Abstract
Corynebacterium glutamicum grew on resorcinol as a sole source of carbon and energy. By genome-wide data mining, two gene clusters, designated NCgl1110-NCgl1113 and NCgl2950-NCgl2953, were proposed to encode putative proteins involved in resorcinol catabolism. Deletion of the NCgl2950-NCgl2953 gene cluster did not result in any observable phenotype changes. Disruption and complementation of each gene at NCgl1110-NCgl1113, NCgl2951, and NCgl2952 indicated that these genes were involved in resorcinol degradation. Expression of NCgl1112, NCgl1113, and NCgl2951 in Escherichia coli revealed that NCgl1113 and NCgl2951 both coded for hydroxyquinol 1,2-dioxygenases and NCgl1112 coded for maleylacetate reductases. NCgl1111 encoded a putative monooxygenase, but this putative hydroxylase was very different from previously functionally identified hydroxylases. Cloning and expression of NCgl1111 in E. coli revealed that NCgl1111 encoded a resorcinol hydroxylase that needs NADPH as a cofactor. E. coli cells containing Ncgl1111 and Ncgl1113 sequentially converted resorcinol into maleylacetate. NCgl1110 and NCgl2950 both encoded putative TetR family repressors, but only NCgl1110 was transcribed and functional. NCgl2953 encoded a putative transporter, but disruption of this gene did not affect resorcinol degradation by C. glutamicum. The function of NCgl2953 remains unclear.
Various resorcinol compounds are produced in nature as secondary plant products (4). Early studies indicated that resorcinol was degraded via three different pathways in bacteria: In Azotobacter vinelandii, resorcinol was converted into pyrogallol, and subsequently the aromatic ring was cleaved by a pyrogallol 1,2-dioxygenase (9). Pseudomonas putida apparently adopted two different pathways: Resorcinol was converted into hydroxyquinol, and hydroxyquinol was subsequently degraded by (i) 2,3,5-trihydroxytoluene 1,2-dioxygenase (meta cleavage) (3) and (ii) hydroxyquinol 1,2-dioxygenase (ortho cleavage) (4). For all three degradative pathways, degradation of resorcinol was initiated by hydroxylation, although the hydroxylation happened at different positions of the aromatic ring: C-2 for A. vinelandii and C-4 or C-6 for P. putida. Two resorcinol hydroxylases from P. putida were purified, but they were not characterized at the genetic level (16, 17). Evidence supporting the conversion of resorcinol into pyrogallol by resorcinol-induced cells of A. vinelandii was obtained, but attempts to detect resorcinol 2-hydroxylase activity failed (9). To the best of our knowledge, no amino acid sequence of any resorcinol hydroxylase has been reported.
Not only is hydroxyquinol involved in resorcinol degradation; it is also the key intermediate during microbial degradation of a range of aromatic compounds, such as chlorophenol (15), 2,4,6-trichlorophenol (14), dibenzo-p-dioxin (1), 4-aminophenol (27), and 2-aminobenzoate (24). Consequently, the degradation of hydroxyquinol and its derivatives is of importance for the understanding of microbial processes that govern the metabolism of aromatic compounds and for the understanding of the geobiochemical cycling of aromatic compounds. Recently, advances have been made in the understanding of aromatic compound degradation by Corynebacterium glutamicum (7, 25, 26). Here we describe the genetic characterization of the resorcinol catabolic pathway in C. glutamicum.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown aerobically on a rotary shaker (150 rpm) at 37°C in Luria-Bertani (LB) broth or on an LB plate with 1.5% (wt/vol) agar. C. glutamicum was routinely grown at 30°C in LB or in mineral salts medium (MM) (31), which was adjusted to pH 8.4 and supplemented with 0.05 g of yeast extract liter −1 on a rotary shaker (150 rpm). Aromatic compounds were added at final concentrations of 2 mM (sterilized by filtration through 0.2-μm-pore-size filters) when they served as carbon and energy sources. Cellular growth was monitored by measuring the turbidity at 600 nm. For generation of mutants and maintenance of C. glutamicum, BHIS (brain heart broth with 0.5 M sorbitol) medium was used. When needed, antibiotics were used at the following concentrations: kanamycin, 50 μg ml−1 for E. coli and 25 μg ml−1 for C. glutamicum; chloramphenicol, 20 μg ml−1 for E. coli and 10 μg ml−1 for C. glutamicum; ampicillin, 100 μg ml−1 for E. coli.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristicsa | Source, reference, or note | ||
|---|---|---|---|---|
| Strains | ||||
| E. coli | ||||
| JM109 | recAl supE44 endAI hsdR17 gyrA96 relA1 thi Δ(lac-proAB) [F′ traD36 proAB lacIqlacZΔM15] | Stratagene (catalog no. 200235) | ||
