Abstract
Cancer cells contain numerous clonal mutations, i.e., mutations that are present in most or all malignant cells of a tumor and have presumably been selected because they confer a proliferative advantage. An important question is whether cancer cells also contain a large number of random mutations, i.e., randomly distributed unselected mutations that occur in only one or a few cells of a tumor. Such random mutations could contribute to the morphologic and functional heterogeneity of cancers and include mutations that confer resistance to therapy. We have postulated that malignant cells exhibit a mutator phenotype resulting in the generation of random mutations throughout the genome. We have recently developed an assay to quantify random mutations in human tissue with unprecedented sensitivity. Here, we report measurements of random single-nucleotide substitutions in normal and neoplastic human tissues. In normal tissues, the frequency of spontaneous random mutations is exceedingly low, less than 1 × 10−8 per base pair. In contrast, tumors from the same individuals exhibited an average frequency of 210 × 10−8 per base pair, an elevation of at least two orders of magnitude. Our data document tumor heterogeneity at the single-nucleotide level, indicate that accelerated mutagenesis prevails late into tumor progression, and suggest that elevation of random mutation frequency in tumors might serve as a novel prognostic indicator.
Keywords: genetic instability, random mutation frequency, tumor heterogeneity, point mutation instability (PIN), carcinogenesis
Somatic mutations are a hallmark of human cancer (1). Cancer cells contain numerous clonal mutations, such as those in p53 (2, 3) and ras (4), as well as chromosomal aberrations involving transposition, deletion, or insertion of millions of nucleotides (5). Underlying these well recognized genomic alterations may be an even higher frequency of randomly distributed unselected mutations that would be present in only one or a few cells of a tumor. Large numbers of random mutations could contribute to the heterogeneity of cancer cells in a tumor, the rapid emergence of resistance to radiation and chemotherapy, and the ability of cancer cells to invade adjacent tissues and to metastasize. Endeavors to elucidate the frequency of genetic changes in cancer have been largely restricted to the documentation of clonally expanded mutations in tumor populations (6–8). Unfortunately, because of the limited sensitivity of mutational assays, the measurement of random mutations in normal and tumor tissues has not been feasible.
We have recently established a method for quantifying random mutations in cell populations, called the random mutation capture (RMC) assay (9). The RMC assay is >100-fold more sensitive than previous methods that employ genomic selection, permits analysis of a large number of nucleotides, and can identify one mutant base pair among 109 wild-type nucleotides. We have used the RMC method to show that mutations in cultured normal human diploid fibroblasts are very infrequent (1.6 × 10−8 mutations per base pair) (9). Here, we use the RMC assay to measure the frequency of random mutations in normal and neoplastic human tissue. We report that, in addition to chromosomal instability (CIN) (10, 11) and microsatellite instability (MIN) (12–14), the genomes of cancer cells display genetic instability in the form of greatly elevated frequencies of random single-nucleotide substitutions. Our findings are in accord with the proposal that cancers exhibit a mutator phenotype (15, 16) and indicate that the phenotype is ongoing late in tumor evolution.
Results
The RMC Assay Quantifies Random Point Mutations in Single DNA Molecules from Human Tissue.
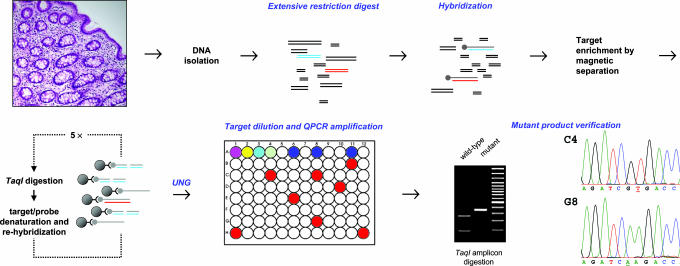
We have adapted the RMC assay for application to human tissue, as illustrated in Fig. 1. The assay involves initial enrichment of the mutational target sequence, a TaqI restriction site (TCGA) in intron VI of p53, by repeated hybridization to a biotin-labeled probe and magnetic bead separation. Nucleotide substitutions in the enriched target sequence that render it resistant to TaqI cleavage are then quantified by dilution to single molecules followed by real-time quantitative PCR (QPCR) amplification. By avoiding the limitations associated with sequencing of large populations of DNA molecules and misincorporation during PCR amplification, the RMC assay provides greater sensitivity than previous methods. We have shown that mutations in the target sequence are genetically neutral, i.e., that they impart neither positive nor negative selection to cells in culture (9).
