Abstract
A putative iron- and Fur-regulated hemin uptake gene cluster, composed of the transport genes chuABCD and a putative heme oxygenase gene (Cj1613c), has been identified in Campylobacter jejuni NCTC 11168. Mutation of chuA or Cj1613c leads to an inability to grow in the presence of hemin or hemoglobin as a sole source of iron. Mutation of chuB, -C, or -D only partially attenuates growth where hemin is the sole iron source, suggesting that an additional inner membrane (IM) ABC (ATP-binding cassette) transport system(s) for heme is present in C. jejuni. Genotyping experiments revealed that Cj1613c is highly conserved in 32 clinical isolates. One strain did not possess chuC, though it was still capable of using hemin/hemoglobin as a sole iron source, supporting the hypothesis that additional IM transport genes are present. In two other strains, sequence variations within the gene cluster were apparent and may account for an observed negative heme utilization phenotype. Analysis of promoter activity within the Cj1613c-chuA intergenic spacer region revealed chuABCD and Cj1613c are expressed from separate iron-repressed promoters and that this region also specifically binds purified recombinant FurCj in gel retardation studies. Absorbance spectroscopy of purified recombinant His6-Cj1613c revealed a 1:1 heme:His6-Cj1613c binding ratio. The complex was oxidatively degraded in the presence of ascorbic acid as the electron donor, indicating that the Cj1613c gene product functions as a heme oxygenase. In conclusion, we confirm the involvement of Cj1613c and ChuABCD in heme/hemoglobin utilization in C. jejuni.
The gram-negative microaerophilic bacterium Campylobacter jejuni is a common zoonotic commensal and the most frequently isolated causative agent of severe bacterial enteritis in humans (25). In addition, previous infection with C. jejuni is implicated in the serious neurological conditions Guillain-Barré syndrome and Miller-Fischer syndrome (69). The virulence mechanisms involved in infection of the human intestine remain unclear; however, chemotactic motility (22), toxin production (42), and host cell invasion (13) are likely to be important, with the latter contributing to the formation of bloody diarrhea. Iron acquisition is another important mechanism involved in survival and persistence in the intestine (48, 59), and the iron regulon is widely regarded as a virulence-associated gene network used by pathogenic bacteria to coordinate gene expression on entry into the host environment (44).
The large redox potential of the Fe2+/Fe3+ couple makes iron ideally suited as a redox cofactor, and as such it can be found complexed to a wide variety of enzymes in virtually all cell types (1). In the host, free iron is maintained at very low levels in order to restrict microbial growth, and invading microorganisms must possess the means for acquiring sufficient levels of this nutrient in order to survive and persist (19). However, the cellular toxicity of iron, resulting from participation as a Haber-Weiss-Fenton redox catalyst in the formation of toxic oxygen species, chiefly the highly deleterious hydroxyl radical and superoxide anion, requires that the uptake and storage of iron must be tightly regulated. In bacteria, this regulation is primarily coordinated by the ferric uptake repressor (Fur) protein, an iron-dependent transcriptional repressor of iron acquisition and storage genes (19). Fur has been most extensively studied in Escherichia coli, although homologues have now been described in many other species, including C. jejuni (60, 64). Fur-regulated genes possess a promoter-operator sequence, termed the Fur box, to which the Fe2+:Fur dimer binds under iron-replete conditions, preventing RNA polymerase binding to the promoter and thus repressing transcription (10). Common bacterial iron-scavenging strategies involve uptake systems that employ high-affinity extracellular iron-binding siderophores and their cognate membrane transport systems, ferrous iron transport proteins, transferrin and lactoferrin receptors, and systems involved in the acquisition of iron in the form of heme (1).
Due to the insolubility and toxicity of free Fe3+, intracellular iron in the host is mostly complexed to proteins in the form of hemin (15). For bacteria to acquire iron from hemoproteins, heme must first be removed from the protein complex. This process cannot be fulfilled by siderophores and may involve specific degradative enzymes. In some systems, a heme-sequestering protein, termed a hemophore, delivers heme to the cell surface receptor (16, 30). Examples of receptors for heme or major circulating hemoproteins include the hemoglobin/hemoglobin-haptoglobin receptor complex HpuAB from Neisseria meningitidis (30); the HasR heme receptor of Serratia marcescens, which can function with or without the cognate hemophore HasA (29); and the hemopexin receptor HxuA of Haemophilus influenzae (8). In gram-negative bacteria, heme transport across the outer membrane is energized by a TonB-ExbB-ExbD complex, whereas transport across the inner membrane proceeds by ATP hydrolysis involving an ABC (ATP-binding cassette) complex in conjunction with a periplasmic shuttle protein, and both the inner and outer membrane transport genes are frequently genetically linked (15). In eukaryotes, iron must be liberated from heme by a heme oxygenase (HO) in conjunction with an aerobic electron donor in order to oxidatively cleave the porphyrin backbone via successive α-meso-hydroxyheme and verdoheme intermediates to yield ferric biliverdin and carbon monoxide (CO). The subsequent release of iron from ferric biliverdin in eukaryotes requires a biliverdin reductase (31). Recently, the biochemistry of HO-dependent heme degradation was described for several important gram-negative pathogens. The HemO protein from Neisseria spp. yields ferric biliverdin and CO as end products although the mechanism of iron release from this complex is unknown (71). Unlike eukaryotic HOs and HemO, a study of the ChuS HO from E. coli O157:H7 suggested that the iron-free form of biliverdin together with CO were formed as end products of heme degradation (53). A third type of gram-negative HO homologue from Vibrio cholerae, termed HutZ, was shown to bind heme but not degrade it and may instead be involved in heme storage to prevent cellular toxicity rather than degradation (67).
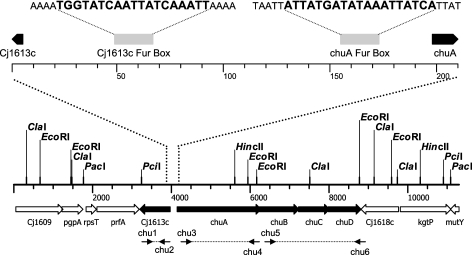
Transcriptional profiling of the C. jejuni iron regulon has identified several iron-responsive genes under the control of the global iron-dependent repressor Fur, including a cluster of five genes, Cj1613c-Cj1617 (Fig. 1), proposed to be involved in heme iron acquisition (23, 39, 59). On the basis of sequence similarity to known outer membrane heme receptors, Cj1614 was designated chuA, while the Cj1615-17 genes show homology to the cognate inner membrane ABC transport and periplasmic binding components and were designated chuBCD (40). The hypothetical protein encoded by Cj1613c is arranged divergently to chuABCD. Previously, chuA was shown to be regulated by iron and Fur (60). Analysis of the intergenic region between Cj1613c and chuA reveals two 19-bp sites which align perfectly to the C. jejuni consensus Fur box sequences proposed by van Vliet et al. (59) and Palyada et al. (39). Here, we describe the iron- and Fur-dependent regulation of the Cj1613c-17 genes and their role in heme utilization in C. jejuni. We conclude that chuABCD likely represents the major transport genes, although additional loci may be involved in iron uptake from heme/hemoproteins. Furthermore, we demonstrate that degradation of heme requires the Cj1613c gene product, which functions as a heme oxygenase, and propose the redesignation of Cj1613c as chuZ.
FIG. 1.