| BL21(DE3) | hsdS gal (λcIts857 ind-l Sam7 nin-5 lacUV5-T7 gene 1) | Novagen (catalog no. 69387-3) | ||
| C. glutamicum | ||||
| RES167 | Restriction-deficient mutant of ATCC 13032; Δ(cglIM-cglIR-cglIIR) | University of Bielefeld | ||
| RES167ΔNCgl1110 | A fragment of DNA coding for amino acids 105 to 215 of NCgl1110 was deleted | This study | ||
| RES167ΔNCgl1111 | A fragment of DNA coding for amino acids 123 to 375 of NCgl1111 was deleted | This study | ||
| RES167ΔNCgl1112 | A fragment of DNA coding for amino acids 114 to 222 of NCgl1112 was deleted | This study | ||
| RES167ΔNCgl1113 | A fragment of DNA coding for amino acids 122 to 180 of NCgl1113 was deleted | This study | ||
| RES167ΔNCgl(2950-2953) | A fragment of DNA coding for amino acids 170 to 1288 of NCgl(2950-2953) was deleted | This study | ||
| RES167ΔNCgl(2950-2953)/ΔNCgl1112 | Fragments of DNA coding for amino acids 114 to 222 of NCgl1112 and 170 to 1288 of NCgl(2950-2953) were deleted | This study | ||
| RES167ΔNCgl(2950-2953)/ΔNCgl1113 | Fragments of DNA coding for amino acids 122 to 180 of NCgl1113 and 170 to 1288 of NCgl(2950-2953) were deleted | This study | ||
| Plasmids | ||||
| pK18mobsacB | Mobilizable vector, allows for selection of double crossover in C. glutamicum | Schäfer et al. (22) | ||
| pK18mobsacB-ΔNCgl1110 | Carrying NCgl1110 deletion (see RES167ΔNCgl1110) | This study | ||
| pK18mobsacB-ΔNCgl1111 | Carrying NCgl1111 deletion (see RES167ΔNCgl1111) | This study | ||
| pK18mobsacB-ΔNCgl1112 | Carrying NCgl1112 deletion (see RES167ΔNCgl1112) | This study | ||
| pK18mobsacB-ΔNCgl1113 | Carrying NCgl1113 deletion (see RES167ΔNCgl1113) | This study | ||
| pK18mobsacB-ΔNCgl(2950-2953) | Carrying NCgl(2950-2953) deletion [see RES167ΔNCgl(2950-2953)] | This study | ||
| pXMJ19 | Shuttle vector (CamrPtac lacIqpBL1 oriVC. glutamicumpK18 oriVE. coli) | Jakoby et al. (11) | ||
| pXMJ19-NCgl1110 | Carrying NCgl1110 (to generate complementation for NCgl1110) | This study | ||
| pXMJ19-NCgl1111 | Carrying NCgl1111 (to generate complementation for NCgl1111) | This study | ||
| pXMJ19-NCgl1112 | Carrying NCgl1112 (to generate complementation for NCgl1112) | This study | ||
| pXMJ19-NCgl1113 | Carrying NCgl1113 (to generate complementation for NCgl1113) | This study | ||
| pXMJ19-NCgl2951 | Carrying NCgl2951 (to generate complementation for NCgl2951) | This study | ||
| pXMJ19-NCgl2952 | Carrying NCgl2952 (to generate complementation for NCgl2952) | This study | ||
| pET28a | Expression vector with N-terminal hexahistidine affinity tag | Novagen | ||
| pET28a-NCgl1111 | pET28a derivative for expression of NCgl1111 | |||
| pET28a-NCgl1112 | pET28a derivative for expression of NCgl1112 | This work | ||
| pGEM-T Easy | Cloning of PCR products | Promega | ||
| Primers | ||||
| 1110F(a) | GATTCTAGAAAAGGAGGACACATATGCCCACGCCTTCG (XbaI) | To generate pK18mobsacB-ΔNCgl1110 and pXMJ19-NCgl1110 | ||
| 1110R | AGCAAGCTTGAGCTCCCTGAGTCTGGTGCTT (HindIII, SacI) | |||
| 1111F(a) | GATTCTAGAAAAGGAGGACCATATGTCACCCAATAACTTCG (XbaI, NdeI) | To generate pK18mobsacB-ΔNCgl1111, pXMJ19-NCgl1111, and pET28a-NCgl1111 | ||
| 1111R | AGCAAGCTTCCCGGGCCTCACCTGAACCGAAC (HindIII, SmaI) | |||
| 1112F(a) | GATTCTAGAAAAGGAGGACAACCATGTCTTTACAGTTCGAT (XbaI) | To construct pK18mobsacB-ΔNCgl1112 and pXMJ19-NCgl1112 | ||
| 1112R | ACAGAATTCACACGAGCGTTTATACC (HindIII, SacI) | |||
| 1112EF | GTCGCTAGCATGTCTTTACAGTTCGATC (NheI) | |||
| 1112ER | AGCAAGCTTGAATTCCGTAGTTCCACTCTTCTT (HindIII, SacI) | |||
| 1113a | GATTCTAGAAAAGGAGGACAACCATGACTATTTCAGCACAA (XbaI) | To construct pK18mobsacB-ΔNCgl1113, pXMJ19-NCgl1113, and RT-PCR | ||
| 1113b | GGTATTCGCCGTTTTCTGTCATCTCCGTTTTG | |||
| 1113c | GAAAACGGCGAATACC | |||
| 1113d | AGCAAGCTTGAGCTCTCTCAAATTGCCAAAA (HindIII, SacI) | |||
| 2950-2953a | GATTCTAGAAAATGCCCCTCGCTTGAAAA (XbaI) | To generate pK18mobsacB-ΔNCgl(2950-2953) | ||
| 2950-2953b | AGCAAGCTTGTCCGCACCCCTGTCCTAAA (HindIII) | |||
| 2950-2953c | TTTAGGACAGGGGTGCGGACAAGCTTATCCTGGTCCGTAGAACTGC (HindIII) | |||
| 2950-2953d | AGCGAATTCACAGGGCTAAATGCTCCAA (EcoRI) | |||
| 1110b | AATCCGCCCCAAATCC | To perform RT-PCR on the fragments of the related genes | ||
| 1111b | AATGATGGCGGAGTTTT | |||
| 1112b | AACCATTGAGACCAGA | |||
| 2950a | GATTCTAGAAAAGGAGGACACATATGAATCTGAAAGATCTC (XbaI, NdeI) | |||
| 2950b | TCATTGAAAGCCACTC | |||
| 2951a | GATTCTAGAAAAGGAGGACACATATGACAACCACCACCG (XbaI, NdeI) | |||
| 2951b | TTCCTTCAATCCACGC | |||
| 2952a | GATTCTAGAAAAGGAGGACACATATGAACAACTCACTCGCATT (XbaI, NdeI) | |||
| 2952b | GAGCACCTTCAGATCAACA | |||
Restriction enzyme sites are underlined. Ribosome binding sites are boldfaced. Nucleotides that are complementary to primer 2950-2953b are italicized.