Fig. 1.
The RMC applied to human tissue. Genomic DNA is isolated from intact tissue and digested with restriction enzymes that do not cut the mutational target sequence, a 4-bp TaqI restriction site (TCGA) in intron VI of p53; blue lines represent the wild-type target sequence, and red lines represent mutant target sequence. A complementary probe (gray lines) that contains dUMP in place of dTMP and a biotinylated nucleotide at the 5′ terminus is hybridized to the mutational target. The hybridized target is isolated by complexing to magnetic beads, digested with TaqI (cleaving the TCGA target site in the wild-type sequence and failing to cleave if a nucleotide substitution is present at that site), and denatured. Rehybridization and TaqI digestion are carried out four times. The probe is then disabled for further hybridization by digestion with uracil-DNA glycosylase, and the mutational target is diluted in 96-well plates so that 1 in ≈10 wells contains a PCR-amplifiable product (red wells) as measured with SYBR green by using real-time QPCR. The mutation frequency is quantified by QPCR amplification and is calculated as the number of wells containing a mutant sequence divided by the product of the total number of target molecules screened and the restriction site length (bp). The mutant sequence of the amplified product in all positive wells is verified by DNA sequencing; C4 and G8 represent the mutant sequences found in wells C4 and G8, respectively, of the 96-well plate shown. In some cases, preliminary verification was carried out by redigestion with TaqI.
Random Mutation Frequency in Normal Human Tissue.
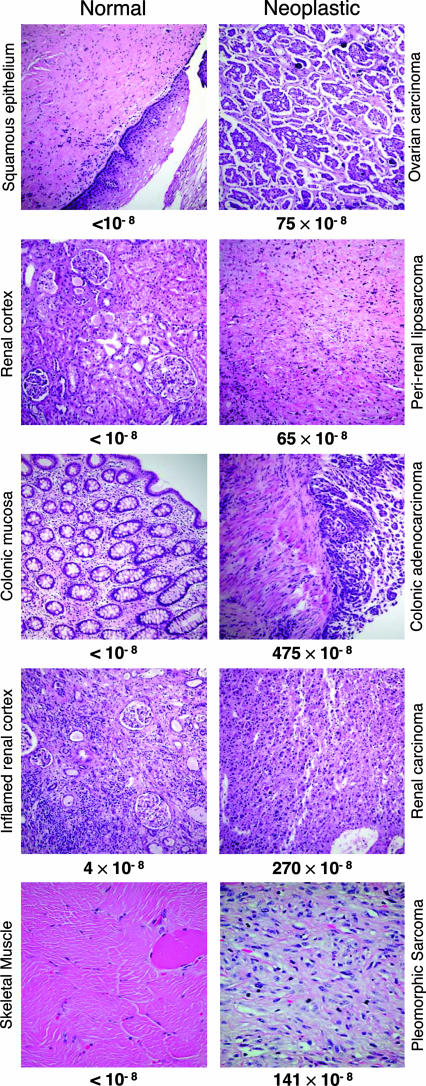
We analyzed a set of tissue pairs, each pair consisting of matching normal and tumor tissue from a different patient who had not been treated with radiation or chemotherapy. The normal tissues were examined histologically to confirm their identity, as illustrated in Fig. 2. They were then microdissected to ensure that the samples subjected to mutational analysis were free of detectable tumor cells. All samples (including the concurrently processed paired tumors, see below) were then coded so that their identity was unknown to all investigators conducting the mutation assays. The mutation frequency we observed in the normal samples (squamous epithelium, renal cortex, colon epithelium, skeletal muscle) is less than 1 in 108 (Table 1), consistent with the frequency we measured (1.6 × 10−8) in cultured human diploid fibroblasts (9). In total, we assayed >500 megabases of DNA from normal tissues and detected mutations in only one sample. This exceptional sample (the inflamed renal cortex in Fig. 2) was distinguished by lymphocytic infiltration, consistent with the concept that inflammation may be a key factor in the neoplastic process (17, 18). The low frequency of random mutations we measured at intron VI of p53 is consistent with the frequencies observed in circulating human lymphocytes at the HPRT locus by selection for 6-thioguanine resistance (19).