Map of the heme utilization gene cluster of C. jejuni NCTC 11168. Restriction sites used in Southern hybridization and primer binding sites used in PCR mapping are indicated. (Top) An expanded view shows the positions, distances (in base pairs), and sequences of the putative Fur boxes of Cj1613c and chuA.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All chemicals were purchased from Sigma-Aldrich (United Kingdom), and all media were purchased from Oxoid (United Kingdom) unless otherwise stated. Bacterial strains and plasmids used in this study are presented in Table 1. E. coli strains were cultured aerobically at 37°C on Luria-Bertani (LB) medium supplemented where necessary with kanamycin (50 μg/ml), chloramphenicol (20 μg/ml), or ampicillin (100 μg/ml). C. jejuni strains were routinely cultured on either Mueller-Hinton (MH) medium or blood agar base no. 2 plates containing 7% defibrinated horse blood, supplemented with vancomycin (10 μg/ml) and trimethoprim (5 μg/ml) at 37°C under microaerobic conditions (10% CO2, 85% N2, 5% O2) in a variable-atmosphere incubator (Don Whitley, Shipley, United Kingdom). Antibiotic selection was provided where necessary by addition of kanamycin (50 μg/ml) or chloramphenicol (10 μg/ml). Iron-replete conditions were achieved by addition of Fe2(SO4)3 to a final concentration of 40 μM (60). Iron-restricted conditions were achieved by addition of the iron chelator desferrioxamine mesylate (Desferal) to a final concentration of 20 μM (60). Clinical isolates of C. jejuni were obtained as a gift from J. Frost, Laboratory of Enteric Pathogens, Health Protection Agency. Growth of C. jejuni strains in the presence of hemin or hemoglobin as sole iron source at equivalent ferric molarities was achieved by addition of porcine hemin, to a final concentration of either 1 or 50 μM, or human hemoglobin, to a final concentration of 0.25 or 12.5 μM, to iron-restricted MH broth. C. jejuni strains were initially cultured on MH agar plates overnight, harvested in a suitable volume of MH broth, and used to inoculate 5 ml MH broth to an optical density at 600 nm (OD600) of 0.05. Cultures were incubated microaerobically with shaking, and the optical density was monitored at regular time intervals.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Description | Reference |
|---|---|---|
| E. coli strains | ||
| DH5α | General cloning host strain; F−endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG φ80dlacZΔM15 Δ(lacZYA-argF)U169 hsdR17(rK− mK+) λ− | 18 |
| BL21(DE3) | Host for recombinant protein expression; F−ompT gal dcm lon hsdSB(rB− mB−) λ(DE3) | 52 |
| BL21(pTrc1613c) | BL21(DE3) harboring plasmid pTrc1613c | This study |
| C. jejuni strains | ||
| 480 (NCTC 12744) | Host strain for reporter gene studies | NCTCa |
| NCTC 11168 | Wild type | NCTC |
| 81-176 | 27 | |
| JDR5 | NCTC 11168 allelic replacement with pJDR5 (ΔchuA::cat) | This study |
| JDR6 | NCTC 11168 allelic replacement with pJDR6 (ΔchuB::cat) | This study |
| JDR7 | NCTC 11168 allelic replacement with pJDR7 (ΔchuC::cat) | This study |
| JDR8 | NCTC 11168 allelic replacement with pJDR8 (ΔchuD::cat) | This study |
| KAR1 | NCTC 11168 allelic replacement with pKAR1 (Cj1613c::kan) | This study |
| Clinical strainsb | ||
| Plasmids | ||
| pMW10 | E. coli-C. jejuni shuttle reporter vector; lacZ Kanr | 65 |
| pAV35 | pBluescript containing C. coli cat | 60 |
| pAV201 | pMW10::pkatA reporter construct | 62 |
| pmetK | pMW10::pmetK reporter construct | 62, 65 |
| pJDR13 | pMW10::pchuA reporter construct; insert genomic region 1540370-1540988 | This study |
| pJDR14 | pMW10::pCj1613c reporter construct; insert genomic region 1540988-1540370 | This study |
| pUC19 | General cloning vector | Lab collection |
| pJDR1 | pUC19 harboring C. jejuni NCTC 11168 genomic fragment bases 1540262-1543554 (chuA) via KpnI and PstI sites | This study |
| pJDR2 | pUC19 harboring C. jejuni NCTC 11168 genomic fragment bases 1542113-1544467 (chuB) via KpnI and PstI sites | This study |
| pJDR3 | pUC19 harboring C. jejuni NCTC 11168 genomic fragment bases 1543075-1545245 (chuC) via KpnI and PstI sites | This study |
| pJDR4 | pUC19 harboring C. jejuni NCTC 11168 genomic fragment bases 1543998-1546079 (chuD) via KpnI and PstI sites | This study |
| pcam114b5c | pUC19 harboring C. jejuni NCTC 11168 genomic fragment bases 1539603-1540584 via SmaI site | 40 |
| pJDR5 | pJDR1 catA::cat; deletion of 1,940 bp of chuA (1546096-1540980) and insertion of cat | This study |
| pJDR6 | pJDR2 catB::cat; deletion of 650 bp of chuB (1543744-1543094) and insertion of cat | This study |
| pJDR7 | pJDR3 ΔcatC::cat; deletion of 479 bp of chuC (1544498-1544019) | This study |
| pJDR8 | pJDR4 ΔcatD::cat; deletion of 659 bp of chuD (1545408-1544749) and insertion of cat | This study |
| pKAR1 | pcam114b5::kan; insertion of kan at BsaBI site at base position 619 of Cj1613c | This study |
| pTrcHisB | Vector for expression of N-terminally His-tagged recombinant protein expression; Ampr | Invitrogen Corporation |
| pTrc1613c | His6-Cj1613c expression vector | This study |
NCTC, National Collection of Type Cultures, London, United Kingdom.
C. jejuni clinical isolates for genotyping (strains 50612, 60238, 53486, 51566, 61666, 37531, 44406, 53305, 35305, 54950, 34565, 58766, 45283, 38608, 45600, 38577, 41803, 45385, 44253, 37895, 35503, 54386, 59653, 57073, 39271, 51585, 43771, 60584, and 41999) were obtained from the Health Protection Agency, London, United Kingdom.
A gift from J. Parkhill, PSU, Sanger Centre, Cambridge, United Kingdom.
A method adapted from Wyckoff et al. (67) was used to test the sensitivity of C. jejuni strains to hydrogen peroxide. Briefly, cells were harvested from MH agar plates and used to inoculate molten iron-restricted MH agar containing various concentrations of hemin. Following solidification, filter disks containing 10 μl of 1 M H2O2 were placed on the agar plates and incubated in the variable-atmosphere incubator for 24 h, after which the diameter of the zone of growth inhibition was measured.
Molecular biology procedures.
All restriction enzymes were purchased from New England Biolabs, and all DNA modification enzymes were obtained from Promega. DNA manipulation was performed using standard molecular biology techniques as described previously (43), unless otherwise stated. PCR was performed using an Eppendorf Mastercycler, and the oligonucleotide primers used are described in Table 2. High-fidelity PCR or amplification of target sequences greater than 3 kbp in length was performed using the Expand High Fidelity system (Roche). The isolation of plasmid DNA was performed using QIAGEN Qiaprep Spin Mini- and Midi-prep kits in accordance with the manufacturer's instructions. Genomic DNA was isolated from C. jejuni strains using the method described by Ausubel et al. (2). For Southern hybridization experiments, probes of approximately 200 to 300 bp for each of the Cj1613c-chuD genes were amplified and labeled with digoxigenin (DIG)-11-dUTP using the Roche PCR DIG labeling kit in accordance with the manufacturer's instructions. Hybridization and detection were performed as described in the Roche DIG applications manual. DNA sequencing was performed using the BigDye V3.1 Terminator kit on an ABI 377 DNA sequencer (Applied Biosystems). DNA sequences were analyzed using Clone Manager Suite version 8 (Scientific and Educational Software). Transformation of E. coli and C. jejuni strains was achieved by electroporation using the methods of Ausubel et al. (2) and van Vliet et al. (58), respectively. Phylogenetic comparison of outer membrane receptor amino acid sequences was performed using the ClustalW algorithm (56) and an unrooted tree assembled from pair distances using PhyloDraw version 0.8 (Graphics Application Laboratory, Pusan National University; http://pearl.cs.pusan.ac.kr/phylodraw/).
TABLE 2.
Primers
| Technique and primer | Nucleotide sequence (5′-3′)a | Product or target |
|---|---|---|
| Mutagenesis | ||
| Hpo318 | CGGGGTACCGTGCATACGAGCAAACAAC | pJDR1 insert |
| ChuBR | AAACTGCAGCTAGCTTCATCATCTCCGC | |
| ChuAF | CGGGGTACCGACCTACTATCATAGACTC | pJDR2 insert |
| ChuCR | AAACTGCAGGCAAGATAGCAACAACGGC | |
| ChuBF | CGGGGTACCACTAGATGGACGCTTACCAC | pJDR3 insert |
| ChuDR | AAACTGCAGCTGCTGATATAACAGGTA | |
| ChuCF | CGGGGTACCGCCAAATGGCTCAGGCAAAAGC | pJDR4 insert |
| Hpo586 | AAACTGCAGGAAATCACTACAAGTGGC | |
| Inverse PCR | ||
| ChuAF2 | CGGGGATCCGATACTACACCTATCAAGAC | pJDR5 |
| ChuAR2 | CGCGGATCCGCCTTCTATGCTGATTAC | |
| ChuBF2 | CGCGGATCCATGTACAGCATTTGGAGGA | pJDR6 |
| ChuBR2 | CGCGGATCCGTGGTAAGCGTCCATCTATG | |
| ChuCF2 | CGCGGATCCTAGCTTCTATATTCTGCGAT | pJDR7 |
| ChuCR2 | CGCGGATCCGCTTTTGCCTGAGCCATTTGGC | |
| ChuDF2 | CGCGGATCCCGTCTTAGTCCTAAGATAATAG | pJDR8 |
| ChuDR2 | CGCGGATCCGCAGGATCAAGCACTACAAGGC | |
| PCR mapping | ||
| chu1 | CATGTGCAACATTTC | Cj1613c |
| chu2 | TTTGGTGGTATTGAG | Cj1613c |
| chu3 | AAATGCACAAGAATC | chuA |
| chu4 | ATACTGTCTTGATAG | chuA |
| chu5 | GAAATGGCAGAAACTACTATG | chuBCD |
| chu6 | CTTGCTGTTTGGATACTAAAG | chuBCD |
| Southern hybridization | ||
| 1613probeF | AGTTTTAATGCCTCCTTC | Cj1613c |
| 1613probeR | AAGTTGTTTGCTCGTATG | Cj1613c |
| ChuAprobeF | ACCAGCAGTGGCTATCTAAC | chuA |
| ChuAprobeR | ATCCCTGTAAGCGTGTCTTC | chuA |
| ChuBprobeF | ATAGTCGCTTGGCTTATGG | chuB |
| ChuBprobeR | GCAACCGCAAAAGATACAG | chuB |
| ChuCprobeF | AAAACACGCTTTTAGTTC | chuC |
| ChuCprobeR | GGATTTTATCGCAGAATATAG | chuC |
| ChuDprobeF | TCCTGCAAGCATAGAAAC | chuD |
| ChuDprobeR | TTGGTGATTTGGCCTAAG | chuD |
| EMSA | ||
| EMSACHUF | TGAGAATTCATATGAGAAATAATGCTTTC | |
| EMSACHUR | GCTGGATCCTTTGGGTGCAAATTTTACTC | |
| Cj1613c expression | ||
| 1613TrcF | ATGCCTGCAGCTATGAATTTTGAAAGCATTATTTCTC | |
| 1613TrcR | ATGCGAATTCTTAATGCTTATGTAGGAATTTATG | |
| Promoter analysis | ||
| Cj1613R2 | CGCGGATCCGCTCGCTCTTTGCACTCATGC | |
| ChuAR4 | CGCGGATCCAGATCTTTGCCTTCTATGC |
Restriction enzyme sites are underlined.
