Abstract
Regulatory genes hepK, hepN, henR, and hepS are required for heterocyst maturation in Anabaena sp. strain PCC 7120. They presumptively encode two histidine kinases, a response regulator, and a serine/threonine kinase, respectively. To identify relationships between those genes, we compared global patterns of gene expression, at 14 h after nitrogen step-down, in corresponding mutants and in the wild-type strain. Heterocyst envelopes of mutants affected in any of those genes lack a homogeneous, polysaccharide layer. Those of a henR mutant also lack a glycolipid layer. patA, which encodes a positive effector of heterocyst differentiation, was up-regulated in all mutants except the hepK mutant, suggesting that patA expression may be inhibited by products related to heterocyst development. hepS and hepK were up-regulated if mutated and so appear to be negatively autoregulated. HepS and HenR regulated a common set of genes and so appear to belong to one regulatory system. Some nontranscriptional mechanism may account for the observation that henR mutants lack, and hepS mutants possess, a glycolipid layer, even though both mutations down-regulated genes involved in formation of the glycolipid layer. HepK and HepN also affected transcription of a common set of genes and therefore appear to share a regulatory pathway. However, the transcript abundance of other genes differed very significantly from expression in the wild-type strain in either the hepK or hepN mutant while differing very little from wild-type expression in the other of those two mutants. Therefore, hepK and hepN appear to participate also in separate pathways.
Anabaena sp. strain PCC 7120 is a filamentous cyanobacterium. In response to nitrogen deprivation, about 10% of its cells differentiate into semiregularly spaced cells called heterocysts. Heterocysts are micro-oxic because they produce no oxygen (1, 60), respire rapidly (50), and have an envelope, composed of glycolipids and polysaccharides, that impedes the entry of oxygen (50, 61). Nitrogen fixation, an oxygen-sensitive process, takes place in the heterocysts and is thereby protected from environmental oxygen, including the oxygen produced by vegetative cells (18, 23, 67).
Relatively few genes are known to regulate heterocyst differentiation. HetR, the principal regulator of differentiation (8), is encoded by an autoregulatory gene (5, 34). In the absence of HetR no heterocysts form, whereas overexpression of hetR leads to the formation of multiple contiguous heterocysts and to differentiation in the presence of fixed nitrogen, whose presence normally inhibits differentiation (8). ntcA, which encodes a nitrogen-controlled transcription factor that is regulated by 2-oxoglutarate (39) and potentially by the signal transduction protein PII (21, 31, 40), is also autoregulatory. Its product, NtcA, regulates the transcription of hetR and of other genes that are involved in differentiation (21, 31, 39). PatS (69) and HetN (3, 6) inhibit heterocyst development, and overexpression of hetN inhibits heterocyst formation even in a strain that overexpresses hetR (10). A dimer of HetR binds upstream from hetR and from patS and regulates their expression (34). PatA, a presumptive response regulator that lacks a known DNA-binding domain (43), attenuates the inhibition by PatS and HetN by an unknown mechanism (53). The deletion of nrrA, a presumed two-component response regulator whose expression increases significantly within 3 h after removal of combined nitrogen, greatly delays the increased expression of ntcA, slows the formation of a pattern of semiregularly spaced heterocysts, and leads to a reduced frequency of heterocysts in a nitrogen-free medium (13). Current knowledge of the regulation of heterocyst differentiation and of the formation of patterns of spaced heterocysts has been reviewed (70, 71).
Cells that have visibly initiated differentiation are referred to as proheterocysts. Proheterocysts deposit a protective, enveloping layer of polysaccharide (Hep). Deposition depends upon a cluster of genes (33), hepA among them (32, 64, 74), whose expression is up-regulated 8 h after nitrogen step-down (14). Mutations in other genes, e.g., hepB (alr3698), alr3699, and all4160 (46, 64; Q. Fan et al., unpublished data), that presumptively encode glycosyl transferases (GTs) also result in a Hep− phenotype. Regulation of Hep deposition depends upon a two-component system comprising, at least, the products of hepK (all4496) and devR from Anabaena (devRA) (alr0442) (72, 74) and upon hepN (alr0117), hepS (all2760), and henR (alr1086), which presumptively encode a histidine kinase, a serine/threonine kinase, and a response regulator with limited similarity to Ser/Thr phosphatases of type PP2C, respectively (17, 51, 62).
An envelope layer of glycolipids (Hgl) is deposited between the cell wall and the Hep and limits the penetration of oxygen into the heterocyst (26, 50, 61, 63). A cluster of genes that are involved in Hgl biosynthesis and deposition has been identified by transposon mutagenesis (16); its expression is up-regulated 24 h after nitrogen deprivation (14). Genes involved in Hgl deposition and found elsewhere on the chromosome include devBCA and hglK (4, 19, 20). A transcriptional regulator, DevH (All3952), is required for Hgl synthesis; hglEA (alr5351) and the related gene hglE2 (all1646) were expressed at significantly lower levels in a devH mutant than in wild-type Anabaena sp. strain PCC 7120 (27, 56).
In both prokaryotes and eukaryotes, transcriptional regulation of particular genes in response to environmental signals is mediated by phosphotransfer cascades. Two-component systems comprising a histidine kinase as a sensor and transmitter and a response regulator as a receiver are common in bacteria (54), whereas serine/threonine or tyrosine phosphorylation is more common in eukaryotes (24). The genome of Anabaena sp. strain PCC 7120 presumptively encodes up to 131 histidine kinases, 80 response regulators, and 52 serine/threonine kinases (52, 62), most of whose mutant phenotypes and functions are unknown. Complex interactions between histidine kinases and response regulators in cyanobacteria are illustrated by experiments of Murata and coworkers, who systematically mutated nearly all of the histidine kinases and response regulators of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. They showed that a histidine kinase (Hik33) may respond to different growth conditions by phosphorylating different response regulators (Rre26 and Rre31) and that a response regulator (Rre1) may be phosphorylated by different kinases (Hik2 and Hik34) under different growth conditions and affect a different set of genes (49).
The term “expression island” was used by Ehira et al. (14) to describe physically clustered genes that have similar patterns of expression. Those authors recognized three major expression islands in Anabaena sp. strain PCC 7120: around Mb 1.70 on the chromosome, genes related to nitrogen fixation; around Mb 3.45, genes related to synthesis of heterocyst envelope polysaccharide; and around Mb 6.39, genes related to synthesis of heterocyst envelope glycolipids. Near Mb 3.72 on the chromosome is a cluster of open reading frames (ORFs), alr3057 to alr3074, that presumptively encode GTs and proteins involved in the biosynthesis and export of polysaccharides. The ORFs in this cluster are significantly up-regulated in wild-type Anabaena sp. strain PCC 7120 at 3 h after nitrogen step-down (13). We refer to these clusters as the Nif island, the Hep island, the Hgl island, and the Pol (for polysaccharide) island, respectively.
Using hepK, hepN, henR, and hepS regulatory mutants that are defective in the formation of Hep or of both Hep and Hgl, we have studied regulatory networks that lead to the formation of Hep and Hgl. Whereas a henR mutant has a Hep− Hgl− phenotype, hepK, hepS, and hepN mutants all have a Hep− Hgl+ phenotype (17, 51, 74). We have used oligonucleotide microarrays to compare the global gene expression in each of these mutants with that in wild-type Anabaena sp. strain PCC 7120 as of 14 h after nitrogen step-down.
MATERIALS AND METHODS
Strains, growth, and extraction of RNA.