Fig. 2.
Matched normal and neoplastic tissues analyzed in the RMC assay. Hematoxylin/eosin-stained sections of the paired normal (Left) and tumor (Right) tissues listed in Table 1 are shown. The tissues are normal squamous vaginal epithelium and high-grade papillary serous ovarian carcinoma with psammoma bodies; normal renal cortex and dedifferentiated sclerosing perirenal liposarcoma; normal colonic mucosa and invasive colonic adenocarcinoma; renal cortex with lymphocytic inflammation and malignant renal epithelioid angiomyolipoma; and normal skeletal muscle and high-grade malignant fibrous histiocytoma pleomorphic sarcoma. Immunohistochemical analysis of MLH1 and MSH2 proteins involved in DNA mismatch repair showed the colonic adenocarcinoma to lack MLH1 expression and to have normal expression of MSH2. These results suggest that the tumor is defective in mismatch repair. The mutation frequencies measured in the RMC assay are indicated below each section.
Table 1.
Random mutation frequency in human tissues and cells
| Normal | Nucleotides analyzed ×10−6 | Mutation frequency* ×108 | Neoplastic | Nucleotides analyzed ×10−6 | Mutation frequency* ×108 Tissues† |
|---|---|---|---|---|---|
| Squamous epithelium | 115 | <1 | Ovarian carcinoma | 18 | 75 |
| Renal cortex | 108 | <1 | Perirenal liposarcoma | 24 | 65 |
| Colonic mucosa | 115 | <1 | Colonic adenocarcinoma | 10 | 475 |
| Inflamed renal cortex | 55 | 4 | Renal carcinoma | 15 | 270 |
| Skeletal muscle | 110 | <1 | Pleomorphic sarcoma | 15 | 141 |
| — | Non-Hodgkin's lymphoma | 27 | 300 | ||
| Cultured fibroblasts‡ | |||||
| Untreated | 218 | 2 | ENU-treated§ | 24 | 175 |
*Measured in the RMC assay.
†Normal and neoplastic tissues listed in the same row are paired samples from the same individual.
‡Data for cultured normal dermal fibroblasts are from ref. 9.
§Treated with 1 mg/ml N-ethyl-N-nitrosourea for 1 h.
Elevated Random Mutation Frequency in Cancers.
To assess the prevalence of random mutations in human cancers, we initially examined a lymph node involved by non-Hodgkin's lymphoma. The mutation frequency was 300 × 10−8, 190-fold greater than that observed in cultured human fibroblasts (1.6 × 10−8) (Table 1). For comparison, Table 1 records the mutation frequency of 175 × 10−8 that we found for normal human diploid fibroblasts treated with an extremely high dose (1 mg/ml) of the potent mutagen N-ethyl-N-nitrosourea (9).
The well demarcated, sporadic tumors belonging to the matched tissue pairs were examined histologically to confirm the diagnosis (Fig. 2). All tumors were highly anaplastic, containing cells that displayed differences in morphology, size, and nuclear staining (Fig. 2). In total we screened >100 megabases of target sequence in tumor DNA (Table 1). In contrast to the paucity of mutations in normal tissues, all tumors exhibited high levels of mutation (Table 1), ranging from 65 × 10−8 for a perirenal liposarcoma to 475 × 10−8 for a colon adenocarcinoma. The mean frequency in the tumors was 210 × 10−8, representing a >200-fold elevation relative to the matching normal tissues. Assuming that the “less than” values for normal tissue samples are equal to 1 × 10−8 (the upper limit), the difference in the median mutation frequencies of the tumor and normal cell populations is statistically significant (P = 0.009, Wilcoxon rank-sum test). The large elevation provides strong evidence that at least some human cancers are genetically unstable at the single-nucleotide level, in accord with a mutator phenotype (15).
The Spectrum of Random Mutations in Human Cancers.