Mutant construction.
Construction of deletion/insertion mutants in chuA, chuB, chuC, and chuD was achieved as follows. Each target gene was amplified from C. jejuni NCTC 11168 genomic DNA to include approximately 500 bp of DNA flanking the site of deletion using the following primer pairs incorporating terminal 5′-KpnI and 3′-PstI sites: Hpo318-ChuBR (chuA), ChuAF-ChuCR (chuB), ChuBF-ChuDR (chuC), and ChuCF-Hpo586 (chuD) (Table 2). Each amplified product was cloned into pUC19 to yield plasmids pJDR1, pJDR2, pJDR3, and pJDR4, respectively (Table 1). Partial deletions within the cloned target genes were achieved by inverse PCR using the following primers incorporating terminal BamHI sites: ChuAR2-ChuAF2 (pJDR1), ChuBR2-ChuBF2 (pJDR2), ChuCR2-ChuCF2 (pJDR3), and ChuDF2-ChuDR2 (pJDR4) (Table 2). A chloramphenicol resistance cassette (cat) was excised from pAV35 using BamHI and ligated to the similarly cut inverse PCR products to yield plasmids pJDR5, pJDR6, pJDR7, and pJDR8, respectively (Table 1). These constructs were then used to electroporate C. jejuni NCTC 11168, and transformant colonies were screened for replacement of the wild-type gene with the deletion/insertion alleles by amplification of chromosomal DNA using oligonucleotide primers flanking the regions present in the plasmid constructs (Table 2). The mutant strains constructed were designated JDR5 (ΔchuA::cat), JDR6 (ΔchuB::cat), JDR7 (ΔchuC::cat), and JDR8 (ΔchuD::cat) (Table 1). Mutagenesis of Cj1613c was performed by insertional inactivation. The region encompassing Cj1613c from C. jejuni NCTC 11168 and flanking DNA had previously been cloned in pUC19 (pcam114b5) (40). A kanamycin resistance marker was excised from pJMK30 with BamHI and blunt-end treated with Pfu polymerase prior to ligation to BsaBI-digested pcam114b5 (Table 1). The resulting construct (pKAR1) was then used to transform C. jejuni NCTC 11168, and transformants were screened for allelic replacement of the wild-type gene as described above. The mutant strain generated (Cj1613c::kan) was designated C. jejuni KAR1 (Table 1).
Overexpression and purification of recombinant His(6)-Cj1613c.
The Cj1613c coding region was amplified with proofreading polymerase from C. jejuni NCTC 11168 genomic DNA using primer pair 1613TrcF and 1613TrcR (Table 2). The amplified product and vector pTrcHisB (Table 1) were simultaneously digested with EcoRI and PstI and ligated. The ligation was used to transform E. coli DH5α and clones harboring recombinant plasmids verified by sequencing prior to propagation in the expression host strain E. coli BL21. An overnight culture of strain BL21(pTrc1613) was diluted 1:100 into 500 ml of LB broth containing ampicillin and grown to an OD600 of 0.6. Recombinant protein expression was induced by addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM for 3 h at 30°C with shaking. Cells were then harvested and resuspended in 10 ml of 20 mM Tris-HCl (pH 7.5), containing mini-complete (EDTA) protease inhibitors (Roche). Lysis was achieved by addition of chicken egg white lysozyme to a final concentration of 100 μg/ml and incubation at 37°C for 1 h, followed by three rounds of sonication for 30 s each on ice. The cell lysate was centrifuged at 10,000 × g for 20 min and filter sterilized using a 0.45-μm-pore-size filter (Sartorius). Imidazole was added to a final concentration of 5 mM, and the recombinant protein was purified using the Ni2+-NTA system (Invitrogen) in accordance with the manufacturer's standard protocol. The purity of the recombinant protein was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (43), and identification was confirmed by Western blot analysis using an anti-His antibody (Invitrogen) and sequencing (Protein and Nucleic Acid Chemistry Laboratory, University of Leicester). Purified protein fractions were pooled and dialyzed three times using Slide-A-Lyzer dialysis cassettes (molecular weight cutoff, 3,000; Pierce) in 1:500 volumes against 20 mM Tris-HCl (pH 7.8) at 4°C and quantified by colorimetry (4). The protein was finally concentrated using Microcon YM-10 microconcentrator columns (Millipore), and yields were typically in the range of 300 to 500 μg/ml.
Spectrophotometric analysis of heme binding and degradation by His6-Cj1613c.
Binding studies based on the method of Wilks and Schmitt (63) were performed using 1-ml volumes of 20 μM His6-Cj1613c in 20 mM Tris-HCl (pH 7.8) at 25°C. Hemin (2.5 mM in 20 mM NaOH) was titrated in 2.5 μM increments to a final twofold molar excess of hemin to protein, and the absorbance spectrum between 350 and 800 nm was recorded using a Cary 300 spectrophotometer (Varian) at a sampling rate of 300 nm/min. Prior to investigating the heme oxygenase capacity of Cj1613c, the heme-His6-Cj1613c complex formed as the end product of the previous binding studies was purified to remove excess heme by filtration using Microcon YM-10 columns and eluted in 1 ml of 20 mM Tris-HCl (pH 7.8). To rule out the involvement of nonenzymatic H2O2-mediated conversion of heme to biliverdin, which has been reported for other heme-binding proteins, catalase (bovine liver) was added to a final concentration of 2 μM. Ascorbic acid was added as an electron donor to a final concentration of 20 mM, and spectra were recorded between 350 and 800 nm every 2 min for up to 1 h (6).
EMSAs and promoter activity assays.
The target promoter region between Cj1613c and chuA was amplified using primers EMSACHUF and EMSACHUR (Table 2). Electrophoretic mobility shift assays (EMSAs) were conducted as described previously (23). The dissociation constant was calculated as follows: the change in intensity of the unshifted molecular species with increasing protein concentration was quantified by densitometry (GeneTools; Syngene), and the equilibrium dissociation constant (KD) was defined as the slope of the linear transformation plotted from log(bound/unbound) against log(unbound). Promoter activity experiments using a lacZ reporter were performed using the E. coli-C. jejuni shuttle plasmid pMW10 as described previously (62).
RESULTS
Phenotypic characterization of hemin utilization gene cluster mutants.