Analysis of sequence data.
The genome sequence of C. glutamicum ATCC 13032 (accession no. NC 003450) was retrieved from GenBank. Sequence comparisons and database searches were carried out using BLAST programs at the BLAST server of the NCBI website.
DNA extraction and manipulation.
The total genomic DNA of C. glutamicum was isolated according to the procedure of Tauch et al. (28). Plasmid isolation, DNA manipulation, and agarose gel electrophoresis were carried out as described by Sambrook et al. (21). DNA restriction enzymes, ligase, and DNA polymerase were used as recommended by the manufacturer. Restricted DNA fragments were separated by agarose gel electrophoresis and were purified by using the agarose gel DNA fragment recovery kit (TIANGEN, Beijing, China).
Amplification of DNA fragments and construction of plasmids.
Target DNA fragments were PCR amplified by using Pyrobest DNA polymerase (TaKaRa) or Taq DNA polymerase (Promega, Madison, Wis.). PCR products were purified using an agarose gel DNA fragment recovery kit. PCR fragments were cloned using the pGEM-T Easy vector system (Promega, Madison, Wis.).
Various plasmids (Table 1) were constructed with pK18mobsacB (22), pXMJ19 (11), or pET28a. The primers used are listed in Table 1. Genes on plasmids were deleted either by gene splicing by overlap extension (10) [NCgl1113 and NCgl(2950-2953)] or by deletions of regions of each gene (NCgl1110, NCgl1111, or NCgl1112) through restriction enzyme digestion (see Fig. 1B). The disrupted genes were fused into pK18mobsacB to generate plasmids pK18mobsacBΔNCgl1110, pK18mobsacBΔNCgl1111, pK18mobsacBΔNCgl1112, pK18mobsacBΔNCgl1113, and pK18mobsacBΔNCgl(2950-2953). For complementation, plasmids pXMJ19-NCgl1110, pXMJ19-NCgl1111, pXMJ19-NCgl1112, pXMJ19-NCgl1113, pXMJ19-NCgl2951, and pXMJ19-NCgl2952 were created by insertion of each PCR-amplified intact gene into pXMJ19 (11). Plasmids for expression of the target genes in E. coli were constructed from each PCR-amplified gene and pET28a.
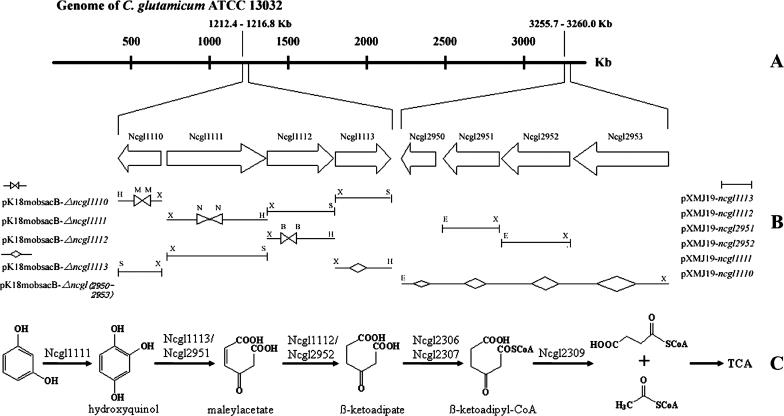
FIG. 1.
Physical map and genetic organization (A) of the resorcinol pathway (C) in C. glutamicum and construction of plasmids for gene disruption and complementation (B). Restriction sites are abbreviated as follows: X, XbaI; H, HindIII; E, EcoRI; S, SacI; B, BanI. Right and left triangles with points touching represent deletion by restriction enzyme digestion; wide diamonds represent deletion by gene splicing by overlap extension.
Genetic disruption and complementation in C. glutamicum.
The pK18mobsacB derivatives for gene disruption and the pXMJ19 derivatives for complementation described above were transformed into C. glutamicum RES167 and its mutants by electroporation (29). Screening for the first and second recombination events, as well as confirmation of the chromosomal deletion, was performed as described by Schäfer et al. (22). The resulting strains were designated C. glutamicum RES167ΔNCgl1110 to RES167ΔNCgl1113 and RES167ΔNCgl(2950-2953) (Table 1). The deletions of the target genes in pK18mobsacB derivatives and in C. glutamicum mutants were verified by PCR and DNA sequencing. The expression of each gene in C. glutamicum was induced by addition of 0.6 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to the culture broth.
Heterologous expression of genes in E. coli, preparation of cellular lysate, and enzymatic activity assays.
Plasmids (pET28a-NCgl1111 and pET28a-NCgl1112, described above) were electroporated into E. coli BL21(DE3). Synthesis of recombinant proteins was initiated by addition of 0.6 mM IPTG, and cultivation was continued for an additional 3 h. Cells were harvested by centrifugation at 10,000 × g, washed twice with ice-cold 50 mM Tris-HCl buffer (pH 8.0), resuspended in the same buffer, and disrupted by sonification in an ice-water bath. After centrifugation at 20,000 × g for 30 min at 4°C, the clear supernatant was used for enzymatic assays and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. All enzymatic assays were performed in 50 mM Tris-HCl buffer (pH 8.0). Resorcinol hydroxylase activity was measured spectrophotometrically by measuring the decrease in absorption at 340 nm due to oxidation of NADPH and was calculated with an extinction coefficient of 6,220 M−1 · cm−1 (17). Maleylacetate reductase activities were determined by monitoring NADH/NADPH oxidation as described by Schlömann et al. (23). Hydroxyquinol 1,2-dioxygenase activity was measured by determination of the increase in absorbance at 243 nm that resulted from the formation of maleylacetate, according to the method of Travkin et al. (30). An extinction coefficient of 4,440 M−1 · cm−1 was used for the calculation of enzymatic activity.