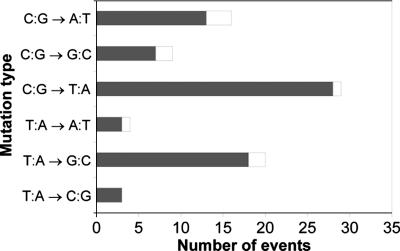
Every mutation that rendered DNA resistant to TaqI cleavage was verified by sequencing. Sequence analysis of all tumor mutations indicates that the majority were not extensively expanded and suggests that they arose from distinct mutational events that occurred after the last clonal expansion (Fig. 3). In every case, multiple occurrences of the same mutation were scored as one single mutational event, as in Fig. 3. All mutations were single-base substitutions, the most frequent being C·G to T·A transitions and T·A to G·C transversions. The transitions in particular are consistent with, but not necessarily diagnostic of or limited to, misincorporation by replicative DNA polymerases in the absence of DNA damage (20). In contrast to these results, T·A to A·T transversions were the most frequent substitutions detected in N-ethyl-N-nitrosourea-treated human fibroblasts by using the same procedure (9).
Fig. 3.
Tumor mutation spectrum. DNA sequencing of all mutants recovered from tumors showed that C·G to T·A transitions were the most common mutation, and the sequencing permitted distinction between independent random mutational events (gray bars, mutation observed only once) and expansion of mutant clones (white bars, same mutation recovered more than once in the same tumor). Identical mutations observed more than once (expanded mutations) are recorded as one single event; the number of these minority events is indicated by the relatively short length of the white bar extending past the gray bar.
Discussion
Our understanding of human cancer, and our ability to treat and prevent it, depends critically on knowledge of the mechanisms and pathways of tumor evolution. It has become apparent that both genetic and epigenetic changes underlie tumorigenesis (21). Genetic instability in cancer cells is evidenced by chromosome aberrations (chromosomal instability or CIN); extensive chromosomal microheterogeneity has been observed among single metastatic cells that arose from the same tumor (22). Genetic instability is further manifested by changes in the length of microsatellite sequences (microsatellite instability or MIN), as well as by clonal mutations, including mutations in oncogenes and tumor suppressor genes. We report here that the genomes of human cancer cells also display greatly elevated frequencies of random nucleotide point mutations (point mutation instability or PIN, in parallel with the above designations).
Our results provide strong support for the hypothesis that cancer cells express a mutator phenotype at the single-nucleotide level (16). The mutator phenotype hypothesis states that normal mutation rates are insufficient to account for the multiple mutations observed in human tumors, and that cancer cells must therefore incur increased rates of mutagenesis. The increased mutation would occur genome-wide, and would affect genes that are required for genomic stability. In principle then, an increase in PIN could contribute to both CIN and MIN, in accord with the observations that clonal single-nucleotide substitutions in many genes have been associated with both phenotypes (23–25).
Estimates of the frequency of nucleotide substitutions in human cancers have been largely restricted to documentation of clonally expanded mutations in tumor cell populations (26, 27). These studies used sequencing reactions containing populations of DNA molecules, and they did not score mutations present in only one or a few cells, because conventional sequencing technology does not permit detection of mutations present in <10% of cells. Thus, the reported consensus sequence is not informative of mutations that were not subject to extensive clonal expansion. Accordingly, conventional DNA sequencing vastly underestimates the number of mutations present in a tumor. To unmask the full extent of heterogeneity, it is necessary to measure random mutations, and to do this, it is necessary to analyze single DNA molecules.
We present here an analysis of mutation frequency in single DNA molecules from human tissues. Our estimate of the frequency of random mutation in normal tissues, <1 × 10−8 mutations per base pair (Table 1), represents an upper limit based on screening of 500 megabases of DNA. The mean mutation frequency we observed in tumors (210 × 10−8 mutations per base pair) is elevated at least 200-fold relative to matching normal tissue (Table 1). Sequence analysis of all mutations detected in tumors indicates that most of the mutants are either not expanded or are not extensively expanded, but rather are present in only one or a few cells that constitute an individual tumor (Fig. 3). We note that, in determining mutation frequency, we scored multiple occurrences of the same mutation (expanded mutations) as a single mutational event, equivalent to a mutation we observed only once. Unexpanded mutations represent genetic changes that occurred after the last round of clonal expansion that included the mutated cells (28). The mutants that we detected multiple times (from 2 to 27 times) represent modest expansions affecting a small fraction of tumor cells. Because our mutational target is genetically neutral [i.e., confers neither a selective growth advantage nor a disadvantage (9)], expansion of cells harboring mutant sequences could reflect a proliferative advantage conferred by another mutation(s) elsewhere in the genome; in other words, expanded mutant sequences could be “passenger” or “piggy-back” mutations. Alternatively, they could occur simply by chance.