Wild-type Anabaena sp. strain PCC 7120 and the transposon mutant derivatives FQ671 (hepN), FQ1487 (hepS), FQ1227 (henR; complemented by plasmid pRL3194, which also complemented henR mutant FQ621 [17]), and Y7 (hepK) (74) were used in this study. PCC 7120 was maintained in AA/8 liquid medium as described previously (33); wild-type cultures to be used as inocula for experiments were supplemented with nitrate. Mutant cultures were supplemented with nitrate plus 20 μg/ml neomycin sulfate. Protocols for growth conditions and RNA extractions were modified from unpublished protocols by R. Mella and J. W. Golden (Texas A&M University, College Station, TX). Inocula grown to an optical density at 750 nm (OD750) of 0.3 to 0.7 (Lambda 3B spectrophotometer; Perkin-Elmer, Wellesley, MA) and then diluted to an OD750 of 0.035 to 0.045 were grown in 400 ml AA/8 with nitrate but without antibiotic in 2.8-liter Pyrex Fernbach culture flasks with aeration and shaking. Cultures were washed three or four times with sterile distilled water 20 to 24 h after inoculation, when the OD750 of the culture reached 0.1 to 0.2. The washed pellet was resuspended in 400 ml of AA/8 medium without NO3− and incubated with aeration and shaking, as before, for 14 h. One-hundred-milliliter portions of cultures were sedimented in the presence of 100 g of crushed ice (48) in 500-ml flat-bottom centrifuge bottles in an SA rotor in a Sorvall RC-5B centrifuge (DuPont Instruments, Wilmington, DE) at 6,000 rpm for 20 min. Each pellet was transferred to a microcentrifuge tube and centrifuged at 13,000 × g for 5 min, and the supernatant solution was removed. Pellets were frozen in liquid nitrogen and stored at −80°C until RNA extraction. RNA was extracted with the RiboPure-Bacteria kit (Ambion, Austin, TX) according to the manufacturer's instructions with a minor modification: cell suspensions were vortexed with zirconia beads and RNAWiz (provided in the kit) for 30 min rather than only 10 min. The integrity of rRNA was monitored with a 2% agarose gel, and the absence of DNA was tested by PCR as described (16).
Microarray characteristics.
Oligonucleotide probes on the array were designed by Xeotron (subsequently purchased by Invitrogen, Carlsbad, CA) and represent 6,126 of the 6,129 annotated ORFs and 57 structural RNA genes. The probes (a list is available upon request) were ca. 45-mers with a melting point of about 65°C. A total of 95 ORFs and RNA genes, among them 79 transposase genes that did not have a unique probe, were probed with an oligonucleotide that appeared elsewhere on the array. Thus, 22 probes appeared between 2 and 12 times on the array. Each of 23 housekeeping and heterocyst-related genes was represented on the array by four or five different probes. Probes were synthesized on spots on the chip, in situ, by Xeotron as described previously (42). The array also included 20 spots that were designated “background” and 1,762 empty and production quality control spots.
RNA labeling, hybridization, and scanning.
We used 10 μg of RNA for each labeling reaction. cDNA samples were prepared with incorporation of amino-allyl dUTP to which a dye, Cy3 or Cy5, was then coupled (28). On each chip, wild-type Anabaena sp. strain PCC 7120 was compared with a mutant. For each mutant four biological replicates were used, and for two of the replicates, a dye swap was performed; thus, a total of six chips were used for each mutant. Hybridization was carried out in an M-2 microfluidic station (Xeotron) according to the instructions of the manufacturer. Hybridized chips were scanned with a 4000A laser scanner (Axon Instruments, Sunnyvale, CA) and the signal intensities were quantified using a GenePix Pro 5.0 (Axon Instruments). Raw intensity data have been deposited in the KEGG Expression database (http://www.genome.jp/kegg/expression/).
Data analysis.
The median foreground intensity in each spot was used for analysis, and the average intensity of 20 background-designated spots on the chip was considered background and subtracted from the intensity values. Spots in which more than 50% of the pixels were saturated and spots that had a signal intensity less than 1.5 times that of the background in at least one color channel were removed from the analysis. The spots that were removed were later checked manually to determine whether there was a significant difference between the signals of the two channels, suggesting differential expression. Empty spots and internal chip controls were also removed from the analysis. The signals from the remaining spots were normalized by comparison of the total signal intensity in the red channel to that in the green channel (13) and also by Global LOWESS normalization, performed using the “normalizeWithinArrays” function in the limma R package (59). Ratios for dye swap experiments for the same biological replicate were averaged. Log2-transformed ratios were used for identification of significantly differentially expressed genes by two different methods of statistical analysis. In the iterative outlier analysis method (7, 44), spots that had values 2.5 standard deviations distant from the mean for all spots were considered significantly differentially expressed; this procedure was repeated two additional times with omission of spots that had already been identified as differentially expressed. A second technique to identify differentially expressed genes was based on a regularized t test with a Bayesian statistical framework, using the Cyber-T web interface (http://visitor.ics.uci.edu/genex/cybert/index.shtml). We estimated the variability of the expression of each gene by averaging the signal intensity of the corresponding spot in each channel relative to the total intensity and then determined the Bayesian standard deviation for the gene using a sliding window of 101 genes. A Bayes confidence estimate value of 10 was used to determine P values. A posterior probability of differential expression [PPDE (<P)] threshold of 0.99 was chosen to identify differentially expressed genes (2, 35, 57). When the expression of a gene or ORF is described as up- or down-regulated, that means relative to the wild-type strain and, unless “without statistical support” is specifically stated, with a PPDE (<P) value of ≥0.99.
Only spots that were identified as differentially expressed in at least one mutant by both normalization methods and both methods of statistical analysis were used for cluster analysis. Cluster analysis was carried out using Hierarchical Clustering Explorer version 3.0 (58). Log2-transformed LOWESS-normalized ratios of significant spots were used for cluster analysis. Distance was estimated using the uncentered Pearson's correlation coefficient, and the dendrogram was constructed by a complete linkage (55, 58). The seven clusters illustrated and their subclusters are also supported by different clustering techniques: Euclidean distance and average linkage (55) (data not shown).
RESULTS AND DISCUSSION
General overview of differentially expressed genes.
Nitrogen deprivation elicits heterocyst formation. From 8 to 24 h of nitrogen deprivation, expression of Hep genes generally decreases, whereas expression of Hgl genes generally increases (13, 32, 38, 66). We assessed the effects of mutations in regulatory genes hepK, hepN, henR, and hepS by measuring global gene expression as of 14 h of nitrogen deprivation. Our working hypothesis was that if two or more of those four genes encode products that belong to the same regulatory phosphocascade, their mutants would show common effects on a group of regulated genes. The henR mutant, the only one that lacks the heterocyst envelope layer of glycolipid, was expected to affect an additional group of regulated genes related to glycolipid deposition. Additional, presumptively regulatory genes found to be differentially expressed in one or more of the mutants relative to the wild-type strain would be candidates for a downstream role in a possible developmental transcriptional cascade.
ORFs that were identified by both the iterative outlier and Cyber-T methods of statistical analysis (see Materials and Methods) as differentially expressed in at least one mutant were further analyzed. The 344 spots on the array that were identified as significantly differentially expressed represented 329 ORFs. Among those ORFs, 320 were probed with a single probe specific to that single ORF, 4 ORFs that encoded the transposase of copies of insertion sequence IS892 (9) were probed with a single oligonucleotide, and 5 genes were probed with more than one specific probe. These five genes were patA (all0521), probed with five oligonucleotides, all of whose spots showed up-regulation in all mutants but the hepK mutant; hepK, hepC (alr2834), and hepA (alr2835), each probed with five oligonucleotides, with only four of the five differentially expressed in at least one mutant; and fdxH (all1430), probed with four oligonucleotides, only three of which were down-regulated in henR and hepS mutants. Different probes for the same gene always clustered together in the cluster analysis that is described below, and they were considered jointly in the following discussion. Log2-transformed ratios, Bayes P values, and PPDE values (see Materials and Methods) for all 344 spots are presented in Table S1 in the supplemental material.