Conversion of resorcinol into maleylacetate via hydroxyquinol.
E. coli BL21(DE3)/pET28-NCgl1111 and E. coli BL21(DE3)/pET28-NCgl1113 were cultivated and induced as described above. Cells were harvested by centrifugation, washed twice with Tris-HCl (50 mM; pH 8.0), and suspended in the same buffer at a final optical density at 600 nm of 2.5. Resorcinol at a final concentration of 2 mM was added to the cell suspensions of E. coli BL21(DE3)/pET28-NCgl1111 and E. coli BL21(DE3)/pET28-NCgl1113, and the mixtures were incubated at 30°C. After 0.5, 1, 2, and 3 h, samples (500 μl each time) were collected, acidified to pH 2.0, extracted with ethyl acetate, and finally dried by N2 blowing. In parallel, cell suspensions of E. coli BL21(DE3)/pET28a and E. coli BL21(DE3)/pET28a-NCgl1113 were used for this conversion (as negative controls).
Operation of MS.
The extracted samples described above were subjected to mass spectrometry (MS) analysis. Mass spectrometry was performed on an electrospray ionization ion trap mass spectrometer (LCQ Deca XPplus; Thermo-Finnigan, San Jose, Calif.) operated in negative-ion mode. Typical parameters were as follows: temperature for ion transfer tube, 160°C; spray voltage, 3.3 kV. The samples were dissolved in methanol. For elucidation of the structure of the product, tandem MS (MS/MS) was performed with 35% normalized collision energy.
SDS-PAGE.
SDS-PAGE was conducted with a 5% stacking gel and a 12% resolving gel and was run in a Mini-PROTEIN II electrophoresis cell (Bio-Rad) according to the manufacturer's instructions. After electrophoresis, the protein bands were visualized by Coomassie brilliant blue staining. Protein concentrations were determined according to the method of Bradford (2), with bovine serum albumin as the standard.
Enzymatic preparation of maleylacetate.
Maleylacetate, the substrate for maleylacetate reductase, was freshly prepared by cleavage of 2 ml of 10 mM hydroxyquinol with hydroxyquinol 1,2-dioxygenase (10 μl) in 50 mM Tris-HCl buffer (pH 6.3) and was monitored by determination of UV absorption at 243 nm. After the conversion was completed (∼2 min), the pH of the buffer was adjusted to 8.0. This preparation was used as a substrate without further purification.
RT-PCR.
C. glutamicum was cultivated either with MM broth containing resorcinol or with LB broth. Total RNA was isolated with a TRNzol extraction kit (TIANGEN, Beijing, China). The RNA preparation was first treated with DNase I and then used as a template for reverse transcription with Moloney murine leukemia virus reverse transcriptase (TIANGEN, Beijing, China). The reverse transcription-PCR (RT-PCR) products (cDNAs) were used to amplify approximately 350- to 500-bp fragments of target genes. The primers used in RT-PCR are listed in Table 1. In order to exclude the possibility that residual DNA was amplified, negative controls were run in parallel, except that the Moloney murine leukemia virus reverse transcriptase was omitted from the reaction mixtures.
RESULTS AND DISCUSSION
Genome data-mining and bioinformatic analyses.
By use of the hydroxyquinol 1,2-dioxygenase of Arthrobacter chlorophenolicus (15) and BLAST searches, the NCg11113 and NCgl2951 genes, encoding putative hydroxyquinol 1,2-dioxygenases, were identified from the sequenced genome of C. glutamicum (Fig. 1). NCgl1113 and NCgl2951, encoding hydroxyquinol 1,2-dioxygenases, had been confirmed previously in recombinant E. coli (25), and their in vivo involvement in resorcinol assimilation was confirmed by gene disruption and complementation in this study (Fig. 2D). Analyses of their neighbor genes led to the discovery that two sets of genes at positions 1212.4 to 1216.8 (NCgl1110-NCgl1113) and 3255.7 to 3260.0 (NCgl2950-NCgl2953) were possibly involved in resorcinol degradation (Fig. 1).
FIG. 2.