In the clinically manifest tumors we studied, unexpanded mutations are indicative of late occurrence in tumor evolution, the high random mutation frequency is indicative of ongoing expression of a mutator phenotype, and expanded mutations may be indicative of ongoing selection of advantageous mutations elsewhere in the genome. Importantly, the elevated frequency of mutations cannot be explained by increased rounds of proliferation alone, but must include an enhanced rate of mutation in tumors. This is so because (i) mutations at the target site are genetically neutral [there is no selection (9)] and (ii) expansion of distinct mutations by proliferation is scored as a single mutational event. In other words, because we report the same genetic change in multiple DNA molecules as one mutation, the calculated frequency of distinct unselected mutational events (Table 1) is unaffected by increased rounds of proliferation.
It is plausible that a mutator phenotype could be detrimental late in tumor evolution when the tumor is relatively well established and adapted, and that persistence of accelerated mutagenesis might therefore be selected against. However, mathematical models do not indicate that negative clonal selection would mitigate against a mutator phenotype (29, 30). Moreover, we find clear evidence for ongoing elevation of mutagenesis in at least some clinically detected tumors. Our evidence that enhanced mutagenesis is an ongoing process in at least some clinically manifest tumors carries the implication that intervention to inhibit this process may impede progression and metastasis after diagnosis.
Emerging data underscore the heterogeneity of mutations in tumors among different individuals and in cancer cells within each tumor. In colorectal cancer, for example, the progression from adenoma to carcinoma has been associated with sequential mutations in APC, K-ras, and p53 (31). Yet, only 7% of these tumors have mutations in all three genes, implying that multiple pathways are involved in colorectal tumorigenesis (32). Early DNA sequencing studies indicated that a limited number of cancer-related genes were mutated in individual tumors, and that these might provide targets for drug development (33). However, in breast and colon cancer, recent work (34) has revealed increasing complexity of clonally expanded mutations. Sequencing of 13,023 genes in 11 breast and 11 colorectal cancers yielded 89 and 126 different genes that were mutated, respectively (34). Moreover, only a small subset of these genes was found to be mutated at significant frequency in either cancer. The diversity of clonal mutations among tumors, theorized to be generated early in tumorigenesis by a mutator phenotype (35), together with the large number of late-arising random substitutions demonstrated here, emphasizes that the heterogeneity of mutations in tumors may be greater than has been appreciated.
The presence of large numbers of random mutations within tumors could limit the efficacy of targeted therapies. By the time a tumor is clinically detected it contains ≈109 cancer cells. The average frequency of random mutations in tumor samples we analyzed was 2.2 × 10−6 per base pair. Thus, each cell would contain more than a thousand random mutations, and the entire tumor could harbor as many as 1012 different single-nucleotide substitutions. Many of these mutations would alter the properties of the encoded proteins, including mutations that confer resistance to radio-, chemo-, and/or immunotherapy (36). Thus, increased genetic variability in newly diagnosed cancers could encompass a reservoir of mutations available for immediate clonal expansion upon initiation of treatment with any given agent, leading to rapid emergence of resistance. This concept provides a molecular basis for the observed clinical efficacy of combination therapy, because any single cell would be unlikely to contain mutations that confer resistance to agents with different mechanisms of cytotoxicity. It can be hypothesized that tumors with fewer random mutations should be treated more conservatively, whereas tumors with a higher frequency of random mutations should be treated more aggressively and with combination therapies. Thus, mutation frequency could provide a new index for stratification of tumors. One possibility is that mutation frequency will exhibit an overall positive association with tumor stage and grade, but that there will be significant variability within defined stages and grades. This variability, which may contribute to differences in within-group outcome, could help to guide therapy for individual patients.
Materials and Methods
Tissues and DNA Isolation.