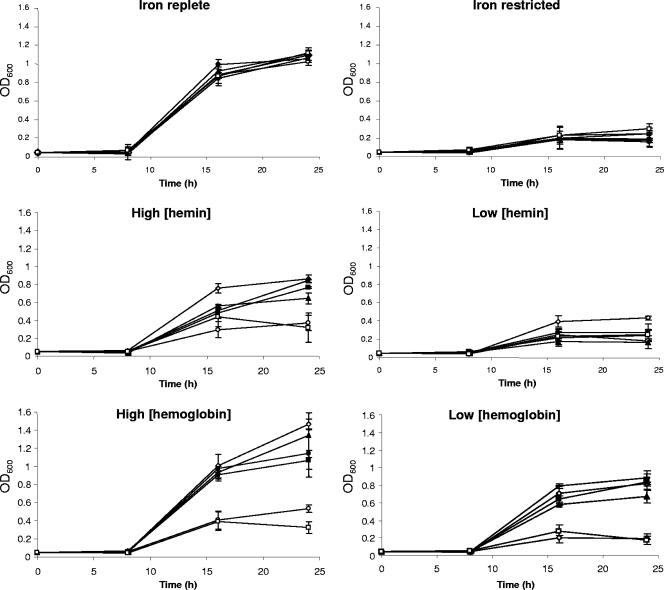
In order to ascertain the role of each gene in the putative heme utilization gene cluster (Fig. 1), a panel of mutants was constructed: JDR5 (ΔchuA::cat), JDR6 (ΔchuB::cat), JDR7 (ΔchuC::cat), JDR8 (ΔchuD::cat), and KAR1 (Cj1613c::kan) (Table 1). A phenotypic test was designed to determine the requirement for each gene during growth of C. jejuni where iron is present solely in the form of hemin or hemoglobin (Fig. 2). Control experiments using iron-replete and iron-restricted conditions showed no difference between the mutant and wild-type strains, and iron restriction severely inhibited growth of all strains. In the presence of a low (1 μM) concentration of hemin, all strains grew poorly, although the wild type appeared to grow slightly better than the mutant strains. In the presence of a high (50 μM) concentration of hemin or both low (0.25 μM) and high (12.5 μM) concentrations of hemoglobin, a clear difference between the mutant phenotypes could be observed: strains JDR5 and KAR1 (chuA and Cj1613c mutants) grew poorly, while strains JDR6, JDR7, and JDR8 (chuB, chuC, and chuD mutants) appeared to grow almost as well as the wild type. Statistical comparison (unpaired t test) of the chuA and Cj1613 mutants to the wild type showed both mutants to be significantly different from the wild type at 16 and 24 h in all assays where hemin or hemoglobin was added. These results demonstrate that growth in the presence of hemin or hemoglobin as a sole source of iron is highly dependent on chuA and Cj1613c; however, these genes may not be essential, as indicated by a small level of growth in the respective mutant strains in the absence of an alternative iron source. Most, but not all, comparisons of the chuB, -C, or -D mutants with the wild type are statistically different at 16 and 24 h (unpaired t test) in the presence of hemin or hemoglobin. However, by comparison with the chuA and Cj1613c mutants, mutation of chuBCD only partially compromises growth in the presence of hemin or hemoglobin as sole iron sources. Given the phenotype of the chuB, chuC, and chuD mutants in comparison to that of chuA, polarity is unlikely to be an issue; however, despite using a variety of genetic strategies, we have not been able to carry out a complementation analysis for the chuA mutant. Comparison of the growth of all strains in the presence of low hemin or hemoglobin concentrations (1 ferric molarity) reveals that hemin is less effective than hemoglobin at stimulating growth, and this may be due to either differences in substrate affinity or minor differences in substrate solubility in the growth media used.
FIG. 2.
Growth assays of heme utilization gene mutants. Samples were tested in triplicate, and the data plotted are the means of two independent experiments together with the sample error. Symbols: ⋄, wild-type strain NCTC 11168; ○ JDR5; ▴, JDR6; •, JDR7; ▪, JDR8; □, KAR1 grown in MH broth supplemented with 40 μM Fe2(SO4)3 (iron replete), 20 μM desferrioxamine mesylate (iron restricted), 1 μM or 50 μM hemin, or 0.25 μM or 12.5 μM hemoglobin.
Heme utilization gene cluster content and phenotyping of C. jejuni clinical isolates.
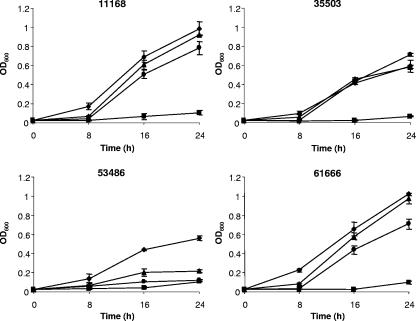
To ascertain the prevalence of heme utilization genes in clinical isolates of C. jejuni, a PCR-based approach was initially used. Six oligonucleotide primers (chu1 to chu6) were designed to yield amplicons spanning the Cj1613-chu locus (Table 2), and 31 strains of clinical origin (Table 1) were compared with NCTC 11168. Gene-specific PCR mapping showed conservation of Cj1613c in all strains tested, whereas three strains (61666, 53486, and 35503) tested negative for the presence of chuA, and two strains (61666 and 35503) had differences in chuBCD (Table 3). Comparison of the amino acid sequences predicted for ChuA orthologues among several C. jejuni strains, Campylobacter species, and non-Campylobacter species (data not shown) reveals sequence variation in the N-terminal region, which for the former may account for failed amplification. To clarify these observations, Southern hybridization experiments were performed using gene-specific probes for all five genes of strains 11168, 35503, 53486, and 61666 (data not shown). The presence of Cj1613c, chuA, chuB, and chuD was confirmed in all strains (Table 3). Interestingly, chuC could not be identified by hybridization in strain 35503, and a polymorphism for one enzyme (ClaI) was also observed in this strain (data not shown). To compare the heme utilization phenotypes, strains 35503, 53486, and 61666 were tested in comparison to NCTC 11168 for their ability to grow using hemin (50 μM) or hemoglobin (0.25 μM) as sole iron sources (Fig. 3). Strains 35503 and 61666 showed growth profiles similar to that of the reference strain, whereas strain 53486 failed to grow well in the presence of hemoglobin or hemin as the sole iron source (Table 3).
TABLE 3.
C. jejuni genotyping and heme utilization phenotyping studies
| Test | Result for indicated strain
|
|||
|---|---|---|---|---|
| 11168 | 35503 | 53486 | 61666 | |
| PCR mapping for: | ||||
| chu1-2 (Cj1613c) | + | + | + | + |
| chu3-4 (chuA) | + | − | − | − |
| chu5-6 (chuBCD) | + | − | + | − |
| Southern hybridization | All genes present | chuC absent | All genes present | All genes present (rearrangement?) |
| Heme utilization phenotype test | + | + | − | + |
FIG. 3.
Growth assays of C. jejuni clinical isolates. C. jejuni strains NCTC 11168, 35503, 58436, and 61666 were tested for their ability to grow in iron-restricted (20 μM desferrioxamine mesylate) MH broth under iron-replete [40 μM Fe2(SO4)3] (⧫) and iron-restricted (▪) conditions or in the presence of 50 μM hemin (•) or 12.5 μM hemoglobin (▴) as sole iron sources. Cultures were performed in triplicate, and the data plotted are the means of two independent experiments together with the sample error.
Cj1613c and Cj1614-17 are Fur regulated and expressed from two separate iron-repressible promoters.
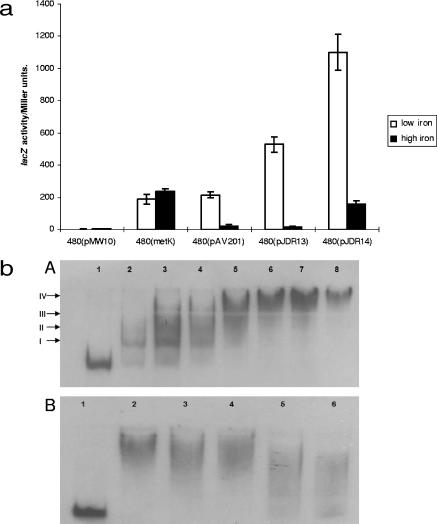
Previous transcriptional profiling studies have indicated that the Cj1613c-chuD genes are repressed under iron-replete conditions and that this regulation is affected by Fur (23, 39). The intergenic spacer region between Cj1613c and chuA contains two putative Fur box operator motifs as determined by comparison with the consensus sequences described by van Vliet et al. (59) and Palyada et al. (39). In order to verify promoter activity and iron responsiveness the Cj1613c-chuA intergenic region was amplified with primer pair Cj1613R2-ChuAR4 (Table 2), which incorporates terminal BamHI sites. This fragment was cloned into the complementary BamHI site upstream of the lacZ gene of the reporter plasmid pMW10 (65) to test for promoter activity in both orientations (pJDR13, a chuA::lacZ transcriptional fusion, and pJDR14, a Cj1613c::lacZ transcriptional fusion) (Table 1). Additional control plasmids bearing the promoter regions of the non-iron-regulated housekeeping gene metK (pmetK) and the iron-repressed gene katA (pAV201) as well as the vector alone (pMW10) were also included (Table 1) (62). The lacZ activity associated with each plasmid in host strain C. jejuni 480 was compared under iron-restricted or iron-replete conditions (65) (Fig. 4a). The results show that the fragments cloned into pJDR13 and pJDR14 possess promoter activity, and this activity is induced under iron-restricted conditions and reduced under iron-replete conditions.
FIG. 4.