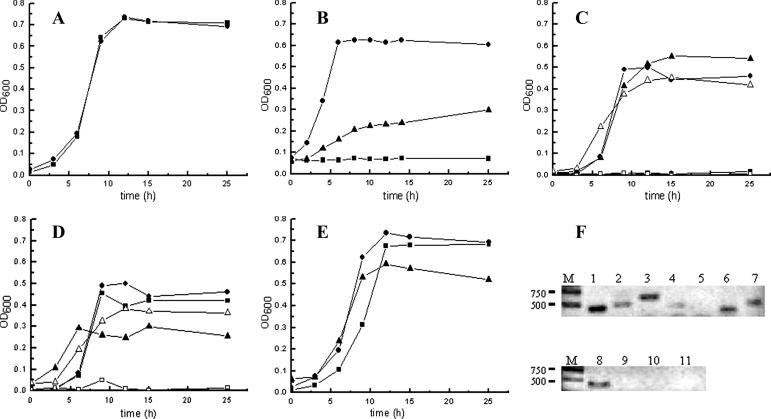
(A through E) Growth curves of various C. glutamicum mutants cultivated with resorcinol; (F) transcriptional analysis by RT-PCR of gene expression during growth in MM broth with resorcinol or in LB broth. According to genome data analysis, NCgl1110 and NCgl2950 encode putative TetR family regulators; NCgl1111 encodes a putative hydroxylase; NCgl1112 and NCgl2952 encode putative maleylacetate reductases; NCgl1113 and NCgl2951 encode putative hydroxyquinol 1,2-dioxygenases; and NCgl2953 encodes a putative transporter. Symbols: •, RES167; (A) ▪, RES167ΔNCgl(2950-2953); (B) ▪, RES167ΔNCgl1111; ▴, RES167ΔNCgl1111/pXMJ19-NCgl1111; (C) ▪, RES167ΔNCgl1112; □, RES167ΔNCgl(2950-2953)/ΔNCgl1112; ▴, RES167ΔNCgl(2950-2953)/ΔNCgl1112/pXMJ19-NCgl1112; ▵, RES167ΔNCgl(2950-2953)/ΔNCgl1112/pXMJ19-NCgl2952; (D) ▪, RES167ΔNCgl1113; □, RES167ΔNCgl(2950-2953)/ΔNCgl1113; ▴, RES167ΔNCgl(2950-2953)/ΔNCgl1113/pXMJ19-NCgl1113; ▵, RES167ΔNCgl(2950-2953)/ΔNCgl1113/pXMJ19-NCgl2951; (E) ▪, RES167ΔNCgl1110; ▴, RES167ΔNCgl1110/pXMJ19-NCgl1110. (F) Strain RES167 was grown in MM with resorcinol (lanes 1 to 7) or in LB broth (lanes 8 to 11). Total RNAs extracted were used as templates for RT-PCR to detect the transcription of each gene as follows: lane 1, NCgl1110 (420 bp); lane 2, NCgl1111 (464 bp); lane 3, NCgl1112 (550 bp); lane 4, NCgl1113 (410 bp); lane 5, NCgl2950 (no corresponding fragment); lane 6, NCgl2951 (430 bp); lane 7, NCgl2952 (464 bp); lane 8, NCgl1110; lane 9, NCgl1112; lane 10, NCgl2950; lane 11, NCgl2952.
The NCgl1111 gene encodes a resorcinol hydroxylase.
The mutant RES167ΔNCgl1111 lost the ability to grow on resorcinol (Fig. 2B), but its ability to grow on hydroxyquinol was the same as that of the wild-type strain (data not shown). A reasonable explanation for these phenotypes was that the NCgl1111 gene was involved in the conversion of resorcinol into hydroxyquinol through hydroxylation. When the mutant RES167ΔNCgl1111 was complemented with the NCgl1111 gene, the ability to grow on resorcinol was partially restored (Fig. 2B). RT-PCR showed that NCgl1111 was transcribed in wild-type RES167 when resorcinol was provided in the culture medium (Fig. 2F, lane 2).
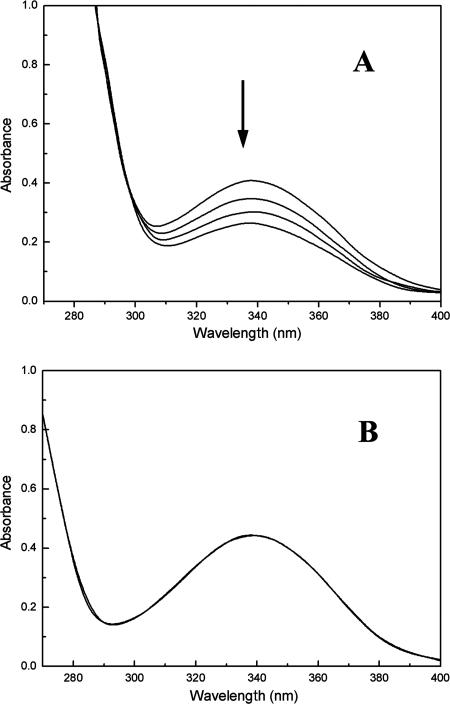
The translational product of the NCgl1111 gene showed only limited identity to the putative monooxygenase and hydroxylase (Table 2). In order to understand its biochemical properties, NCgl1111 was cloned into E. coli BL21(DE3)/pET28a. The recombinant E. coli cells actively synthesized a protein with a molecular mass of ca. 63 kDa. The cellular lysate of the recombinant E. coli cells showed resorcinol hydroxylase activity (Fig. 3), and this activity depended solely on the presence of NADPH. In addition to the enzyme activity, the cellular conversion of resorcinol also supported the hypothesis that NCgl1111 encodes a hydroxylase (Ncgl1111) that is involved in the conversion of resorcinol to hydroxyquinol. The recombinant cells of E. coli BL21(DE3)/pET28a-NCgl1111 converted resorcinol into hydroxyquinol. When Ncgl1111 was coupled with a hydroxyquinol 1,2-dioxygenase [Ncgl1113, encoded by NCgl1113 and expressed in E. coli BL21(DE3)/pET28a-NCgl1113] (25), resorcinol was converted to a product that was identified as maleylacetate by MS and MS/MS (data not shown).
TABLE 2.