The distribution of genes into functional categories in Table 1 was based primarily on the Kazusa annotation (37) but was modified on the basis of current knowledge related to heterocyst differentiation and of gene ontology identification that was downloaded from BioBike (http://nostoc.stanford.edu:8002/biologin). The major functional groups shown in Table 1 included, as expected, genes that are involved in heterocyst differentiation (7% of the 329), most of which are known to function in formation of the heterocyst envelope; genes involved in nitrogen fixation (6%) that were down-regulated in all four mutants, although not always with statistical significance in the hepN mutant; genes possibly related to the cell envelope (5%; see “Genes possibly related to the cell envelope” below); regulatory genes (4%; see “Regulatory relationships” below); hypothetical and unknown proteins (44%); and transposase genes (4%; see “Cluster 7” below). Because nine transposase genes were probed with a common probe (see Materials and Methods), the number of those genes that were differentially expressed may be inaccurate. Of ORFs that had a saturated signal in one color channel and a relatively low signal in the other, we could confidently identify only alr3816 and alr3817 as differentially expressed (data not shown). These two ORFs encode two unknown proteins that share 57% amino acid similarity and are nitrogen responsive (13). Their role, if any, in heterocyst maturation is unknown.
TABLE 1.
Distribution of 329 ORFs that were differentially expressed in at least one mutant compared to in the wild-type strain as of 14 h of nitrogen deprivation, in terms of cluster analysis and functional categorya
| Functional category | No. of genes in cluster:
|
Total no. of genes | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| Biosynthesis of cofactors, prosthetic groups, and carriers | 3 | 1 | 1 | 5 | ||||
| Cell envelope | 14 | 1 | 1 | 1 | 1 | 18 | ||
| Energy metabolism | 6 | 1 | 7 | |||||
| Glycosyl transferases | 20 | 1 | 3 | 24 | ||||
| Heterocyst differentiation | 3 | 5 | 8 | 2 | 1 | 19 | ||
| Hypothetical and unknown | 81 | 9 | 5 | 7 | 34 | 2 | 10 | 148 |
| Nitrogen fixation and metabolism | 22 | 22 | ||||||
| Other | 23 | 14 | 1 | 38 | ||||
| Photosynthesis and respiration | 4 | 5 | 1 | 10 | ||||
| Regulatory functions | 6 | 1 | 5 | 1 | 13 | |||
| Transcription and translation | 2 | 1 | 1 | 4 | ||||
| Transport and binding proteins | 3 | 1 | 4 | 1 | 9 | |||
| Transposase | 2 | 2 | 8 | 12 | ||||
| Total | 189 | 18 | 13 | 9 | 70 | 7 | 23 | 329 |
Cluster analysis was carried out as described in Materials and Methods. The functional classification is based on that of the Kazusa DNA Research Institute (http://www.kazusa.or.jp/cyanobase/Anabaena/) with minor modifications as described in the text.
False positives and false negatives.
As described in Materials and Methods, various Anabaena sp. strain PCC 7120 genes were represented on the array by multiple probes. These genes include some that are known to play a role in the differentiation or function of heterocysts: devB, devC, devA, fdxH, hepA, hepC, hepK, hetP, hetC, hetR, and patA. One out of five probes for each of the genes devB, hetC, and hetP was up-regulated twofold or less in one mutant compared to in the wild-type strain, and this differential expression was found to be statistically significant. Nonetheless, because the other four probes for each of those genes did not show a statistically significant change in expression, those genes were considered false positives and were therefore removed from further analysis. Similarly, one probe for each of the genes fdxH, hepA, hepC, and hepK was identified as not significantly changed in any of the mutants by at least one combination of the techniques of normalization and statistical analysis (data not shown). The latter observations suggest that the filtering was sometimes too rigorous, so that not all differentially expressed genes may have been identified. For comparison, patA was probed with five different oligonucleotides, and all of them were found to be significantly up-regulated in at least one mutant, whereas 12 other genes that were tested with four or five probes were not found to be differentially expressed in any of the mutants (data not shown). Probes were not tested for performance before the array was produced, and it is possible that some of the oligonucleotides did not reliably estimate the original quantities of target RNA in the samples. The finding of false positives and false negatives using multiple probes suggests that false discovery occurred also among the ORFs that were probed with only a single oligonucleotide. However, no attempt was made to estimate the false discovery rate in the current study.
Cluster analysis.
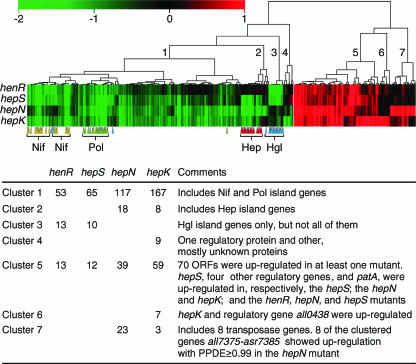
An overview of clustering methods for microarray experiments was presented by Quackenbush (55). Hierarchical clustering was used to cluster 344 spots that were identified as significantly differentially expressed in at least one mutant. We anticipated that coregulated genes with related functions might cluster together. So as to separate hep and hgl genes with a minimum number of clusters (Fig. 1 and Table 1; see Table S1 and Materials and Methods), we arbitrarily set the number of clusters at seven. Genes in clusters 1 to 4 were down-regulated in at least one mutant compared to in the wild-type strain, and genes in clusters 5 to 7 were up-regulated in at least one mutant. Other characteristics of the clusters are described in the tabular portion of Fig. 1. We next describe the clusters in greater detail, and we then consider other relationships between differentially expressed ORFs and mutants.
FIG. 1.
Cluster analysis and heat map of 344 spots differentially expressed in at least one mutant compared to in wild-type Anabaena sp. strain PCC 7120 at 14 h of nitrogen deprivation. Green and red represent down- and up-regulation, respectively, relative to the wild type. Values on the color scale are log2-transformed ratios. Clusters were generated using the program Hierarchical Clustering Explorer, distances were calculated using the uncentered Pearson's correlation coefficient, and the branching order was determined by complete linkage. Cluster numbers corresponding to Table 1 and the text are shown on branches. Colored triangles point to genes in the following expression islands: yellow, Nif; green, Pol; red, Hep, and blue, Hgl. The tabular portion summarizes major characteristics of each cluster and the number of ORFs differentially expressed in each mutant in each cluster.
Cluster 1.
Most of the genes in cluster 1 were down-regulated in hepK and hepN mutants (Fig. 1). This cluster includes genes in the Nif and Pol expression islands (see “Grouping of expression island genes by cluster analysis” below). Most ORFs in two additional groups of physically linked ORFs found in cluster 1, all4420 to alr4428 and all5221 to all5247, were down-regulated in hepK and hepN mutants and were up-regulated following nitrogen step-down of wild-type Anabaena sp. strain PCC 7120 (13), although not always with statistical support. A few ORFs in each of these two groups putatively encode GTs (see “Genes possibly related to the cell envelope” below).
Cluster 2.
