Abstract
We have created a hybrid i-motif composed of two DNA and two peptide nucleic acid (PNA) strands from an equimolar mixture of a C-rich DNA and analogous PNA sequence. Nano-electrospray ionization mass spectrometry confirmed the formation of a tetrameric species, composed of PNA–DNA heteroduplexes. Thermal denaturation and CD experiments revealed that the structure was held together by C-H+-C base pairs. High resolution NMR spectroscopy confirmed that PNA and DNA form a unique complex comprising five C-H+-C base pairs per heteroduplex. The imino protons are protected from D2O exchange suggesting intercalation of the heteroduplexes as seen in DNA4 i-motifs. FRET established the relative DNA and PNA strand polarities in the hybrid. The DNA strands were arranged antiparallel with respect to one another. The same topology was observed for PNA strands. Fluorescence quenching revealed that both PNA–DNA parallel heteroduplexes are intercalated, such that both DNA strands occupy one of the narrow grooves. H1′–H1′ NOEs show that both heteroduplexes are fully intercalated and that both DNA strands are disposed towards a narrow groove, invoking sugar–sugar interactions as seen in DNA4 i-motifs. The hybrid i-motif shows enhanced thermal stability, intermediate pH dependence and forms at relatively low concentrations making it an ideal nanoscale structural element for pH-based molecular switches. It also serves as a good model system to assess the contribution of sugar–sugar contacts in i-motif tetramerization.
INTRODUCTION
Oligonucleotides having tandem repeats of cytosine have been shown to associate under acidic conditions to form a tetrameric structure called the i-motif (1). The i-motif consists of two parallel stranded duplexes that are intercalated in an antiparallel orientation. Each parallel duplex is held together via C-H.C+ base pairs that are formed by hemi-protonation of the cytosine bases at acidic pH. Oligonucleotide fragments from naturally occurring sequences in the human genome have been shown to form the i-motif suggesting that this unique structural motif could possibly be biologically relevant. Several such sequences form intramolecular and intermolecular i-motifs in vitro. Tetrahymena thermophila and human telomeric repeats form tetramolecular i-motifs (2,3). Double and quadruple repeats of the tetrahymena and human telomeric C-rich strand also form bimolecular and unimolecular i-motifs, respectively (4–7). In addition, centromeric sequences have been shown to form bimolecular i-motifs (8). The human insulin minisatellite (9) and the human centromeric satellite (10) also form unimolecular i-motif structures. Select non-telomeric and non-centromeric sequences, such as the fragile X repeat, also fold into intramolecular i-motifs (11) (Chart 1).
Chart 1.
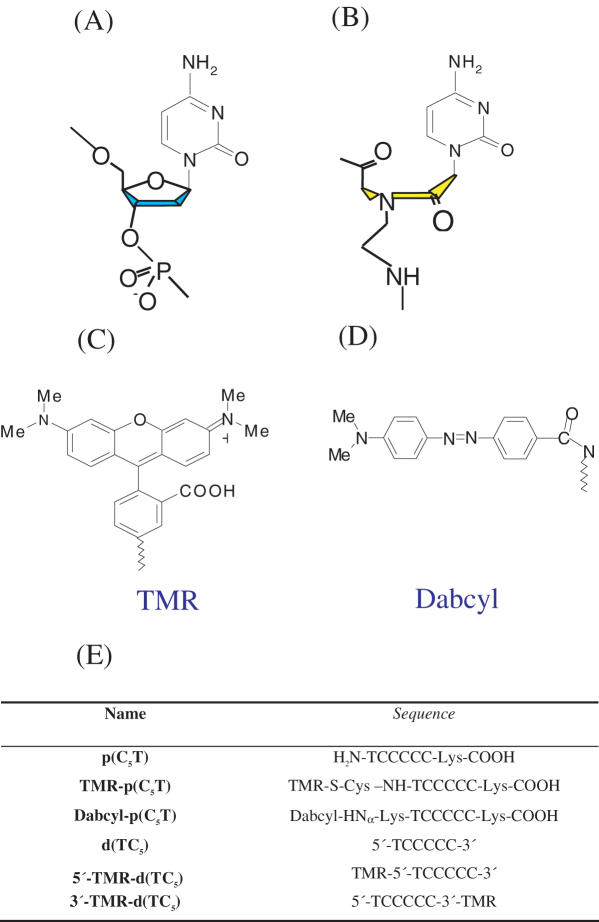
Molecular structures of (A) DNA, (B) PNA backbones, (C) fluorophore, (D) quencher and (E) sequences of PNA and DNA oligomers used in this study
Non-DNA based C-rich sequences have also been shown to form i-motifs. DNA phosphorothioate analogues, but not methyl phosphonate analogues, were also capable of i-motif formation (12). C-rich RNA-based i-motifs had gone undetected till recently due to the low stability of RNA-based i-motifs (13). Along these lines, i-motifs from peptide nucleic acids (PNAs) had also gone undetected until recently due to its stringent pH requirements for i-motif formation (14). Another analogue of PNA, called Alanyl PNA was also capable of forming i-motifs in the presence of an appropriate chiral partner (15). Such studies using structural analogues of the DNA backbone have provided useful information regarding the key backbone structural requirements for i-motif formation. The investigations on modified backbones have mainly implicated the role of DNA sugar–sugar interactions in stabilizing the i-motif. However, there has been no direct measurement yet of the effect of the deoxyribose phosphate backbone in bringing about tetramerization in solution.