Blast search results and identified functions of genes involved in resorcinol catabolism with Corynebacterium glutamicum
| Gene | Identified function of gene product | Sequence identitya | Related gene product (source, accession no.) |
|---|---|---|---|
| NCgl1110 | Transcriptional regulator | 70/218 (32) | Putative TetR family transcriptional regulator (Streptomyces avermitilis, BAC68276.1) |
| 75/232 (32) | Putative TetR family transcriptional regulator (Streptomyces avermitilis, BAC68295.1) | ||
| NCgl1111 | Resorcinol hydroxylase | 210/534 (39) | Putative FAD-dependent monooxygenase, geldanamycin biosynthesis gene cluster (Streptomyces hygroscopicus, AAO06914.1) (19) |
| 163/548 (29) | 3-(3-Hydroxyphenyl) propionate hydroxylases (E. coli, CAA70747.1) (8) | ||
| NCgl1112 | Maleylacetate reductase | 257/355 (72) | Putative maleylacetate reductase (Arthrobacter chlorophenolicus, AAO46999.1) (15) |
| 189/351 (53) | Maleylacetate reductase (Pimelobacter simplex, ABA61848.1) (41) | ||
| NCgl1113 | Hydroxyquinol 1,2-dioxygenase | 216/290 (74) | Hydroxyquinol 1,2-dioxygenase (Arthrobacter chlorophenolicus, AAN08758.1) (15) |
| 204/290 (70) | Hydroxyquinol 1,2-dioxygenase (Arthrobacter chlorophenolicus, AAN08760.1) (15) | ||
| NCgl2950 | Transcriptional regulator | 125/185 (67) | Putative transcription regulator (Corynebacterium efficiens, BAC19720.1) |
| 52/180 (28) | Putative TetR family transcriptional regulator (Streptomyces avermitilis, BAC72413.1) | ||
| NCgl2951 | Hydroxyquinol 1,2-dioxygenase | 220/294 (74) | Hydroxyquinol 1,2-dioxygenase (Arthrobacter chlorophenolicus, AAN08760.1) (15) |
| 209/299 (69) | Hydroxyquinol 1,2-dioxygenase (Arthrobacter chlorophenolicus, AAN08758.1) (15) | ||
| NCgl2952 | Maleylacetate reductase | 251/354 (70) | Putative maleylacetate reductase (Arthrobacter chlorophenolicus, AAN08761.1) |
| 197/351 (56) | Maleylacetate reductase (Pimelobacter simplex, ABA61848.1) (30) | ||
| NCgl2953 | Unknown | 268/480 (55) | Metabolic transport protein (Corynebacterium glutamicum, CAF18749.1) |
| 187/468 (39) | Putative sugar permease gene (Lactobacillus casei, AAF74348.1) |
Expressed as number of identical amino acids/number of amino acids in the related gene product (percent).
FIG. 3.
(A) Resorcinol hydroxylase activity in recombinant E. coli/pET28a-NCgl1111. (B) E. coli/pET28a was used as a negative control. The absorbances of the reaction mixture at wavelengths of 270 to 400 nm were recorded at time zero (top line) and at 60, 120, and 180 s (bottom line), Hydroxylase activity was also determined with 3-hydroxybenzoate, 3-aminophenol, 3,5-dihydroxytoluene, and 2,4-dihydroxybenzoate, but no activity was detected.
Ncgl1111 is the first resorcinol hydroxylase that has been characterized at the genetic level. Although a similar resorcinol hydroxylase (68 kDa) had been purified from P. putida previously, neither the DNA sequence nor the amino acid sequence of this resorcinol hydroxylase was disclosed (17). Attempts to find an Ncg11111 homolog in the P. putida genome failed, suggesting that the P. putida resorcinol hydroxylase might not be homologous to the C. glutamicum resorcinol hydroxylase. Moreover, the C. glutamicum Ncgl1111 did not show significant identities to previously identified 4-hydroxybenzoate 3-hydroxylases and 3-hydroxybenzoate 6-hydroxylases, which catalyze hydroxylation at the ortho and para positions, respectively. Since Ncgl111 is the first functionally identified hydroxylase that takes resorcinol as a substrate, we propose that it represents a new type of hydroxylase that is involved in aromatic compound catabolism.
Both the NCgl1112 gene and the NCgl2952 gene code for maleylacetate reductases.
Disruption of NCgl1112 resulted in the mutant RES167ΔNCgl1112, which was not able to grow on resorcinol (Fig. 2C). The mutant RES167Δ(NCgl2950-NCgl2953), in which the NCgl2952 gene was deleted, did not show disturbed assimilation of resorcinol. The double mutant RES167Δ(NCgl2950-NCgl2953)/ΔNCgl1112 was created and was used for complementation with NCgl1112 or NCgl2952. The results indicated that either NCgl1112 or NCgl2952 restored the ability to grow on resorcinol (Fig. 2C). Based on BLAST results showing high identities of Ncgl1112 and Ncgl2952 to maleylacetate reductases that had been functionally identified previously (Table 2), both NCgl1112 and NCgl2952 were assumed to code for maleylacetate reductases. When NCgl1112 was heterologously expressed in E. coli, the cellular lysate showed significant maleylacetate reductase activity. The results also indicated that the maleylacetate reductase preferred NADH (0.072 U/mg) to NADPH (0.047 U/mg) as a cofactor. It was also found that both NCgl1112 and NCgl2952 were transcribed in wild-type RES167 when resorcinol was provided in the culture broth (Fig. 2F, lanes 3 and 7).
NCgl1110, but not NCgl2950, codes for a negative transcriptional regulator.