The 18 genes in cluster 2 (Fig. 1 and Table 1) were all down-regulated in the hepN mutant; 8 were down-regulated in the hepK mutant. Of the 18, 12 are in the Hep island, 10 of which were down-regulated also in the hepS mutant, but without statistical support (see Table S1). The only two ORFs in cluster 2 that are outside of the Hep island and have products with a presumed, annotated functional identity are all2367, annotated as encoding a thioredoxin, and all2368, annotated as encoding translation initiation factor 3 and further discussed below. Both were down-regulated only in the hepN mutant. The 324 nucleotides that separate these two ORFs contain several stop codons in every possible reading frame and show strong secondary structure based on application of Mfold (75), Thus, the two ORFs are unlikely to be cotranscribed.
Cluster 3.
The 13 genes in cluster 3 all belong to the Hgl island and were down-regulated in the henR mutant and usually also in the hepS mutant (Fig. 1 and Table 1; see Table S1 and “Grouping of expression island genes by cluster analysis” below).
Cluster 4.
The nine genes in cluster 4 were significantly down-regulated only in the hepK mutant (Fig. 1 and Table 1). However, only the coclustering of the ORFs asl0550, alr3932, and all2883 was supported by other clustering methods (Euclidean distance and average linkage) (data not shown). ORFs asl0550 and alr3932 presumptively encode unknown proteins. Presumptive regulatory gene all2883 is discussed below.
Cluster 5.
Cluster 5 contains 70 ORFs that were up-regulated in at least one mutant (Fig. 1 and Table 1); most of the 70 were up-regulated in hepK and/or hepN mutants (see the tabular portion of Fig. 1). These genes include patA, which was up-regulated in all of the mutants except the hepK mutant, and hepS, which was up-regulated only in the hepS mutant. Presumptive regulatory genes all1072, alr1869, and asl2551 are not known to be involved in heterocyst differentiation (Table 2) (see “Regulatory relationships” below). Cluster 5 also includes physically linked genes all0916 through all0919, three of which are up-regulated in the wild-type strain at 8 h of nitrogen deprivation (13) and were up-regulated in all four tested mutants. all0916 and all0917 predict components of an ABC transporter, and all0918 and all0919 predict an unknown protein and a probable GT, respectively.
TABLE 2.
Regulatory genes and genes related to transcription and translation that are, at 14 h of nitrogen deprivation, differentially expressed in at least one mutant relative to in wild-type Anabaena sp. strain PCC 7120
| ORF | Gene | Description of product | Cluster | Regulation in mutant:
|
|||
|---|---|---|---|---|---|---|---|
| henR | hepS | hepN | hepK | ||||
| all0648 | Probable anti-sigma factor antagonist | 1 | Down | Down | |||
| all2164 | Two-component system, regulatory protein | 1 | Down | Down | |||
| all2165 | Two-component response regulator with PATAN domain | 1 | Down | Down | |||
| all3447 | Unknown protein with a DNA-binding HTHdomain | 1 | Down | Down | |||
| alr5251 | Two-component response regulator | 1 | Down | Down | |||
| alr1381 | prcA | Ca2+-dependent protease | 1 | (Down)a | Down | ||
| alr3170 | Hypothetical protein containing PAS, PAC, and GGDEF domains | 1 | Down | Down | Down | ||
| all4927 | Two-component response regulator | 1 | Down | ||||
| all2368 | Translation initiation factor 3 | 2 | Down | ||||
| all2883 | Two-component sensor histidine kinase | 4 | Down | ||||
| all0521 | patA | Two-component response regulator with PATAN domain | 5 | Up | Up | Up | |
| asl0125 | Transcriptional regulator | 5 | Up | Up | |||
| all1072 | Two-component response regulator with PATAN domain | 5 | Up | Up | |||
| all1173 | DNA-binding protein, starvation inducible | 5 | Up | Up | |||
| alr1869 | Serine/threonine kinase | 5 | Up | Up | |||
| asl2551 | Transcriptional regulator | 5 | (Up)b | Up | |||
| all2760c | hepS | Serine/threonine kinase | 5 | Up | |||
| all0438c | Serine/threonine kinase with WD-40 repeats | 6 | Up | ||||
| all4496d | hepK | Two-component system sensory histidine kinase | 6 | Up | |||
| all0844 | Peptidyl-tRNA hydrolase | 7 | Up | ||||
Down-regulated more than twofold, a ratio not statistically significant because of a large variation among experimental replicates. alr1381 is not a Fox gene (47).
Up-regulated 1.98-fold in the hepN mutant, with a PPDE (<P) value of 0.989.
Up-regulated at 8 h in wild-type Anabaena sp. strain PCC 7120 (13).
Up-regulated at 3 and 8 h in wild-type Anabaena sp. strain PCC 7120 (13).
Cluster 6.
The seven ORFs in cluster 6, one of which is hepK, were all up-regulated only in the hepK mutant (Fig. 1). (The sequence of one of the five hepK probes was based on a region of hepK downstream from the position of the transposon in the hepK mutant. Predictably, that probe did not show differential expression in the hepK mutant.) The other six ORFs are all0438, which predicts a serine/threonine kinase with WD-40 repeats (Table 2); ORFs that predict two unknown proteins and two transposases; and furA (all1691), whose product regulates the uptake of ferric ions (29, 30). At 3 and 8 h of nitrogen deprivation, gene furA was up-regulated also in wild-type Anabaena sp. strain PCC 7120 (13), specifically in proheterocysts and heterocysts (44a). Iron is important in heterocyst function because of its presence in nitrogenase; in ferredoxin, which conveys electrons to nitrogenase; in uptake hydrogenase, which recovers electrons from hydrogen produced by nitrogenase; and in photosystem I, which provides ATP for nitrogen fixation and can enhance the transfer of electrons, to nitrogenase, from metabolites in heterocysts. Why the expression of furA was up-regulated only in the hepK mutant is unclear.
Cluster 7.
Cluster 7 includes 23 ORFs, all of them up-regulated in the hepN mutant and 3 of them also up-regulated in the hepK mutant (Fig. 1 and Table 1). Eight ORFs in the region of all7375 through asr7385 on the α-plasmid, two encoding transposases (one from IS892), were up-regulated only in the hepN mutant. Three other ORFs in cluster 7 are copies of the same transposase gene from IS892. Of the 329 ORFs that responded differentially in at least one of the mutants, 7 others also predict transposase proteins.
Grouping of expression-island genes by cluster analysis.
Anabaena sp. strain PCC 7120 ORFs belonging to expression islands identified in response to nitrogen step-down by analysis of microarray experiments (13, 14) were clustered together in our experiments (Fig. 1). Most ORFs in the Pol island were grouped in cluster 1. Nearly all were down-regulated in all four mutants studied and up-regulated in the wild type as of 3 h after nitrogen deprivation (13). Two observations support the suggestion that they may participate in a stress response more general than one related just to heterocysts. (i) DNA segments containing part or all of ORFs alr3062 through alr3068 in the Pol island were down-regulated in wild-type Anabaena sp. strain PCC 7120 as of 2 h of temperature downshift (15). (ii) In our near-saturation transposon mutagenesis experiments (16, 17, 33, 41), the locus of transposition in only one of 1,076 Fox− transposon mutations (i.e., mutants unable to fix dinitrogen in the presence of oxygen) was localized to the Pol island, a frequency consistent with the frequency of false-positive mutants observed (Fan et al., unpublished observations).
Nif island genes, grouped in cluster 1, were down-regulated in henR, hepK, and hepS mutants, and their expression in the hepN mutant was variable (see Table S1). We consider it likely that down-regulation of nifHDK in the mutants may have resulted from oxygen that penetrated into the heterocyst due to defects in the heterocyst envelope (25).