Reporter gene assay and EMSA of the Cj1613c-chuA intergenic region. (a) Reporter gene assay of the chuA and Cj1613c promoter regions under differential iron conditions. LacZ activity was determined after strain 480-containing reporter constructs (Table 1) were cultured in MH broth under iron-restricted (20 μM desferrioxamine mesylate, low iron) or iron-replete [40 μM Fe2(SO4)3, high iron] conditions. Data presented are the means of triplicate sampling from two independent experiments with the standard error. (b) EMSA of the Cj1613c-chuA intergenic region with purified Fur protein. (Panel A) DIG-labeled DNA was present at 0.0775 nM. Lanes: 1, no FurCj protein; 2 to 8, labeled fragment incubated with 0.25 nM, 0.5 nM, 0.75 nM, 1 nM, 1.25 nM, 1.5 nM, and 1.75 nM FurCj, respectively. Bands were labeled I, II, III, and IV as described in the text. (Panel B) Self-competitive EMSA. Lanes: 1, labeled probe alone; 2, labeled probe with 1.5 nM FurCj; 3 to 6, labeled probe with 1.5 nM FurCj and 100-fold, 500-fold, 1,000-fold, or 1,500-fold excess unlabeled probe, respectively.
To experimentally confirm Fur binding within the Cj1613c-chuA intergenic region, EMSA was conducted using a 218-bp DNA probe fragment encompassing both putative Fur boxes and purified recombinant FurCj (23). Figure 4b, panel A, shows that in the presence of Fur, a concentration-dependent shift can be seen with four consistently observed individual protein:DNA complex species. The first two shift species (I and II) are proposed to arise from the differential affinity of Fur for the two separate Fur box sites within the target DNA fragment, and thus successive binding events occur in a concentration-dependent manner. Additional shift species (III and IV) may be due to Fur polymerization along the DNA fragment; a process which has been previously observed by other investigators (14, 28). The KD value for Fur binding to the probe DNA fragment was calculated to be 0.17 μM ± 0.03 μM, indicating a high binding affinity. To confirm the specificity of binding, competitive EMSA using isogenic unlabeled competitor DNA was performed (Fig. 4b, panel B). An approximately 1,000- to 1,500-fold excess concentration of competitor DNA was required, indicating that Fur binding proceeds specifically. Specificity of Fur binding was verified using unlabeled, nonspecific control DNA in place of the Cj1613c-chuA intergenic region (data not shown).
Biophysical analysis of the role of Cj1613c in hemin utilization.
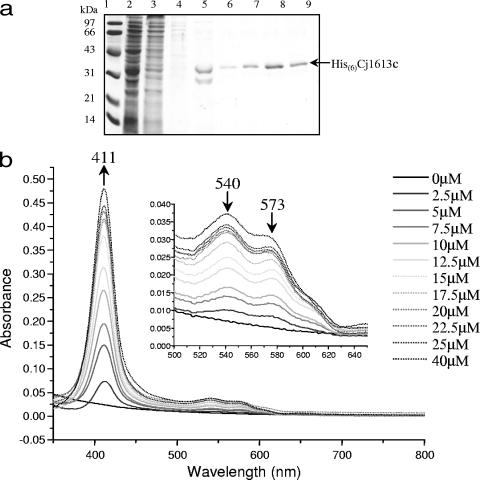
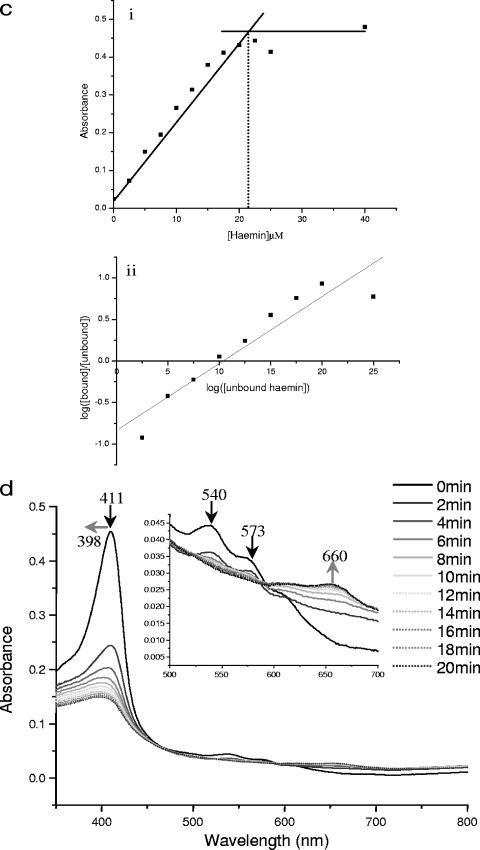
Recently, bacterial HOs have been implicated as the major class of enzymes involved in degradation of heme by oxidative cleavage of the protoporphyrin ring to release iron via successive α-meso-hydroxyheme and verdoheme intermediates, finally yielding biliverdin and CO as by-products (6, 71). However, biochemical testing of the putative HO HutZ from V. cholerae indicated that although the purified recombinant protein could bind heme, it could not degrade it in the presence of an electron donor, and it was therefore suggested that this protein is involved in heme sequestration (67). The phenotype of the Cj1613 mutant indicates a role in heme utilization, and therefore the role of Cj1613c in heme binding and degradation was examined by investigating the interaction of Cj1613c with heme. Cj1613c protein was overexpressed as an N-terminal hexahistidine-tagged recombinant fusion protein in E. coli, and SDS-PAGE analysis showed that the protein could be purified to homogeneity by standard Ni2+-affinity chromatography with average yields of ∼1 to 2 mg ml−1 (Fig. 5a). The ability of His6-Cj1613c to bind heme in vitro was determined by using absorbance spectroscopy. Titration with hemin resulted in the formation of a prominent Soret peak at 411 nm with a smaller peak at 573 nm and a shoulder at 540 nm corresponding to the α- and β-porphyrin bands of the heme:His6-Cj1613c complex, respectively (Fig. 5b). The absorbance maxima at these wavelengths were achieved following addition of hemin to approximately 20 μM and did not increase significantly beyond this concentration up to 40 μM, indicating a 1:1 binding stoichiometry (Fig. 5c). The presence of this complex was apparent by the formation of a pale yellow coloration compared to the control buffer (data not shown). The calculated molecular affinity of His6-Cj1613c for heme gave a KD value of 8.3 μM ± 1.7, which is similar to, though slightly weaker than, affinities reported for other bacterial HOs (46, 53, 63).
FIG.5.
Purification of recombinant His6-Cj1613c and absorbance spectroscopy of hemin binding and the degradation of heme:His6-Cj1613c complex. (a) SDS-PAGE of His6-Cj1613c purification. Lanes: 1, molecular weight markers; 2, column flowthrough; 3, 5 mM imidazole wash; 4, 60 mM imidazole wash; 5 to 10, elution fractions. Arrow indicates the band identified by Western analysis and sequencing as recombinant His6-Cj1613c. (b) Hemin was added incrementally (0 to 40 μM) to 20 μM His6-Cj1613c protein. Absorbance changes are indicated by the position and direction of the arrows. (Inset) The region between 500 and 650 nm has been enlarged to show the peaks at 540 and 573 nm. (c) Heme:His6-Cj1613c binding stoichiometry and affinity. Values were plotted as (i) the change in absorbance at 411 nm against heme concentration and (ii) log(unbound) against log(bound/unbound) hemin. (d) Degradation of the heme:His6-Cj1613c complex in the presence of ascorbic acid. Spectra show changes in absorbance at 2 min intervals up to 20 min (no significant change was observed after 1 h). (Inset) The region between 500 and 700 nm has been enlarged. Arrows indicate the positions and directions of absorbance changes. Black arrows indicate spectral changes indicating conversion of heme:His6-Cj1613c to the ferric biliverdin:His6-Cj1613c complex. Gray arrows correspond to spectral shifts indicative of iron-free biliverdin formation.
FIG. 5—
Continued.
Heme oxygenase activity may be assessed in vitro by providing the heme:enzyme complex with a suitable electron donor, such as ascorbic acid, for participation in oxidative porphyrin cleavage. In these experiments, the HO activity of the purified heme:His(6)Cj1613c complex was investigated (Fig. 5d). Following addition of ascorbic acid in the presence of catalase to rule out the involvement of H2O2-mediated hemin degradation, absorbance at 411, 540, and 573 nm decreased, suggesting the formation of a ferric biliverdin:His6-Cj1613c complex. Over 20 min, the spectrum shifted to give final broad maxima centered around 398 and 660 nm, suggestive of iron-free biliverdin formation. No further changes were observed beyond 20 min. In vitro biliverdin formation was also visually apparent by a change in coloration of the samples from pale yellow to pale green (data not shown).
The by-products of hemin degradation are not involved in protection from peroxide stress.