According to the BLAST search results, both NCgl1110 and NCgl2950 encode putative TetR family transcriptional regulators. To determine whether the two genes were functional, NCgl1110 and NCgl2950 were individually disrupted. The resulting mutants, RESΔNCgl1110 and RESΔNCgl(2950-2953), were not phenotypically affected when resorcinol was provided as a source of carbon and energy. However, when these mutants were cultivated in LB broth, significant hydroxyquinol 1,2-dioxygenase activity (0.305 U/mg) was observed for the mutant RESΔNCgl1110 but not for the mutant RESΔNCgl(2950-2953) or wild type RES167. When the NCgl1110 gene was introduced (complementation) into the mutant RES167ΔNCgl1110, hydroxyquinol 1,2-dioxygenase activities disappeared. RT-PCR indicated that only NCgl1110 was transcribed (Fig. 2F, lane 1), and NCgl2950 was not transcribed (Fig. 2F, lane 5), when RES167 was cultivated with resorcinol. Similarly, when RES167 was cultivated in LB broth, only NCgl1110 was transcribed (Fig. 2F, lane 8). These results clearly demonstrate that Ncgl1110 functioned as a repressor and that its repression was neutralized by resorcinol induction. The function of Ncgl2950 was not clear.
The TetR family regulators are usually transcriptional repressors for the biosynthesis of antibiotics, efflux pumps, and osmotic stress (18). There are 15 TetR family members on the genome of C. glutamicum, and 3 were functionally identified (12, 13, 20). But none of the three functioned in aromatic degradation. Our results confirmed that Ncgl1110 was a transcriptional repressor of resorcinol degradation in C. glutamicum. Previously, a TetR family regulator, CymR, was determined to be involved in the regulation of p-cymene (cym) and p-cumate (cmt) degradation in P. putida (6). As far as we know, Ncgl1110 is the second example of a TetR family regulator that is involved in the regulation of aromatic compound catabolism.
The NCgl2953 gene is not necessary for resorcinol assimilation.
According to the results of sequence analysis and BLAST searches, the NCgl2953 gene encodes a protein that is possibly involved in membrane transportation of metabolic substrates or products, and we had deduced that it encoded a resorcinol or hydroxyquinol transporter. However, genetic disruption of the NCgl2953 gene or the whole NCgl2950-NCgl2953 genetic cluster did not affect aromatic compound assimilation. The mutants RES167ΔNCgl2953 and RES167ΔNCgl(2950-2953) grew as well as wild-type RES167 did, and this excluded the necessity of NCgl2953 for resorcinol assimilation. The function of this gene needs more investigation.
The two genetic clusters NCgl2950-NCgl2953 and NCgl1110-NCgl1113 function differently for resorcinol assimilation.
As shown above, all the genes at the NCgl1110-NCgl1113 genetic cluster were involved in resorcinol assimilation, but the genes at the NCgl2950-NCgl2953 genetic cluster were different: the mutant RES167ΔNCgl(2950-2953), in which in the NCgl2950-NCgl2953 genetic cluster was deleted, grew as well as the wild-type strain, RES167, when resorcinol was provided (Fig. 2A), indicating that this genetic cluster was not necessary for resorcinol assimilation. Interestingly, the RT-PCR results showed that NCgl1112 and NCgl1113 at the NCgl1110-NCgl1113 gene cluster, and NCgl2951 and NCgl2952 at the NCgl2950-NCgl2953 gene cluster, were all transcribed when C. glutamicum was cultivated with resorcinol. Apparently, the NCgl2951 and NCgl2952 genes, coding for hydroxyquinol 1,2-dioxygenase and maleylacetate reductase, respectively, were redundant to the NCgl1113 and NCgl1112 genes. Such catabolic redundancy has also been found for A. chlorophenolicus (15) and Pimelobacter simplex (30). One reasonable explanation for this redundancy is that some genes are expressed only under certain conditions: In Burkholderia xenovorans, there are three benzoate-catabolic pathways, and the benzoyl coenzyme A pathways are preferentially active under reduced oxygen tension (5).
Acknowledgments
We are grateful to Luying Xun at Washington State University for critical reading and constructive suggestions.
This work was supported by grants from the National Natural Science Foundation of China (30230010) and the Chinese Academy of Sciences (KSCX2-SW-113).
Footnotes
Published ahead of print on 8 September 2006.
REFERENCES
- 1.Armengaud, J., K. N. Timmis, and R.-M. Wittich. 1999. A functional 4-hydroxysalicylate/hydroxyquinol degradative pathway gene cluster is linked to the initial dibenzo-p-dioxin pathway genes in Sphingomonas sp. strain RW1. J. Bacteriol. 181:3452-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Chapman, P. J., and D. W. Ribbons. 1976. Metabolism of resorcinylic compounds by bacteria: orcinol pathway in Pseudomonas putida. J. Bacteriol. 125:974-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman, P. J., and D. W. Ribbons. 1976. Metabolism of resorcinylic compounds by bacteria: alternative pathways for resorcinol catabolism in Pseudomonas putida. J. Bacteriol. 125:985-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denef, V. J., J. A. Klappenbach, M. A. Patrauchan, C. Florizone, J. L. M. Rodrigues, T. V. Tsoi, W. Verstraete, L. D. Eltis, and J. M. Tiedje. 2006. Genetic and genomic insights into the role of benzoate-catabolic pathway redundancy in Burkholderia xenovorans LB400. Appl. Environ. Microbiol. 72:585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eaton, R. W. 1997. p-Cymene catabolic pathway in Pseudomonas putida T1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. J. Bacteriol. 179:3171-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng, J., Y. Che, J. Milse, Y.-J. Yin, L. Liu, C. Rückert, X.-H Shen, S.-W. Qi, J. Kalinowaski, and S.-J. Liu. 2006. The gene ncgl2918 encodes a novel maleylpyruvate isomerase that needs mycothiol as cofactor and links mycothiol synthesis and gentisate assimilation in Corynebacterium glutamicum. J. Biol. Chem. 281:10778-10785. [DOI] [PubMed] [Google Scholar]