Mutants with mutations in hepA, hepC, and 10 additional ORFs in the Hep island have a Hep− phenotype and are believed to be involved directly in Hep biosynthesis and deposition (32, 33, 64, 65). All of these genes were grouped in cluster 2, consistent with the idea that they are coregulated. We cannot confidently account for the extensive variation in expression of Hep island genes in the henR and hepS mutants compared to the wild type (see Table S1).
Hgl island genes comprise ORFs all5341 to alr5358. Mutants mutated in at least 8 of the 18 genes in this physical cluster have an Hgl− phenotype, at least 3 others are required for normal deposition of Hgls, and yet 3 others show enhanced transcription in response to nitrogen deprivation (3, 10, 13, 16; K. Awai and C. P. Wolk, unpublished data). Sixteen of the ORFs in that island were down-regulated in the henR mutant, and 13 of these were also down-regulated in the hepS mutant (Fig. 1; see Table S1). In henR and hepS mutants, the PPDE (<P) of hglD (alr5354) was just below the cutoff value [PPDE (<P) of ≥0.99] (see Materials and Methods; data not shown), considered differentially down-regulated. ORF alr5348, which encodes a putative protein with a Spo0J-like regulatory motif and is reported to have a Hep− Hgl+ mutant phenotype (16), was not differentially expressed in any of the tested mutants and so is probably transcribed independently from its downstream genes. ORFs related to biosynthesis of the Hgl aglycone and to Hgl deposition were found in cluster 3, except for three ORFs in cluster 1 (Fig. 1). Hgl island ORFs in cluster 1 are hglT (all5341), which encodes the GT that attaches a glucosyl residue to the Hgl aglycone (Awai and Wolk, unpublished data); its neighboring ORF, all5342, whose role is unknown; and hetN (alr5358), which is not required for Hgl synthesis (10). Although these ORFs, like the other Hgl island genes, were significantly down-regulated only in the henR and hepS mutants, their clustering based on Euclidian distance and average linkage also separated these three from the other Hgl genes (data not shown).
Genes possibly related to the cell envelope.
Twenty-six of the differentially expressed ORFs presumptively encode for GTs and 24 for other proteins probably related to cell envelope or other oligo- or polysaccharides. Nineteen of the 26 presumptive GT ORFs are part of the Hep and Pol expression islands and of the ORF groups all4420 through all4428 and all5221 through all5247. Because restructuring of the vegetative cell wall appears to be required for correct Hep formation (33, 41, 68, 73), it is unclear in which kind of cell, heterocysts or vegetative cells, differentially regulated genes related to envelope synthesis normally function.
In contrast to nine transpositions in Fox genes (i.e., genes required for a Fox+ phenotype) rfbP (all4829) and rfbZ (all4830), which are known to affect vegetative cell lipopolysaccharide (68), and 63 transpositions in genes in the Hep island (33), our screen for Fox genes identified no transpositions in the region of all4420 through all4428 and only three in the extensive region of all5221 through all5247. Two of these three were in a gene that proved to be a false positive (the other was not tested) (Fan et al., unpublished data). Therefore, we consider it likely that the latter two regions are not required specifically for heterocyst formation or function, although they may participate in the aforementioned restructuring of the vegetative cell wall. Of two orthologs of each of rfbB and rfbC in the Anabaena sp. strain PCC 7120 genome, only alr0657 in cluster 1 and alr2830 in cluster 2 in the Hep island are nitrogen responsive (13). It remains undetermined whether those two ORFs are required for heterocyst maturation.
Regulatory relationships. (i) General considerations.
We sought by our analysis to identify possible relationships between the effects of four regulatory genes that are required for heterocyst maturation. Our approach was to assess the expression of nearly every known and suspected Anabaena sp. strain PCC 7120 gene. We studied whole filaments, but heterocysts constitute only about 5 to 10% of the total number of cells; thus, a ca. 10- to 20-fold-greater abundance per cell of a heterocyst-specific mRNA would be required to give a hybridization signal equal in strength to that of an mRNA that is present in equal abundance in vegetative cells. As of 14 h of nitrogen deprivation, proheterocysts can be visualized and some genes are expressed cell type specifically (see, e.g., reference 66). Perhaps more important, microarray analyses assess only mRNA levels, and phenotype is not always predictable from transcript level. For example, although mRNA levels of hepA and hepC are extensively decreased in abp2 and abp3 mutants, the mutant strains are Hep+ and Hgl− (38). Moreover, measurements of mRNA levels, as in the seemingly constitutive expression of devR and devRA (12, 13), may obscure possible translational or posttranslational regulation of activity.
(ii) Do hepS and henR belong to one regulatory system?
Despite the common Hep− Hgl+ phenotype of hepK, hepN, and hepS mutants, hepK and hepN mutants share differential expression of numerous ORFs that are not differentially expressed by henR and hepS mutants. Therefore, it seems unlikely that hepK, hepN, and hepS are closely linked elements of a regulatory cascade. The sets of ORFs that are differentially expressed in the henR and hepS mutants are extensively similar. Of the 106 ORFs (out of the 329 total) that were differentially expressed in the henR and/or hepS mutants, 19 were not differentially expressed in the hepS mutant and 27 were not differentially expressed in the henR mutant; 60 were differentially expressed in both mutants. Examination of the 19 and 27 ORFs shows that of those not considered differentially expressed, only 5 had a PPDE of <0.91. With or without statistical support, all 106 ORFs were up- or down-regulated in parallel in both mutants. Because of the similarity of the effects of the hepS and henR mutations, it seems unlikely that henR and hepS acted independently despite their differing Hgl phenotypes.
The major group of ORFs that was down-regulated in henR and hepS mutants and not in hepN and hepK mutants was genes from the Hgl island (Fig. 1; see Table S1). It was, therefore, surprising to find that the hepS mutant had a Hgl+ phenotype. Some nontranscriptional mechanism may account for the observation that a henR mutation, but not a hepS mutation, led to greatly decreased synthesis of glycolipid. If, as appears likely, HepS and HenR form part of a phosphotransfer cascade that regulates transcription, at least a histidine kinase will presumably be required to complete the cascade.
devH, which encodes a DNA-binding protein, is up-regulated soon after nitrogen deprivation in wild-type Anabaena sp. strain PCC 7120 (13, 27). Both PCC 7120 orthologs of hglE (11), hglEA and hglE2, show significantly reduced expression in a devH mutant, whose mutant phenotype is Hep+ Hgl− (56). hepS and henR mutants showed down-regulation of hglEA and of other genes of the Hgl island but of neither hglE2 nor devH. Therefore, devH cannot function downstream of hepS and henR in a presumptive transcriptional cascade (because hglE2 should then be down-regulated in hepS and henR mutants), and if it functions upstream from them, it must independently regulate hglE2.
(iii) Genes hepK and hepN appear to act both jointly and independently.
HepK and HepN are both members of the histidine kinase family. The transfer of a phosphate group from HepK to DevRA was shown biochemically (72). ORFs in the genomic regions all4420 through all4428 and all5221 through all5247, as well as genes in the Hep island, are down-regulated in the hepK and hepN mutants (see “Genes possibly related to the cell envelope” above). In fact, half of the 300 ORFs that were differentially expressed in the hepK and/or hepN mutants were differentially expressed in both. Those 150 ORFs were up- or down-regulated in parallel in both mutants. We conjecture that, as in Synechocystis sp., in which two kinases and a response regulator were shown to regulate a common set of genes (49), HepK and HepN regulate the expression of the Hep island genes plus all4420 through all4428 and all5221 through all5247 and other ORFs via a common response regulator. Because HepK and HepN are required for developmental up-regulation of hepA (51, 74) (see Table S1) and they and DevRA, which interacts directly with HepK, are required for Hep biosynthesis (17, 51, 72, 74), DevRA is a likely candidate for that regulator.