In eukaryotes, a reductase is involved in the conversion of ferric biliverdin to yield bilirubin, and furthermore, this by-product has been implicated in protection against oxidative stress (31). This role may be particularly relevant given the increase in intracellular iron accompanying hemin degradation, resulting in an increase in the Haber-Weiss-Fenton-type iron-catalyzed formation of superoxide and hydroxyl radical from H2O2. Mutation of Cj1613c or chuA may therefore starve the cells of biliverdin and thus prevent generation of the protective metabolite bilirubin. To test whether disruption of Cj1613c or chuA renders C. jejuni more sensitive to the effects of peroxide stress a simple plate assay was conducted. Mutants and wild-type strains were used to inoculate agar under iron-replete or iron-restricted conditions or with hemin as the sole iron source, and the zone of growth inhibition surrounding the filter disk impregnated with H2O2 was measured. No apparent difference was observed (data not shown), indicating that these mutants are no more sensitive to the effects of peroxide than the wild type.
DISCUSSION
Efficient transport of heme across the cell envelope of C. jejuni is dependent on ChuA but not ChuBCD.
Previously, Pickett et al. (41) observed that hemin, hemoglobin, hemin-hemopexin, and hemoglobin-haptoglobin were capable of stimulating growth of C. jejuni under iron-restricted conditions. Comparison of the growth profiles obtained for the wild-type strain (Fig. 2 and 3) indicates that iron in the form of lower concentrations of hemoglobin appears to stimulate C. jejuni growth more rapidly and to a higher final OD600 than it does in the form of hemin. These observations may reflect a preference for heme when supplied in the form of hemoglobin. Indeed, free heme is rapidly absorbed in the small intestine to limit toxicity and bacterial proliferation and therefore is unlikely to be the primary form encountered in vivo. Various degrees of hemolytic activity have been reported for some C. jejuni strains (24, 41). Infection by C. jejuni can result in inflammatory bloody diarrhea (5), and this hemolytic activity may contribute to the release of hemoglobin from erythrocytes during invasion of the human gut epithelium. However, the relative contribution of the major circulating hemoproteins to the iron nutrition of C. jejuni and their dependence on chuABCD for translocation across the bacterial cell envelope require further investigation. On the basis of the findings presented here we can conclude that chuA is required for efficient uptake of heme or heme from hemoglobin. Due to the large size of hemoglobin, we envisage that heme is removed from the globin complex at the cell surface and is transported across the outer membrane alone.
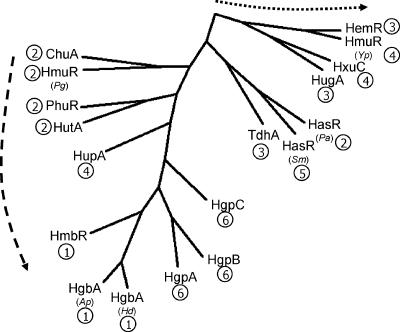
A phylogenetic analysis of the ChuA amino acid sequence with experimentally characterized orthologous proteins from other bacterial species (Fig. 6) reveals that these types of receptors display a pseudocladistic phylogeny. ChuA shows closest relatedness to the heme/hemoglobin- and hemoglobin-specific branches, indicating that ChuA may belong to a clade that contains hemoglobin-specific receptors, and as such the preferential utilization of hemoglobin over heme observed in these studies may reflect an evolutionary trend towards increasing substrate specificity. As the experimentally defined data pool of receptor substrate specificities becomes larger, a more comprehensive phylogenetic analysis, which may reveal a more distinct hemoglobin-specific clade, should be possible. Interestingly, the S. marcescens hemophore receptor HasR shows close relatedness to both heme-specific and lower-specificity hemoprotein receptors, though due to the absence of other hemophore receptors having been described it is not presently possible to determine a distinct lineage.
FIG. 6.
Phylogenetic comparison of ChuA from C. jejuni NCTC 11168 with heme, hemoprotein, and hemophore receptors characterized in other bacterial species. Receptor substrate specificities are coded as hemoglobin only (1), heme and hemoglobin (2), heme only (3), heme and multiple hemoproteins (4), hemophore (5), and hemoglobin/hemoglobin:haptoblobin (6). The dashed arrow indicates a direction of evolutionary conservation towards hemoglobin substrate specificity. The dotted arrow indicates a direction of evolution away from receptor substrate specificity. Abbreviations: HmbR, N. meningitidis HmbR (51); HgbA (Ap), Actinobacillus pleuropneumoniae HgbA (47); HgbA (Hd), Haemophilus ducreyi HgbA (11, 12); HmuR (Pg), Porphyromonas gingivalis HmuR (38, 45); HasR (Pa), Pseudomonas aeruginosa HasR (37); PhuR, P. aeruginosa PhuR (37); HutA, V. cholerae HutA (20, 32); ChuA, C. jejuni ChuA (this study); TdhA, H. ducreyi TdhA (55); HemR, Y. enterocolitica HemR (49); HugA, P. shigelloides HugA (21); HmuR (Yp), Yersinia pestis HmuR (57); HxuC, H. influenzae HxuC (7, 9); HasR (Sm), S. marsescens HasR (16), HgpA, -B, and -C, H. influenzae HgpA, -B, and -C (34); HupA, H. influenzae HupA (33).
By contrast to chuA, the chuBCD genes do not appear to be essential for heme or hemoglobin utilization. Redundancy in inner membrane heme transport genes has been observed with other bacterial heme utilization systems; for example, the Pleisomonas shigelloides inner membrane transport genes hugBCD are not required to restore heme iron utilization in E. coli 1017 in the presence of the hut system (21). The Yersinia enterocolitica ATPase and hemUV permease genes are essential for heme utilization, although the periplasmic binding component hemT is not required (50). Wyckoff and coworkers (66) have suggested that in the absence of a strict requirement for a single inner membrane transport system, alternative ABC complexes may be capable of shuttling heme from the periplasm to the cytoplasm. In C. jejuni, several putative ABC transport systems associated with the iron regulon that may be involved in transport of heme-iron have been identified: Cj0173c-5c, the putative siderophore transport system ceuBCDE, and Cj1661-3 (23, 39, 59).
Analysis of the heme utilization gene cluster in C. jejuni clinical isolates revealed that all strains possess Cj1613c, which on the basis of PCR mapping appears to be highly conserved. By contrast, there appears to be a lower degree of conservation within chuA, though the biological implications of this are unclear at present. The heme-negative phenotype of strain 53486 indicates that chuA, while present, may be nonfunctional, and it seems that this loss of chuA function is not complemented by the presence of an alternative receptor in this strain. Likewise, substrate transport across the inner membrane may not be a role exclusively fulfilled by chuBCD, as indicated by the absence of chuC in the heme/hemoglobin utilization-positive strain 35503. A similar mapping study of the heme utilization gene cluster of Vibrio anguillarum revealed that in most strains lacking the cluster-linked huvA locus, an alternative outer membrane heme receptor gene, which was designated huvS and could complement huvA in E. coli and V. anguillarum, was present (36). We have attempted to identify additional outer membrane heme- or hemoglobin-binding proteins that may serve as alternative receptors to ChuA using a batch affinity chromatography method (54); however, no candidates have yet been identified (data not shown).
A cross-species comparison of the heme biosynthesis pathway of E. coli with C. jejuni NCTC 11168 indicates that the latter possesses all the components required for the de novo synthesis of heme (http://biocyc.org). The close genetic linkage between specific heme transport and heme oxygenase loci and the absence of linkage to other genes associated with the heme metabolic pathway (i.e., coproporhyrinogen oxidase; Cj0992c, hemN) support iron nutrition as the major purpose of heme acquisition in C. jejuni. It may therefore be inferred that acquisition of heme may have evolved in C. jejuni solely to meet the iron requirements of the cell rather than as a source of metabolic heme.
Regulation and organization of the C. jejuni heme utilization gene cluster.
The presence of two putative Fur boxes within the heme utilization gene cluster led us to investigate the possibility that Cj1613c and chuABCD are expressed independently of each other but that their expression is dependent on Fur and iron. The results of the reporter gene studies described here have confirmed this hypothesis. Coordinated expression via a common regulator may ensure that once heme has entered the cytoplasm it can be promptly degraded not only to meet the cells' iron requirements, but also to reduce its potential toxicity through degradation to biliverdin. To date, no other candidate regulators of iron uptake have been identified in C. jejuni.
In attempts to identify additional genes involved in heme utilization it would be prudent to consider attributes characteristic of Cj1613c-chuD, such as classical iron-repressible, Fur-dependent regulation and the possession of a putative Fur box. The potential existence of such genes is indicated by the apparent semiredundancy of chuBCD described here and also by comparison of the Cj1613c-17 gene cluster with known heme utilization gene clusters from other bacterial species (21, 35, 36). Most notable is the absence of a linked TonB-ExbB-ExbD system, which is expected to be required for energizing the outer membrane receptor(s). Although three putative systems are present in the genome of C. jejuni NCTC 11168 (Cj0179-81, Cj1628-30, and Cj0753c), none have yet been specifically associated with heme transport. Further investigation using a panel of single, double, and triple ton mutants would resolve this issue. Other key differences include an unlinked putative coproporphyrinogen oxidase (hemN) and absence of a putative transcriptional activator (66). A relatively small complement of genes required for heme utilization may reflect the apparent genetic minimalism observed in Campylobacter species. However, several putative iron transport systems exist in C. jejuni, and it is reasonable to suggest that this organism utilizes various sources of iron during transmission, colonization, or infection of different host animal intestines.
