioavailable and could be rapidly mobilized for use in the event that the stress signal is released. The iron pool in that case would be well situated to be diverted into synthesis of haem and Fe-S clusters within mitochondria. Studies to identify a matrix GTPase that participates in mitochondrial iron metabolism are currently in progress in our laboratory.
Acknowledgments
We thank Boominathan Amutha (Department of Pharmacology and Physiology, UMDNJ) for helpful discussions on Nm23-H4 activity assays. This work was supported by grants from the American Heart Association to D.P. (0655946T) and D.M.G. (0335473T), and National Institutes of Health (DK 43953) to A.D.
References
- 1.Knight S. A. B., Kim R., Pain D., Dancis A. The yeast connection to Friedreich ataxia. Am. J. Hum. Genet. 1999;64:365–371. doi: 10.1086/302270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Napier I., Ponka P., Richardson D. R. Iron trafficking in the mitochondrion: novel pathways revealed by disease. Blood. 2005;105:1867–1874. doi: 10.1182/blood-2004-10-3856. [DOI] [PubMed] [Google Scholar]
- 3.Rouault T. A., Tong W.-H. Iron–sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat. Rev. Mol. Cell Biol. 2005;6:345–351. doi: 10.1038/nrm1620. [DOI] [PubMed] [Google Scholar]
- 4.Kaplan J., Ward D. M., Crisp R. J., Philpott C. C. Iron-dependent metabolic remodeling in S. cerevisiae. Biochim. Biophys. Acta. 2006;1763:646–651. doi: 10.1016/j.bbamcr.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Lill R., Mühlenhoff U. Iron–sulfur-protein biogenesis in eukaryotes. Trends Biochem. Sci. 2005;30:133–141. doi: 10.1016/j.tibs.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Palmieri F. The mitochondrial transport family (SLC25): physiological and pathological implications. Pflugers Arch. Eur. J. Physiol. 2004;447:689–709. doi: 10.1007/s00424-003-1099-7. [DOI] [PubMed] [Google Scholar]
- 7.del Arco A., Satrústegui J. New mitochondrial carriers: an overview. Cell. Mol. Life Sci. 2005;62:2204–2227. doi: 10.1007/s00018-005-5197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesuisse E., Lyver E. R., Knight S. A. B., Dancis A. Role of YHM1, encoding a mitochondrial carrier protein, in iron distribution of yeast. Biochem. J. 2004;378:599–607. doi: 10.1042/BJ20031387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vozza A., Blanco E., Palmieri L., Palmieri F. Identification of the mitochondrial GTP/GDP transporter in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:20850–20857. doi: 10.1074/jbc.M313610200. [DOI] [PubMed] [Google Scholar]
- 10.Kao L. R., Megraw T. L., Chae C. B. SHM1: a multicopy suppressor of a temperature-sensitive null mutation in the HMG1-like abf2 gene. Yeast. 1996;12:1239–1250. doi: 10.1002/(sici)1097-0061(19960930)12:12<1239::aid-yea17>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 11.Gu Y., Gordon D. M., Amutha B., Pain D. A GTP:AMP phosphotransferase, Adk2p, in Saccharomyces cerevisiae: role of the C-terminus in protein folding/stabilization, thermal tolerance, and enzymatic activity. J. Biol. Chem. 2005;280:18604–18609. doi: 10.1074/jbc.M500847200. [DOI] [PubMed] [Google Scholar]
- 12.Amutha B., Pain D. Nucleoside diphosphate kinase of Saccharomyces cerevisiae, Ynk1p: localization to the mitochondrial intermembrane space. Biochem. J. 2003;370:805–815. doi: 10.1042/BJ20021415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhunia A. K., Han H., Snowden A., Chatterjee S. Lactosylceramide stimulates Ras-GTP loading, kinases (MEK, Raf), p44 mitogen-activated protein kinase and c-fos expression in human aortic smooth muscle cells. J. Biol. Chem. 1996;271:10660–10666. doi: 10.1074/jbc.271.18.10660. [DOI] [PubMed] [Google Scholar]
- 14.Murakami H., Pain D., Blobel G. 70-kD heat shock-related protein is one of at least two distinct cytosolic factors stimulating protein import into mitochondria. J. Cell Biol. 1988;107:2051–2057. doi: 10.1083/jcb.107.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sepuri N. B. V., Gordon D. M., Pain D. A GTP-dependent ‘push’ is generally required for efficient protein translocation across the mitochondrial inner membrane into the matrix. J. Biol. Chem. 1998;273:20941–20950. doi: 10.1074/jbc.273.33.20941. [DOI] [PubMed] [Google Scholar]