One hundred three ORFs were differentially expressed in the hepK mutant and not in the hepN mutant, and 47 ORFs vice versa. The expression of a substantial fraction of the 103 and 47 ORFs was up- or down-regulated relative to expression in the wild type in one of those two mutants and differed very little from wild-type expression in the other (Fig. 1; see Table S1). We conjecture that, as in Synechocystis sp., in which one kinase regulates different sets of genes through interactions with different response regulators (49), HepK and HepN control other ORFs through, presumably, interactions with other response regulators that are specific to one kinase or the other.
Down-regulation of presumptive regulatory genes.
Some of the ORFs whose transcription is down-regulated in common by hepK and hepN mutants (Table 2) presumptively encode regulatory proteins: (i) a response regulator, Alr5251, with a GAF domain; (ii) a hypothetical protein, Alr3170, with domains that appear likely to be involved in signal transduction; (iii) an unknown protein, All3447, with a DNA-binding helix-turn-helix (HTH) domain; (iv) a protein, All0648, that is annotated as a probable anti-sigma factor antagonist; and (v) two response regulators, All2164 and All2165, the predicted products of adjacent genes (Table 2). All2165 contains, in addition to a CheY-like receiver domain, PATAN (PatA-like), α-clip, and HTH domains. It has been suggested that the latter three domains are involved in ligand binding, protein-protein interaction, and DNA binding, respectively (45). However, none of the ORFs just mentioned is necessarily involved specifically in heterocyst differentiation.
Presumptive regulatory ORFs all4927, which predicts a response regulator, and all2883, which predicts a hybrid two-component sensor histidine kinase with transmitter and receiver domains (52, 62), were down-regulated only in the hepK mutant. ORF all2883 was up-regulated in response to temperature down-shift of wild-type Anabaena sp. strain PCC 7120. It may, therefore, be involved primarily in regulation of adaptation to low temperature (15) and perhaps only indirectly in heterocyst differentiation.
The translation initiation factor 3 ortholog, all2368, was down-regulated ca. 22-fold only in the hepN mutant, a greater average change in signal ratio between the wild type and any of the mutants than shown by any other transcript, suggesting that HepN may have a role in regulation of protein translation. A translation initiation factor 3 ortholog is involved in differentiation of another bacterium, Myxococcus xanthus (36).
(iv) Up-regulation of presumptive regulatory genes.
Ten genes with presumptive roles in signal transduction, transcription, or translation were up-regulated in at least one mutant compared to in the wild type (Table 2). Of the 10, none was found to be down-regulated by nitrogen deprivation in the wild-type strain (13), whereas up-regulation of hepK, hepS, and ORF all0438 was reported to occur in the wild-type strain as of 8 h of nitrogen deprivation (13). Up-regulation of hepS and hepK in their corresponding mutants suggests that, like ntcA and hetR (see above), these genes are autoregulated, perhaps indirectly. We observed up-regulation of ORF all0438 only in the hepK mutant, suggesting that, directly or indirectly, HepK represses the expression of all0438. Similar statements may be made for other of the 10 ORFs. A patA mutant forms mostly terminal, and very few intercalary, heterocysts; its expression is slightly increased as of 3 to 6 h of nitrogen step-down (43). Up-regulation of patA in the henR, hepN, and hepS mutants suggests that its expression may be regulated by a product related to heterocyst differentiation.
ORF all1173, which presumptively encodes a DNA-binding protein and is up-regulated as of 24 h of nitrogen deprivation (14), was up-regulated in hepN and hepK mutants relative to in wild-type Anabaena sp. strain PCC 7120 at 14 h. The suggestion (14) that All1173 functions in nucleoid rearrangements to allow transcription of certain physical regions of the chromosome brings to mind Fogg's observation of much more uniform dispersal of DNA in heterocysts than in vegetative cells of Anabaena cylindrica (22).
Supplementary Material
Acknowledgments
We thank R. Mella and J. W. Golden (Texas A&M University) for sharing with us, prior to publication, details about culture densities appropriate for RNA extraction and about modification of the Ambion RiboPure protocol; R. Britton (Michigan State University) for assistance with identification of differentially expressed genes using the iterative outlier analysis method; J. Tiedje (Michigan State University) for allowing us to use his array scanner; and J. Landgraf (Research Technology Support Facility, Michigan State University) for assistance with probe labeling and array analysis.
This work was supported by U.S. National Science Foundation grant MCB-0090232 and U.S. Department of Energy grant DE-FG02-91ER20021.
Footnotes
Published ahead of print on 25 August 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Almon, H., and H. Böhme. 1980. Components and activity of the photosynthetic electron transport system of intact heterocysts isolated from the blue-green alga Nostoc muscorum. Biochim. Biophys. Acta 592:113-120. [DOI] [PubMed] [Google Scholar]
- 2.Baldi, P., and A. D. Long. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509-519. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, C. C., K. S. Ramaswamy, S. Endley, L. A. Scappino, J. W. Golden, and R. Haselkorn. 1997. Suppression of heterocyst differentiation in Anabaena PCC 7120 by a cosmid carrying wild-type genes encoding enzymes for fatty acid synthesis. FEMS Microbiol. Lett. 151:23-30. [DOI] [PubMed] [Google Scholar]
- 4.Black, K., W. J. Buikema, and R. Haselkorn. 1995. The hglK gene is required for localization of heterocyst-specific glycolipids in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 177:6440-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. [DOI] [PubMed] [Google Scholar]
- 6.Black, T. A., and C. P. Wolk. 1994. Analysis of a Het− mutation in Anabaena sp. strain PCC 7120 implicates a secondary metabolite in the regulation of heterocyst spacing. J. Bacteriol. 176:2282-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britton, R. A., P. Eichenberger, J. E. Gonzalez-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buikema, W. J., and R. Haselkorn. 1991. Characterization of a gene controlling heterocyst differentiation in the cyanobacterium Anabaena 7120. Genes Dev. 5:321-330. [DOI] [PubMed] [Google Scholar]
- 9.Cai, Y. 1991. Molecular genetic approaches towards the understanding of heterocyst differentiation and pattern formation in the cyanobacterium Anabaena sp. Thesis. Michigan State University, East Lansing.