Cj1613c is a heme oxygenase.
Foundation studies on the biochemical nature of the human HOs have formed the basis for many of the current studies of bacterial HO orthologues (26, 68). The recent identification of heme oxygenase genes within heme utilization gene clusters in several important gram-negative pathogens bearing sequence homology with Cj1613c prompted the biophysical studies presented here. The purified recombinant protein binds hemin in vitro at a hemin-to-protein ratio approaching 1:1 with moderate affinity (KD value in the low micromolar range) and displaying absorbance maxima at 411, 540, and 573 nm that are similar to those reported for other HOs in other species (53, 67, 71). The HO activity of His6-Cj1613c was confirmed by the appearance of characteristic spectral changes following addition of ascorbic acid as the electron donor. Importantly, the formation of broad absorbance maxima at 395 and 660 nm suggests that the end product of heme degradation is the iron-free form of biliverdin, similar to that observed with ChuS from E. coli O157:H7 (53), rather than ferric biliverdin, which was suggested to be the end product of heme degradation by Neisseria spp. HemO (70).
Conversion of ferric biliverdin to bilirubin in eukaryotes requires a reductase (31), and it is possible that a similar pathway may exist in prokaryotes even though no gene candidate has yet been proposed for C. jejuni. Comparison of the chuA and Cj1613c mutants to the wild-type strain in a peroxide sensitivity assay found no differences, suggesting that bilirubin, if produced as a by-product of hemin metabolism, may not play an important role in protection against oxidative stress in C. jejuni. Alternatively, C. jejuni possesses several enzymes involved in oxidative stress protection, namely catalase (katA) (17), alkyl-hydroperoxide reductase (ahpC) (3), and ferredoxin (fdxA) (61). Additionally, C. jejuni possesses a PerR homologue that regulates katA and ahpC. In other organisms, PerR, a Fur structural homologue, functions in coordinating peroxide stress defense and iron uptake genes using iron as a cofactor in response to intracellular peroxide, and as such a similar mechanism exists in C. jejuni.
The coordinated expression of HO with heme uptake through iron and Fur and the subsequent HO-mediated degradation of heme to biliverdin likely serve to allow access to a valuable iron source in vivo while limiting the toxic effects of intracellular uncomplexed heme. By comparison of the heme utilization gene cluster of C. jejuni with similar known gene clusters from other bacterial species, the confirmed and putative functional homologues of Cj1613c commonly bear the suffix -Z. On this basis we propose the redesignation of this gene as chuZ.
In conclusion, we have experimentally characterized the heme utilization gene cluster in C. jejuni. We have identified ChuA as the major outer membrane transport protein for heme/hemoproteins and Cj1613c (ChuZ) as a highly conserved oxygenase required for heme degradation. ChuBCD are most likely involved in the transport of heme across the inner membrane, although they do not appear essential for this process. The gene cluster is highly conserved among clinical isolates of C. jejuni, though additional heme transport genes may be present in some strains and capable of substituting for chuBCD. Promoter analysis confirms classical Fur-dependent, iron-repressible regulation of the gene cluster, though Cj1613c expression is independent of chuABCD expression.
Acknowledgments
This work was supported by a grant (91/D19661) and a studentship (to J.D.R.) from the Biotechnology and Biological Sciences Research Council, United Kingdom.
We acknowledge Andrew Westlake, Department of Biochemistry, University of Leicester, United Kingdom, for assistance with spectrophotometric analyses and Jenny Frost and Andrew Lawson, Laboratory of Enteric Pathogens, Health Protection Agency Centre for Infections, London, United Kingdom, for C. jejuni strains.
Footnotes
Published ahead of print on 15 September 2006.
REFERENCES
- 1.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinone. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1992. Short protocols in molecular biology, 2nd ed., vol. 1. John Wiley & Sons, New York, N.Y.
- 3.Baillon, M. L., A. H. van Vliet, J. M. Ketley, C. Constantinidou, and C. W. Penn. 1999. An iron-regulated alkyl hydroperoxide reductase (AhpC) confers aerotolerance and oxidative stress resistance to the microaerophilic pathogen Campylobacter jejuni. J. Bacteriol. 181:4798-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Butzler, J. P. 2004. Campylobacter, from obscurity to celebrity. Clin. Microbiol. Infect. 10:868-876. [DOI] [PubMed] [Google Scholar]
- 6.Chu, G. C., K. Katakura, X. Zhang, T. Yoshida, and M. Ikeda-Saito. 1999. Heme degradation as catalyzed by a recombinant bacterial heme oxygenase (HmuO) from Corynebacterium diphtheriae. J. Biol. Chem. 274:21319-21325. [DOI] [PubMed] [Google Scholar]
- 7.Cope, L. D., R. P. Love, S. E. Guinn, A. Gilep, S. Usanov, R. W. Estabrook, Z. Hrkal, and E. J. Hansen. 2001. Involvement of HxuC outer membrane protein in utilization of hemoglobin by Haemophilus influenzae. Infect. Immun. 69:2353-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cope, L. D., S. E. Thomas, Z. Hrkal, and E. J. Hansen. 1998. Binding of heme-hemopexin complexes by soluble HxuA protein allows utilization of this complexed heme by Haemophilus influenzae. Infect. Immun. 66:4511-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cope, L. D., R. Yogev, U. Muller-Eberhard, and E. J. Hansen. 1995. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J. Bacteriol. 177:2644-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crosa, J. H. 1997. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol. Rev. 61:319-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elkins, C. 1995. Identification and purification of a conserved heme-regulated hemoglobin-binding outer membrane protein from Haemophilus ducreyi. Infect. Immun. 63:1241-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elkins, C., C. J. Chen, and C. E. Thomas. 1995. Characterization of the hgbA locus encoding a hemoglobin receptor from Haemophilus ducreyi. Infect. Immun. 63:2194-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everest, P. H., H. Goossens, J. P. Butzler, D. Lloyd, S. Knutton, J. M. Ketley, and P. H. Williams. 1992. Differentiated caco-2 cells as a model for enteric invasion by Campylobacter jejuni and Campylobacter coli. J. Med. Microbiol. 37:319-325. [DOI] [PubMed] [Google Scholar]
- 14.Frechon, D., and E. Le Cam. 1994. Fur (ferric uptake regulation) protein interaction with target DNA: comparison of gel retardation, footprinting and electron microscopy analyses. Biochem. Biophys. Res. Commun. 201:346-355. [DOI] [PubMed] [Google Scholar]
- 15.Genco, C. A., and D. W. Dixon. 2001. Emerging strategies in microbial haem capture. Mol. Microbiol. 39:1-11. [DOI] [PubMed] [Google Scholar]
- 16.Ghigo, J., S. Letoffe, and C. Wandersman. 1997. A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. J. Bacteriol. 179:3572-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant, K. A., and S. F. Park. 1995. Molecular characterization of katA from Campylobacter jejuni and generation of a catalase-deficient mutant of Campylobacter coli by interspecific allelic exchange. Microbiology 141:1369-1376. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 19.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 20.Henderson, D., and S. Payne. 1994. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J. Bacteriol. 176:3269-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson, D. P., E. E. Wyckoff, C. E. Rashidi, H. Verlei, and A. L. Oldham. 2001. Characterization of the Plesiomonas shigelloides genes encoding the heme iron utilization system. J. Bacteriol. 183:2715-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendrixson, D. R., and V. J. DiRita. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52:471-484. [DOI] [PubMed] [Google Scholar]
- 23.Holmes, K., F. Mulholland, B. M. Pearson, C. Pin, J. McNicholl-Kennedy, J. M. Ketley, and J. M. Wells. 2005. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151:243-257. [DOI] [PubMed] [Google Scholar]
- 24.Hossain, A., D. E. S. Tull, and J. H. Freer. 1993. Heat-labile and heat-stable haemolysins of Campylobacter jejuni. FEMS Immunol. Med. Microbiol. 6:331-339. [DOI] [PubMed] [Google Scholar]
- 25.Ketley, J. M. 1997. Pathogenesis of enteric infection by Campylobacter. Microbiology 143:5-21. [DOI] [PubMed] [Google Scholar]
- 26.Kikuchi, G., T. Yoshida, and M. Noguchi. 2005. Heme oxygenase and heme degradation. Biochem. Biophys. Res. Commun. 338:558-567. [DOI] [PubMed] [Google Scholar]
- 27.Korlath, J. A., M. T. Osterholm, L. A. Judy, K. C. Forfang, and R. A. Robinson. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152:592-596. [DOI] [PubMed] [Google Scholar]