Tissues were obtained as anonymous samples from the Department of Pathology, University of Washington. The specimens were frozen in liquid nitrogen and stored at −70°C. Hematoxylin/eosin-stained sections were reexamined by pathologists to confirm tissue identity and tumor diagnoses. Tissues were microdissected to obtain normal tissue samples that were free of detectable tumor cells and tumor samples that were composed of at least 90% tumor cells. Tissue samples (400 mg) were immersed in 20 ml of digestion buffer (800 mM guanidine·HCl/30 mM Tris·HCl, pH 8.0/30 mM EDTA/5% Tween-20/0.5% Triton X-100/2 mg/ml proteinase K/200 μg/ml RNase A) and thoroughly homogenized mechanically with an UltraTurrax T25 homogenizer (IKA Works, Wilmington, NC). The homogenate was incubated at 50°C for 2 h and applied to a prewashed 500/G genomic-tip column (Qiagen, Valencia, CA) for DNA recovery according to the manufacturer's directions.
RMC Assay.
The procedure for quantification of random mutations (9) is outlined here with modifications for DNA from human tissues. Briefly, 400 μg of purified genomic DNA is incubated with the following restriction enzymes that do not cut the target sequence: 100 units each of PvuI and RsaI and 200 units each of EcoRI, EcoRV, and BamHI. The digested DNA is hybridized with a 100-fold excess of the complementary probe that contains dUMP in place of dTMP and a biotinylated nucleotide at the 5′ terminus. This complementary sequence was generated by copying the cloned target in reactions containing Taq DNA polymerase, a 5′-biotin-terminated oligonucleotide, and 200 μM dUTP in place of dTTP. The hybridized target is isolated by magnetic separation after complexing with streptavidin coupled to superparamagnetic polymer spheres (Dynabeads; Dynal Biotech, Lake Success, NY). The total number of target molecules in each sample is determined by dilution and PCR amplification. Mutation in the target site TCGA is determined by digesting the hybridized DNA with TaqI, which cleaves the site in the wild-type sequence and fails to cleave if a nucleotide substitution is present at that site. Incubation is carried out with TaqI at 65°C for 1 h, and denaturation is at 95°C for 5 min. The digested product is heat-denatured and rehybridized to the probe. To cleave all wild-type sequences, the restriction digestion protocol is iterated five times. The probe is disabled for further hybridization by digestion with uracil-DNA glycosylase, and the target molecule is diluted and displayed in a 96-well format.
Mutation Frequency Calculation.
The extent of DNA copy dilution is determined in preliminary experiments so that 1 in ≈10 wells contains a mutant PCR-amplifiable product as measured with SYBR green by using real-time QPCR. The total number of target molecules in each well is precisely established by using a standard QPCR dilution curve by amplification using control primers that flank regions distant from the TaqI restriction site (see Fig. 1, row A of the 96-well plate). The mutation frequency is equal to the number of wells that contain a mutant sequence, as determined by using primers that flank the TaqI site (Fig. 1, rows B–H in the 96-well plate), divided by the total number of target base pairs screened. For example, in an experiment where it was determined that 100,000 TaqI sites were seeded in each of the 84 mutant detection wells, we calculate that a total of 33.6 × 106 bp were screened, as follows: 84 wells × 100,000 sites per well × 4 bp per TaqI site = 33.6 × 106 bp. If 8 mutant TaqI sites were detected by the generation of an amplicon over the restriction site, the mutant frequency is 2.4 × 10−7 per bp (8 mutants ÷ 33.6 × 106 bp).
Acknowledgments
We thank A. Blank for editing the manuscript and A. Blank, G. M. Martin, and N. Fausto for insightful comments. This work was funded by Grants CA78885 and CA102029 from the National Cancer Institute (to L.A.L.). A Postdoctoral Fellowship from the Natural Sciences and Engineering Research Council of Canada (NSERC), followed by a Canadian Institutes of Health Research (CIHR) Fellowship provided support for J.H.B. during the completion of these studies.
Abbreviations
- QPCR
quantitative PCR
- RMC
random mutation capture.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
See Commentary on page 18033.