- 10.Callahan, S. M., and W. J. Buikema. 2001. The role of HetN in maintenance of the heterocyst pattern in Anabaena sp. PCC 7120. Mol. Microbiol. 40:941-950. [DOI] [PubMed] [Google Scholar]
- 11.Campbell, E. L., M. F. Cohen, and J. C. Meeks. 1997. A polyketide-synthase-like gene is involved in the synthesis of heterocyst glycolipids in Nostoc punctiforme strain ATCC 29133. Arch. Microbiol. 167:251-258. [DOI] [PubMed] [Google Scholar]
- 12.Campbell, E. L., K. D. Hagen, M. F. Cohen, M. L. Summers, and J. C. Meeks. 1996. The devR gene product is characteristic of receivers of two-component regulatory systems and is essential for heterocyst development in the filamentous cyanobacterium Nostoc sp. strain ATCC 29133. J. Bacteriol. 178:2037-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehira, S., and M. Ohmori. 2006. NrrA, a novel nitrogen-responsive response regulator facilitates heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 59:1692-1703. [DOI] [PubMed] [Google Scholar]
- 14.Ehira, S., M. Ohmori, and N. Sato. 2003. Genome-wide expression analysis of the responses to nitrogen deprivation in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 10:97-113. [DOI] [PubMed] [Google Scholar]
- 15.Ehira, S., M. Ohmori, and N. Sato. 2005. Identification of low-temperature-regulated ORFs in the cyanobacterium Anabaena sp. strain PCC 7120: distinguishing the effects of low temperature from the effects of photosystem II excitation pressure. Plant Cell Physiol. 46:1237-1245. [DOI] [PubMed] [Google Scholar]
- 16.Fan, Q., G. Huang, S. Lechno-Yossef, C. P. Wolk, T. Kaneko, and S. Tabata. 2005. Clustered genes required for synthesis and deposition of envelope glycolipids in Anabaena sp. strain PCC 7120. Mol. Microbiol. 58:227-243. [DOI] [PubMed] [Google Scholar]
- 17.Fan, Q., S. Lechno-Yossef, S. Ehira, T. Kaneko, M. Ohmori, N. Sato, S. Tabata, and C. P. Wolk. 2006. Signal transduction genes required for heterocyst maturation in Anabaena sp. strain PCC 7120. J. Bacteriol. 188:6688-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fay, P. 1992. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol. Rev. 56:340-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiedler, G., M. Arnold, S. Hannus, and I. Maldener. 1998. The DevBCA exporter is essential for envelope formation in heterocysts of the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 27:1193-1202. [DOI] [PubMed] [Google Scholar]
- 20.Fiedler, G., M. Arnold, and I. Maldener. 1998. Sequence and mutational analysis of the devBCA gene cluster encoding a putative ABC transporter in the cyanobacterium Anabaena variabilis ATCC 29413. Biochim. Biophys. Acta 1375:140-143. [DOI] [PubMed] [Google Scholar]
- 21.Flores, E., and A. Herrero. 2005. Nitrogen assimilation and nitrogen control in cyanobacteria. Biochem. Soc. Trans. 33:164-167. [DOI] [PubMed] [Google Scholar]
- 22.Fogg, G. E. 1951. Growth and heterocyst production in Anabaena cylindrica Lemm. III. The cytology of heterocysts. Ann. Bot. N.S. 15:23-35. [Google Scholar]
- 23.Gallon, J. R. 1992. Reconciling the incompatible: N2 fixation and O2. New Phytol. 122:571-609. [Google Scholar]
- 24.Hanks, S. K., and T. Hunter. 1995. Protein kinases. 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9:576-596. [PubMed] [Google Scholar]
- 25.Haselkorn, R., D. Rice, S. E. Curtis, and S. J. Robinson. 1983. Organization and transcription of genes important in Anabaena heterocyst differentiation. Ann. Microbiol. (Paris). 134B:181-193. [DOI] [PubMed] [Google Scholar]
- 26.Haury, J. F., and C. P. Wolk. 1978. Classes of Anabaena variabilis mutants with oxygen-sensitive nitrogenase activity. J. Bacteriol. 136:688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hebbar, P. B., and S. E. Curtis. 2000. Characterization of devH, a gene encoding a putative DNA binding protein required for heterocyst function in Anabaena sp. strain PCC 7120. J. Bacteriol. 182:3572-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hegde, P., R. Qi, K. Abernathy, C. Gay, S. Dharap, R. Gaspard, J. E. Hughes, E. Snesrud, N. Lee, and J. Quackenbush. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29:548-562. [DOI] [PubMed] [Google Scholar]
- 29.Hernández, J. A., S. López-Gomollón, M. T. Bes, M. F. Fillat, and M. L. Peleato. 2004. Three fur homologues from Anabaena sp. PCC7120: exploring reciprocal protein-promoter recognition. FEMS Microbiol. Lett. 236:275-282. [DOI] [PubMed] [Google Scholar]
- 30.Hernández, J. A., A. M. Muro-Pastor, E. Flores, M. T. Bes, M. L. Peleato, and M. F. Fillat. 2006. Identification of a furA cis antisense RNA in the cyanobacterium Anabaena sp. PCC 7120. J. Mol. Biol. 355:325-334. [DOI] [PubMed] [Google Scholar]
- 31.Herrero, A., A. M. Muro-Pastor, A. Valladares, and E. Flores. 2004. Cellular differentiation and the NtcA transcription factor in filamentous cyanobacteria. FEMS Microbiol. Rev. 28:469-487. [DOI] [PubMed] [Google Scholar]
- 32.Holland, D., and C. P. Wolk. 1990. Identification and characterization of hetA, a gene that acts early in the process of morphological differentiation of heterocysts. J. Bacteriol. 172:3131-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang, G., Q. Fan, S. Lechno-Yossef, E. Wojciuch, C. P. Wolk, T. Kaneko, and S. Tabata. 2005. Clustered genes required for the synthesis of heterocyst envelope polysaccharide in Anabaena sp. strain PCC 7120. J. Bacteriol. 187:1114-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang, X., Y. Dong, and J. Zhao. 2004. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc. Natl. Acad. Sci. USA 101:4848-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hung, S., P. Baldi, and G. W. Hatfield. 2002. Global gene expression profiling in Escherichia coli K12. The effects of leucine-responsive regulatory protein. J. Biol. Chem. 277:40309-40323. [DOI] [PubMed] [Google Scholar]
- 36.Kalman, L. V., Y. L. Cheng, and D. Kaiser. 1994. The Myxococcus xanthus dsg gene product performs functions of translation initiation factor IF3 in vivo. J. Bacteriol. 176:1434-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205-213. [DOI] [PubMed] [Google Scholar]
- 38.Koksharova, O. A., and C. P. Wolk. 2002. Novel DNA-binding proteins in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 184:3931-3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laurent, S., H. Chen, S. Bédu, F. Ziarelli, L. Peng, and C.-C. Zhang. 2005. Nonmetabolizable analogue of 2-oxoglutarate elicits heterocyst differentiation under repressive conditions in Anabaena sp. PCC 7120. Proc. Natl. Acad. Sci. USA 102:9907-9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laurent, S., K. Forchhammer, L. Gonzalez, T. Heulin, C.-C. Zhang, and S. Bédu. 2004. Cell-type specific modification of PII is involved in the regulation of nitrogen metabolism in the cyanobacterium Anabaena PCC 7120. FEBS Lett. 576:261-265. [DOI] [PubMed] [Google Scholar]
- 41.Leganés, F., A. Blanco-Rivero, F. Fernández-Piñas, M. Redondo, E. Fernández-Valiente, Q. Fan, S. Lechno-Yossef, and C. P. Wolk. 2005. Wide variation in the cyanobacterial complement of presumptive penicillin-binding proteins. Arch. Microbiol. 184:234-248. [DOI] [PubMed] [Google Scholar]