- 28.Le Cam, E., D. Frechon, M. Barray, A. Fourcade, and E. Delain. 1994. Observation of binding and polymerization of Fur repressor onto operator-containing DNA with electron and atomic force microscopes. Proc. Natl. Acad. Sci. USA 91:11816-11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letoffe, S., P. Delepelaire, and C. Wandersman. 2004. Free and hemophore-bound heme acquisitions through the outer membrane receptor HasR have different requirements for the TonB-ExbB-ExbD complex. J. Bacteriol. 186:4067-4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis, L. A., M. H. Sung, M. Gipson, K. Hartman, and D. W. Dyer. 1998. Transport of intact porphyrin by HpuAB, the hemoglobin-haptoglobin utilization system of Neisseria meningitidis. J. Bacteriol. 180:6043-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, Y., and P. R. Ortiz de Montellano. 2000. Reaction intermediates and single turnover rate constants for the oxidation of heme by human heme oxygenase-1. J. Biol. Chem. 275:5297-5307. [DOI] [PubMed] [Google Scholar]
- 32.Mey, A. R., and S. M. Payne. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 42:835-849. [DOI] [PubMed] [Google Scholar]
- 33.Morton, D. J., A. Smith, Z. Ren, L. L. Madore, T. M. VanWagoner, T. W. Seale, P. W. Whitby, and T. L. Stull. 2004. Identification of a haem-utilization protein (Hup) in Haemophilus influenzae. Microbiology 150:3923-3933. [DOI] [PubMed] [Google Scholar]
- 34.Morton, D. J., P. W. Whitby, H. Jin, Z. Ren, and T. L. Stull. 1999. Effect of multiple mutations in the hemoglobin- and hemoglobin-haptoglobin-binding proteins, HgpA, HgpB, and HgpC, of Haemophilus influenzae type b. Infect. Immun. 67:2729-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mourino, S., C. R. Osorio, and M. L. Lemos. 2004. Characterization of heme uptake cluster genes in the fish pathogen Vibrio anguillarum. J. Bacteriol. 186:6159-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mourino, S., I. Rodriguez-Ares, C. R. Osorio, and M. L. Lemos. 2005. Genetic variability of the heme uptake system among different strains of the fish pathogen Vibrio anguillarum: identification of a new heme receptor. Appl. Environ. Microbiol. 71:8434-8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochsner, U. A., Z. Johnson, and M. L. Vasil. 2000. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146:185-198. [DOI] [PubMed] [Google Scholar]
- 38.Olczak, T., D. W. Dixon, and C. A. Genco. 2001. Binding specificity of the Porphyromonas gingivalis heme and hemoglobin receptor HmuR, gingipain K, and gingipain R1 for heme, porphyrins, and metalloporphyrins. J. Bacteriol. 183:5599-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palyada, K., D. Threadgill, and A. Stintzi. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186:4714-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 41.Pickett, C. L., T. Auffenberg, E. C. Pesci, V. L. Sheen, and S. S. Jusuf. 1992. Iron acquisition and hemolysin production by Campylobacter jejuni. Infect. Immun. 60:3872-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pickett, C. L., and R. B. Lee. 2005. Cytolethal distending toxin, p. 385-396. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter: molecular and cellular biology. Horizon Bioscience, Wymondham, United Kingdom.
- 43.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 44.Schaible, U. E., and S. H. Kaufmann. 2004. Iron and microbial infection. Nat. Rev. Microbiol. 2:946-953. [DOI] [PubMed] [Google Scholar]
- 45.Simpson, W., T. Olczak, and C. A. Genco. 2000. Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J. Bacteriol. 182:5737-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skaar, E. P., A. H. Gaspar, and O. Schneewind. 2004. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 279:436-443. [DOI] [PubMed] [Google Scholar]
- 47.Srikumar, R., L. G. Mikael, P. D. Pawelek, A. Khamessan, B. F. Gibbs, M. Jacques, and J. W. Coulton. 2004. Molecular cloning of haemoglobin-binding protein HgbA in the outer membrane of Actinobacillus pleuropneumoniae. Microbiology 150:1723-1734. [DOI] [PubMed] [Google Scholar]
- 48.Stintzi, A., D. Marlow, K. Palyada, H. Naikare, R. Panciera, L. Whitworth, and C. Clarke. 2005. Use of genome-wide expression profiling and mutagenesis to study the intestinal lifestyle of Campylobacter jejuni. Infect. Immun. 73:1797-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stojiljkovic, I., and K. Hantke. 1992. Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 11:4359-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stojiljkovic, I., and K. Hantke. 1994. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol. Microbiol. 13:719-732. [DOI] [PubMed] [Google Scholar]
- 51.Stojiljkovic, I., V. Hwa, L. de Saint Martin, P. O'Gaora, X. Nassif, F. Heffron, and M. So. 1995. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol. Microbiol. 15:531-541. [DOI] [PubMed] [Google Scholar]
- 52.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 53.Suits, M. D., G. P. Pal, K. Nakatsu, A. Matte, M. Cygler, and Z. Jia. 2005. Identification of an Escherichia coli O157:H7 heme oxygenase with tandem functional repeats. Proc. Natl. Acad. Sci. USA 102:16955-16960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tai, S. S., T. R. Wang, and C. J. Lee. 1997. Characterization of hemin binding activity of Streptococcus pneumoniae. Infect. Immun. 65:1083-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas, C. E., B. Olsen, and C. Elkins. 1998. Cloning and characterization of tdhA, a locus encoding a TonB-dependent heme receptor from Haemophilus ducreyi. Infect. Immun. 66:4254-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson, J. M., H. A. Jones, and R. D. Perry. 1999. Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmu mutants for hemin and hemoprotein utilization. Infect. Immun. 67:3879-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Vliet, A., A. Wood, J. Henderson, K. Wooldridge, and J. Ketley. 1998. Genetic manipulation of enteric Campylobacter species, p. 407-419. In P. Williams, J. Ketley, and G. Salmond (ed.), Techniques for bacterial pathogenesis. Academic Press, London, United Kingdom.
- 59.van Vliet, A. H., J. M. Ketley, S. F. Park, and C. W. Penn. 2002. The role of iron in Campylobacter gene regulation, metabolism and oxidative stress defense. FEMS Microbiol. Rev. 26:173-186. [DOI] [PubMed] [Google Scholar]
- 60.van Vliet, A. H., K. G. Wooldridge, and J. M. Ketley. 1998. Iron-responsive gene regulation in a Campylobacter jejuni Fur mutant. J. Bacteriol. 180:5291-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Vliet, A. H. M., M. L. A. Baillon, C. W. Penn, and J. M. Ketley. 2001. The iron-induced ferredoxin FdxA of Campylobacter jejuni is involved in aerotolerance. FEMS Microbiol. Lett. 196:189-193. [DOI] [PubMed] [Google Scholar]
- 62.van Vliet, A. H. M., J. D. Rock, L. N. Madeleine, and J. M. Ketley. 2000. The iron-responsive regulator Fur of Campylobacter jejuni is expressed from two separate promoters. FEMS Microbiol. Lett. 188:115-118. [DOI] [PubMed] [Google Scholar]
- 63.Wilks, A., and M. P. Schmitt. 1998. Expression and characterization of a heme oxygenase (HmuO) from Corynebacterium diphtheriae. Iron acquisition requires oxidative cleavage of the heme macrocycle. J. Biol. Chem. 273:837-841. [DOI] [PubMed] [Google Scholar]
- 64.Wooldridge, K. G., P. H. Williams, and J. M. Ketley. 1994. Iron-responsive genetic regulation in Campylobacter jejuni: cloning and characterization of a fur homolog. J. Bacteriol. 176:5852-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wosten, M. M., M. Boeve, M. G. Koot, A. C. van Nuene, and B. A. van der Zeijst. 1998. Identification of Campylobacter jejuni promoter sequences. J. Bacteriol. 180:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wyckoff, E. E., D. Duncan, A. G. Torres, M. Mills, K. Maase, and S. M. Payne. 1998. Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol. Microbiol. 28:1139-1152. [DOI] [PubMed] [Google Scholar]
- 67.Wyckoff, E. E., M. Schmitt, A. Wilks, and S. M. Payne. 2004. HutZ is required for efficient heme utilization in Vibrio cholerae. J. Bacteriol. 186:4142-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshida, T., and G. Kikuchi. 1978. Purification and properties of heme oxygenase from pig spleen microsomes. J. Biol. Chem. 253:4224-4229. [PubMed] [Google Scholar]
- 69.Yuki, N. 2005. Carbohydrate mimicry: a new paradigm of autoimmune diseases. Curr. Opin. Immunol. 17:577-582. [DOI] [PubMed] [Google Scholar]
- 70.Zhu, W., D. J. Hunt, A. R. Richardson, and I. Stojiljkovic. 2000. Use of heme compounds as iron sources by pathogenic neisseriae requires the product of the hemO gene. J. Bacteriol. 182:439-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu, W., A. Wilks, and I. Stojiljkovic. 2000. Degradation of heme in gram-negative bacteria: the product of the hemO gene of neisseriae is a heme oxygenase. J. Bacteriol. 182:6783-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]