- 8.Ferrandez, A., J. L. Garcia, and E. Diaz. 1997. Genetic characterization and expression in heterologous hosts of the 3-(3-hydroxyphenyl) propionate catabolic pathway in Escherichia coli K-12. J. Bacteriol. 179:2573-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groseclose, E. E., and D. W. Ribbons. 1981. Metabolism of resorcinylic compounds by bacteria: new pathway for resorcinol catabolism in Azotobacter vinelandii. J. Bacteriol. 146:460-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 11.Jakoby, M., C.-E. Ngouoto-Nkili, and A. Burkovski. 1999. Construction and application of new Corynebacterium glutamicum vectors. Biotechnol. Tech. 13:437-441. [Google Scholar]
- 12.Jakoby, M., L. Nolden, J. Meier-Wagner, R. Kramer, and A. Burkovski. 2000. AmtR, a global repressor in the nitrogen regulation system of Corynebacterium glutamicum. Mol. Microbiol. 37:964-977. [DOI] [PubMed] [Google Scholar]
- 13.Krug, A., V. F. Wendisch, and M. Bott. 2005. Identification of AcnR, a TetR-type repressor of the aconitase gene acn in Corynebacterium glutamicum. J. Biol. Chem. 280:585-595. [DOI] [PubMed] [Google Scholar]
- 14.Louie, T. M., C. M. Webster, and L. Xun. 2002. Genetic and biochemical characterization of a 2,4,6-trichlorophenol degradation pathway in Ralstonia eutropha JMP134. J. Bacteriol. 184:3492-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordin, K., M. Unell, and J. K. Jansson. 2005. Novel 4-chlorophenol degradation gene cluster and degradation route via hydroxyquinol in Arthrobacter chlorophenolicus A6. Appl. Environ. Microbiol. 71:6538-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohta, Y., I. J. Higgins, and D. W. Ribbons. 1975. Metabolism of resorcinylic compounds by bacteria: purification and properties of orcinol hydroxylase from Pseudomonas putida 01. J. Biol. Chem. 250:3814-3825. [PubMed] [Google Scholar]
- 17.Ohta, Y., and D. W. Ribbons. 1976. Bacterial metabolism of resorcinylic compounds: purification and properties of orcinol hydroxylase and resorcinol hydroxylase from Pseudomonas putida ORC. Eur. J. Biochem. 61:259-269. [DOI] [PubMed] [Google Scholar]
- 18.Ramos, J. L., M. Martínez-Bueno, A. J. Molina-Henares, W. Terán, K. Watanabe, X. D. Zhang, M. T. Gallegos, R. Brennan, and R. Tobes. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69:326-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rascher, A., Z. Hu, N. Viswanathan, A. Schirmer, R. Reid, W. C. Nierman, M. Lewis, and C. R. Hutchinson. 2003. Cloning and characterization of a gene cluster for geldanamycin production in Streptomyces hygroscopicus NRRL 3602. FEMS Microbiol. Lett. 218:223-230. [DOI] [PubMed] [Google Scholar]
- 20.Rey, D. A., A. Puhler, and J. Kalinowski. 2003. The putative transcriptional repressor McbR, a member of the TetR family, is involved in the regulation of the metabolic network directing the synthesis of sulfur containing amino acids in Corynebacterium glutamicum. J. Biotechnol. 103:51-65. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Schäfer, A., A. Tauch, W. Jager, J. Kalinowshi, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 23.Schlömann, M., E. Schmidt, and H.-J. Knackmuss. 1990. Different types of dienelactone hydroxylases in 4-fluorobenzoate-utilizing bacteria. J. Bacteriol. 172:5112-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schühle, K., M. Jahn, S. Ghisla, and G. Fuchs. 2001. Two similar gene clusters coding for enzymes of a new type of aerobic 2-aminobenzoate (anthranilate) metabolism in the bacterium Azoarcus evansii. J. Bacteriol. 183:5268-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen, X.-H., Y. Huang, and S.-J. Liu. 2005. Genomic analysis and identification of catabolic pathways for aromatic compounds in Corynebacterium glutamicum. Microbes Environ. 20:160-167. [Google Scholar]
- 26.Shen, X.-H., C.-Y. Jiang, Y. Huang, Z.-P. Liu, and S.-J. Liu. 2005. Functional identification of novel genes involved in the glutathione-independent gentisate pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 71:3442-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takenaka, S., S. Okugawa, M. Kadowaki, S. Murakami, and K. Aoki. 2003. The metabolic pathway of 4-aminophenol in Burkholderia sp. strain AK-5 differs from that of aniline and aniline with C-4 substituents. Appl. Environ. Microbiol. 69:5410-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tauch, A., F. Kassing, J. Kalinowski, and A. Pühler. 1995. The Corynebacterium xerosis composite transposon Tn5432 consists of two identical insertion sequences, designated IS1249, flanking the erythromycin resistance gene ermCX. Plasmid 34:119-131. [DOI] [PubMed] [Google Scholar]
- 29.Tauch, A., O. Kirchner, B. Loffler, S. Gotker, A. Pühler, and J. Kalinowski. 2002. Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the plasmid pGA1. Curr. Microbiol. 45:362-367. [DOI] [PubMed] [Google Scholar]
- 30.Travkin, V. M., A. P. Jadan, F. Briganti, A. Scozzafava, and L. A. Golovleva. 1997. Characterization of an intradiol dioxygenase involved in the biodegradation of the chlorophenoxy herbicides 2,4-D and 2,4,5-T. FEBS Lett. 407:69-72. [DOI] [PubMed] [Google Scholar]
- 31.Tsuji, H., T. Oka, M. Kimoto, Y. M. Hong, Y. Natori, and T. Ogawa. 1996. Cloning and sequencing of cDNA encoding 4-aminobenzoate hydroxylase from Agaricus bisporus. Biochim. Biophys. Acta 1309:31-36. [DOI] [PubMed] [Google Scholar]