- 16.Przybyla-Zawaislak B., Gadde D. M., Ducharme K., McCammon M. T. Genetic and biochemical interactions involving tricarboxylic acid cycle (TCA) function using a collection of mutants defective in all TCA cycle genes. Genetics. 1999;152:153–166. doi: 10.1093/genetics/152.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Przybyla-Zawaislak B., Dennis R. A., Zakharkin S. O., McCammon M. T. Genes of succinyl-CoA ligase from Saccharomyces cerevisiae. Eur. J. Biochem. 1998;258:736–743. doi: 10.1046/j.1432-1327.1998.2580736.x. [DOI] [PubMed] [Google Scholar]
- 18.Parks R. E., Jr, Agarwal R. P. Nucleoside diphosphokinases. In: Boyer P. D., editor. The Enzymes, vol. 8. New York: Academic Press; 1973. pp. 307–333. [Google Scholar]
- 19.Lascu I., Gonin P. The catalytic mechanism of nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 2000;32:237–246. doi: 10.1023/a:1005532912212. [DOI] [PubMed] [Google Scholar]
- 20.Fukuchi T., Nikawa J., Kimura N., Watanabe K. Isolation, overexpression and disruption of a Saccharomyces cerevisiae YNK gene encoding nucleoside diphosphate kinase. Gene. 1993;129:141–146. doi: 10.1016/0378-1119(93)90710-k. [DOI] [PubMed] [Google Scholar]
- 21.Escobar-Henriques M., Daignan-Fornier B. Transcriptional regulation of the yeast GMP synthesis pathway by its end products. J. Biol. Chem. 2001;276:1523–1530. doi: 10.1074/jbc.M007926200. [DOI] [PubMed] [Google Scholar]
- 22.Lacombe M.-L., Milton A., Mehus M. J. G., Lambeth D. O. The human Nm23/nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 2000;32:247–258. doi: 10.1023/a:1005584929050. [DOI] [PubMed] [Google Scholar]
- 23.Milon L., Meyer P., Chiadmi M., Munier A., Johansson M., Karlsson A., Lascu I., Capeau J., Janin J., Lacombe M.-L. The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase. J. Biol. Chem. 2000;275:14264–14272. doi: 10.1074/jbc.275.19.14264. [DOI] [PubMed] [Google Scholar]
- 24.Sepuri N. B. V., Schülke N., Pain D. GTP hydrolysis is essential for protein import into the mitochondrial matrix. J. Biol. Chem. 1998;273:1420–1424. doi: 10.1074/jbc.273.3.1420. [DOI] [PubMed] [Google Scholar]
- 25.Todisco S., Agrimi G., Castegna A., Palmieri F. Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:1524–1531. doi: 10.1074/jbc.M510425200. [DOI] [PubMed] [Google Scholar]
- 26.Marobbio C. M. T., Di Noia M. A., Palmieri F. Identification of a mitochondrial transporter for pyrimidine nucleotides in Saccharomyces cerevisiae: bacterial expression, reconstitution and functional characterization. Biochem. J. 2006;393:441–446. doi: 10.1042/BJ20051284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKee E. E., Bentley A. T., Smith R. M., Jr, Ciaccio C. E. Origin of guanine nucleotides in isolated heart mitochondria. Biochem. Biophys. Res. Commun. 1999;257:466–472. doi: 10.1006/bbrc.1999.0489. [DOI] [PubMed] [Google Scholar]
- 28.McKee E. E., Bentley A. T., Smith R. M., Jr, Kraas J. R., Ciaccio C. E. Guanine nucleotide transport by atractyloside-sensitive and -insensitive carriers in isolated heart mitochondria. Am. J. Physiol. Cell Physiol. 2000;279:C1870–C1879. doi: 10.1152/ajpcell.2000.279.6.C1870. [DOI] [PubMed] [Google Scholar]
- 29.Foury F., Kucej M. Yeast mitochondrial biogenesis: a model system for humans? Curr. Opin. Chem. Biol. 2002;6:106–111. doi: 10.1016/s1367-5931(01)00276-9. [DOI] [PubMed] [Google Scholar]
- 30.Gordon D. M., Shi Q., Dancis A., Pain D. Maturation of frataxin within mammalian and yeast mitochondria: one step processing by matrix processing peptidase. Hum. Mol. Genet. 1999;8:2255–2262. doi: 10.1093/hmg/8.12.2255. [DOI] [PubMed] [Google Scholar]
- 31.Rassow J., Dekker P. J. T., Wilpe S. V., Meijer M., Soll J. The preprotein translocase of the mitochondrial inner membrane: function and evolution. J. Mol. Biol. 1999;286:105–120. doi: 10.1006/jmbi.1998.2455. [DOI] [PubMed] [Google Scholar]
- 32.Bauer M. F., Rothbauer U., Mühlenbein N., Smith R. J. H., Gerbitz K. D., Neupert W., Brunner M., Hofmann S. The mitochondrial TIM22 preprotein translocase is highly conserved throughout the eukaryotic kingdom. FEBS Lett. 1999;464:41–47. doi: 10.1016/s0014-5793(99)01665-8. [DOI] [PubMed] [Google Scholar]
- 33.Okamoto K., Shaw J. M. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]