References
- 1.Weinberg RA. Biology of Cancer. New York: Garland Science; 2007. [Google Scholar]
- 2.Levine AJ. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 3.Hussain SP, Amstad P, Raja K, Ambs S, Nagashima M, Bennett WP, Shields PG, Ham A-J, Swenberg JA, Marrogi AJ, Harris CC. Cancer Res. 2000;60:3333–3337. [PubMed] [Google Scholar]
- 4.Barbacid M. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 5.Albertson DG, Collins C, McCormick F, Gray JW. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 6.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 7.Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, Bignell G, Teague J, Smith R, Stevens C. Nat Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 8.Wang TL, Rago C, Silliman N, Ptak J, Markowitz S, Willson JK, Parmigiani G, Kinzler KW, Vogelstein B, Velculescu VE. Proc Natl Acad Sci USA. 2002;99:3076–3080. doi: 10.1073/pnas.261714699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bielas JH, Loeb LA. Nature Methods. 2005;2:285–290. doi: 10.1038/nmeth751. [DOI] [PubMed] [Google Scholar]
- 10.Meyn MS. In: Genetic Instability and Tumorigenesis. Kastan MB, editor. Berlin: Springer; 1997. pp. 71–148. [Google Scholar]
- 11.Lengauer C, Kinzler KW, Vogelstein B. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 12.Perucho M. Biol Chem. 1996;377:675–684. [PubMed] [Google Scholar]
- 13.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 14.Fishel R, Lescoe MK, Rao MRS, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 15.Loeb LA, Springgate CF, Battula N. Cancer Res. 1974;34:2311–2321. [PubMed] [Google Scholar]
- 16.Loeb LA, Loeb KR, Anderson JP. Proc Natl Acad Sci USA. 2003;100:776–781. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hussain SP, Hofseth LJ, Harris CC. Nat Rev Cancer. 2003;3:276–285. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- 18.Coussens LM, Werb Z. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albertini RJ, Nicklas JA, O'Neill JP, Robison SH. Annu Rev Genet. 1990;24:305–326. doi: 10.1146/annurev.ge.24.120190.001513. [DOI] [PubMed] [Google Scholar]
- 20.Kunkel TA, Bebenek K. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 21.Jones PA, Baylin SB. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 22.Klein CA, Schmidt-Kittler O, Schardt JA, Pantel K, Speicher MR, Riethmuller G. Proc Natl Acad Sci USA. 1999;96:4494–4499. doi: 10.1073/pnas.96.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markowitz S, Wang J, Myeroff L, Parsons R, Sun LZ, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, et al. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 24.Malkhosyan S, McCarty A, Sawai H, Perucho M. Mutat Res. 1996;316:249–259. doi: 10.1016/s0921-8734(96)90007-7. [DOI] [PubMed] [Google Scholar]
- 25.Eshleman JR, Lang EZ, Bowerfind GK, Parsons R, Vogelstein B, Willson JKV, Veigl ML, Sedwick WD, Markowitz SD. Oncogene. 1995;10:33–37. [PubMed] [Google Scholar]
- 26.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. Nat Rev Cancer. 2004;4:117–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, Ptak J, Stillman N, Peters BA, van der Heijden MS, et al. Science. 2004;304:1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 28.Nowell PC. Adv Cancer Res. 1993;62:1–17. doi: 10.1016/s0065-230x(08)60313-9. [DOI] [PubMed] [Google Scholar]
- 29.Iwasa Y, Michor F, Nowak MA. Genetics. 2004;166:1571–1579. doi: 10.1534/genetics.166.3.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beckman RA, Loeb LA. Genetics. 2005;171:2123–2131. doi: 10.1534/genetics.105.040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fearon ER, Vogelstein B. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 32.Smith G, Carey FA, Beattie J, Wilkie MJV, Lightfoot TJ, Coxhead J, Garner RC, Steele RJC, Wolf CR. Proc Natl Acad Sci USA. 2002;99:9433–9438. doi: 10.1073/pnas.122612899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang TL, Rago C, Silliman N, Ptak J, Markowitz S, Willson JKV, Parmigiani G, Kinzler KW, Vogelstein B, Velculescu VE. Proc Natl Acad Sci USA. 2002;99:3076–3080. doi: 10.1073/pnas.261714699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber T, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 35.Loeb LA. Cancer Res. 2001;61:3230–3239. [PubMed] [Google Scholar]
- 36.Soverini S, Martinelli G, Rosti G, Bassi S, Amabile M, Poerio A, Giannini B, Trabacchi E, Castagnetti F, Testoni N, et al. J Clin Oncol. 2005;23:4100–4109. doi: 10.1200/JCO.2005.05.531. [DOI] [PubMed] [Google Scholar]