- 42.LeProust, E., H. Zhang, P. Yu, X. Zhou, and X. Gao. 2001. Characterization of oligodeoxyribonucleotide synthesis on glass plates. Nucleic Acids Res. 29:2171-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang, J., L. Scappino, and R. Haselkorn. 1992. The patA gene product, which contains a region similar to CheY of Escherichia coli, controls heterocyst pattern formation in the cyanobacterium Anabaena 7120. Proc. Natl. Acad. Sci. USA 89:5655-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loos, A., C. Glanemann, L. B. Willis, X. M. O'Brien, P. A. Lessard, R. Gerstmeir, S. Guillouet, and A. J. Sinskey. 2001. Development and validation of Corynebacterium DNA microarrays. Appl. Environ. Microbiol. 67:2310-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.López-Gomollón, S., J. A. Hernández, C. P. Wolk, M. L. Peleato, and M. F. Fillat. Expression of furA is modulated by NtcA and strongly enhanced in heterocysts of Anabaena sp. PCC 7120. Microbiology, in press. [DOI] [PubMed]
- 45.Makarova, K. S., E. V. Koonin, R. Haselkorn, and M. Y. Galperin. 2006. Cyanobacterial response regulator PatA contains a conserved N-terminal domain (PATAN) with an alpha-helical insertion. Bioinformatics 22:1297-1301. [DOI] [PubMed] [Google Scholar]
- 46.Maldener, I., S. Hannus, and M. Kammerer. 2003. Description of five mutants of the cyanobacterium Anabaena sp. strain PCC 7120 affected in heterocyst differentiation and identification of the transposon-tagged genes. FEMS Microbiol. Lett. 224:205-213. [DOI] [PubMed] [Google Scholar]
- 47.Maldener, I., W. Lockau, Y. P. Cai, and C. P. Wolk. 1991. Calcium-dependent protease of the cyanobacterium Anabaena: molecular cloning and expression of the gene in Escherichia coli, sequencing and site-directed mutagenesis. Mol. Gen. Genet. 225:113-120. [DOI] [PubMed] [Google Scholar]
- 48.Mohamed, A., and C. Jansson. 1989. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol. Biol. 13:693-700. [DOI] [PubMed] [Google Scholar]
- 49.Murata, N., and I. Suzuki. 2006. Exploitation of genomic sequences in a systematic analysis to access how cyanobacteria sense environmental stress. J. Exp. Bot. 57:235-247. [DOI] [PubMed] [Google Scholar]
- 50.Murry, M. A., and C. P. Wolk. 1989. Evidence that the barrier to the penetration of oxygen into heterocysts depends upon two layers of the cell envelope. Arch. Microbiol. 151:469-474. [Google Scholar]
- 51.Ning, D., and X. Xu. 2004. alr0117, a two-component histidine kinase gene, is involved in heterocyst development in Anabaena sp. PCC 7120. Microbiology 150:447-453. [DOI] [PubMed] [Google Scholar]
- 52.Ohmori, M., M. Ikeuchi, N. Sato, P. Wolk, T. Kaneko, T. Ogawa, M. Kanehisa, S. Goto, S. Kawashima, S. Okamoto, H. Yoshimura, H. Katoh, T. Fujisawa, S. Ehira, A. Kamei, S. Yoshihara, R. Narikawa, and S. Tabata. 2001. Characterization of genes encoding multi-domain proteins in the genome of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:271-284. [DOI] [PubMed] [Google Scholar]
- 53.Orozco, C. C., D. D. Risser, and S. M. Callahan. 2006. Epistasis analysis of four genes from Anabaena sp. strain PCC 7120 suggests a connection between PatA and PatS in heterocyst pattern formation. J. Bacteriol. 188:1808-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parkinson, J. S. 1993. Signal transduction schemes of bacteria. Cell 73:857-871. [DOI] [PubMed] [Google Scholar]
- 55.Quackenbush, J. 2001. Computational analysis of microarray data. Nat. Rev. Genet. 2:418-427. [DOI] [PubMed] [Google Scholar]
- 56.Ramírez, M. E., P. B. Hebbar, R. Zhou, C. P. Wolk, and S. E. Curtis. 2005. Anabaena sp. strain PCC 7120 gene devH is required for synthesis of the heterocyst glycolipid layer. J. Bacteriol. 187:2326-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salmon, K., S. P. Hung, K. Mekjian, P. Baldi, G. W. Hatfield, and R. P. Gunsalus. 2003. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J. Biol. Chem. 278:29837-29855. [DOI] [PubMed] [Google Scholar]
- 58.Seo, J., H. Gordish-Dressman, and E. P. Hoffman. 2006. An interactive power analysis tool for microarray hypothesis testing and generation. Bioinformatics 22:808-814. [DOI] [PubMed] [Google Scholar]
- 59.Smyth, G. K., and T. Speed. 2003. Normalization of cDNA microarray data. Methods Enzymol. 31:265-273. [DOI] [PubMed] [Google Scholar]
- 60.Tel-Or, E., and W. D. P. Stewart. 1977. Photosynthetic components and activities of nitrogen-fixing isolated heterocysts of Anabaena cylindrica. Proc. R. Soc. London B 198:61-86. [Google Scholar]
- 61.Walsby, A. E. 1985. The permeability of heterocysts to the gases nitrogen and oxygen. Proc. R. Soc. London B 226:345-366. [Google Scholar]
- 62.Wang, L., Y. P. Sun, W. L. Chen, J. H. Li, and C.-C. Zhang. 2002. Genomic analysis of protein kinases, protein phosphatases and two-component regulatory systems of the cyanobacterium Anabaena sp. strain PCC 7120. FEMS Microbiol. Lett. 217:155-165. [DOI] [PubMed] [Google Scholar]
- 63.Winkenbach, F., C. P. Wolk, and M. Jost. 1972. Lipids of membranes and of the cell envelope in heterocysts of a blue-green alga. Planta 107:69-80. [DOI] [PubMed] [Google Scholar]
- 64.Wolk, C. P. 2000. Heterocyst formation in Anabaena, p. 83-104. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 65.Wolk, C. P., Y. Cai, L. Cardemil, E. Flores, B. Hohn, M. Murry, G. Schmetterer, B. Schrautemeier, and R. Wilson. 1988. Isolation and complementation of mutants of Anabaena sp. strain PCC 7120 unable to grow aerobically on dinitrogen. J. Bacteriol. 170:1239-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolk, C. P., J. Elhai, T. Kuritz, and D. Holland. 1993. Amplified expression of a transcriptional pattern formed during development of Anabaena. Mol. Microbiol. 7:441-445. [DOI] [PubMed] [Google Scholar]
- 67.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 68.Xu, X., I. Khudyakov, and C. P. Wolk. 1997. Lipopolysaccharide dependence of cyanophage sensitivity and aerobic nitrogen fixation in Anabaena sp. strain PCC 7120. J. Bacteriol. 179:2884-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoon, H.-S., and J. W. Golden. 1998. Heterocyst pattern formation controlled by a diffusible peptide. Science 282:935-938. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, C.-C., S. Laurent, S. Sakr, L. Peng, and S. Bédu. 2006. Heterocyst differentiation and pattern formation in cyanobacteria: a chorus of signals. Mol. Microbiol. 59:367-375. [DOI] [PubMed] [Google Scholar]
- 71.Zhao, Y., and C. P. Wolk. Developmental biology of heterocysts. In H. B. Kaplan and D. E. Whitworth (ed.), Multicellularity and differentiation among the myxobacteria and their neighbors, in press. American Society for Microbiology, Washington, D.C.
- 72.Zhou, R., and C. P. Wolk. 2003. A two-component system mediates developmental regulation of biosynthesis of a heterocyst polysaccharide. J. Biol. Chem. 278:19939-19946. [DOI] [PubMed] [Google Scholar]
- 73.Zhu, J., K. Jäger, T. Black, K. Zarka, O. Koksharova, and C. P. Wolk. 2001. HcwA, an autolysin, is required for heterocyst maturation in Anabaena sp. strain PCC 7120. J. Bacteriol. 183:6841-6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu, J., R. Kong, and C. P. Wolk. 1998. Regulation of hepA of Anabaena sp. strain PCC 7120 by elements 5′ from the gene and by hepK. J. Bacteriol. 180:4233-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.