PNA are one of the most widely used synthetic mimics of DNA. PNA can form a variety of non-covalent complexes analogous to their parent DNA complexes. Some of its well characterized non-covalent complexes include PNA–PNA (16), PNA–RNA (17), and PNA–DNA duplexes (18), PNA–PNA–PNA (19) and PNA–DNA–PNA triplexes (20). Recently, it has been shown that PNA can form tetramolecular (21) as well as bimolecular G-quadruplexes (22,23). It has also been shown that PNA can, in association with DNA, form a hybrid G-quadruplex, which has several functional implications (24). We and others have recently shown that PNA can also form the i-motif, and that this structure has very different properties compared to its DNA analogue (14,25). Given the current interest in the i-motif as a structural element in nanotechnology as evidenced by the development of various molecular switches (26,27), we wanted to create an alternative hybrid structure that preserves the gross features of the DNA i-motif, but with different physico–chemical characteristics. Since both DNA and PNA form i-motifs, and since PNA forms a variety of non-covalent hybrid complexes with DNA, we wanted to see if one could create a hybrid i-motif from a mixture of DNA and PNA that would possibly have characteristics different from either of the parent structures.
We have used nano-electrospray ionization mass spectrometry to show that a 1:1 mixture of PNA and DNA form a tetramer and that this tetramer is formed from two hemi-protonated PNA–DNA duplexes. pH dependent stability studies by ultraviolet (UV) and CD spectroscopy showed that the hybrid was most stable at a pH = pKa on N3 of cytosine indicating that the hybrid was held together by C-H+C base pairs. High resolution NMR revealed that there were five such C-H+C base pairs in the complex and that the complex was C2-symmetric. These imino protons were protected from D2O exchange, indicating intercalation of the PNA–DNA duplexes to form a solvent shielded inner core as seen in DNA4 i-motifs. The relative strand polarities of PNA and DNA in the C2-symmetric hybrid i-motif were addressed using Homo-FRET and fluorescence quenching. These suggested that the PNA strands were arranged head-to-tail with respect to each other, as were the DNA strands. Further, the heteroduplexes intercalated such that both DNA backbones were closest to each other. 2D NMR confirmed these findings and indicated that the DNA backbones were close enough to invoke sugar–sugar contacts as seen in DNA4 i-motifs. In order to evaluate the physico–chemical characteristics of the hybrid i-motif, we have performed thermal melting studies and validated these by analogous circular dichroism (CD) studies under a variety of conditions. Our studies show that in comparison with either of the parent i-motifs the hybrid has intermediate pH dependence, greater thermal stability and formed at far lower concentrations.
MATERIALS AND METHODS
PNA synthesis and characterization
High-performance liquid chromatography (HPLC) purified d(TC5), 5′-TMR and 3′-TMR labeled d(TC5) were purchased from Sigma–Genosys and used without further purification. PNA monomers and PNA coupling reagents were obtained from Applied Biosystems (Cheshire, UK). Wang resin pre loaded with ɛ-N(Mtt) protected Lys were obtained from Novabiochem. Flurophores were obtained from Invitrogen, UK. All other reagents were obtained from Sigma–Aldrich unless otherwise stated. PNA synthesis was performed using standard solid phase Fmoc chemistry on Nova Syn® TGA resin using analytical grade reagents and Milli-Q water where applicable. Protecting groups and PNA probes were cleaved from the resin simultaneously using 95:2.5:2.5 TFA/Water/TIS. The cleaved mixture was evaporated under reduced pressure and precipitated using dry ether. The precipitate was dissolved in water and subjected to purification by RP-HPLC using a CH3CN/H2O gradient starting from 5/95 v/v CH3CN/H2O to 100/0 v/v CH3CN/H2O over 42 min. ESI-MS p(C5T): [M+2H]2+ 834.99 (calc. 835.37), [M+3H]3+ 556.91 (calc. 557.25). p(C5T) was stored as a 2 mM aqueous solution at 4°C.
N-TMR-p(C5T): PNA synthesis was carried out using standard solid phase Fmoc chemistry on Nova Syn® TGA resin using the above protocol. S-Trt-protected cysteine was then incorporated at N-terminus. The trityl (Trt) group was removed by treatment with 5:1:44 TFA/TIS/CH2Cl2 for 2 h, followed by washing with CH2Cl2 and dry DMF. 1.5 equiv. of TMR-5-maleimide in presence of 3 equiv. DIPEA was added to the resin and stirred for 2.5 h. The N-terminal Fmoc was removed by treatment with 20% piperidine in DMF, cleaved and purified as described earlier. N-TMR-PNA was characterized by positive ion ESI-MS: N-TMR-p(C5T): [M+4H]4+ 564.18 (calc. 564.9).
N-Dabcyl-p(C5T): PNA synthesis was carried out using standard solid phase Fmoc chemistry as described earlier. ɛ-N(Mtt) protected Lys-Fmoc was incorporated at N-terminus in order to attach the Dabcyl group. Post attachment, the Fmoc group was removed by treatment with 20% piperidine/DMF. Dabcyl succinimidyl ester (3 equiv.) in presence of 10 equiv. of DIPEA was added to the resin and stirred for 6 h. The labeled PNA was cleaved and purified as described earlier. N-Dabcyl-PNA was characterized by positive ion ESI-MS: N-Dabcyl-p(C5T): [M+2H]2+ 1024.98 (calc. 1025.17) and [M+3H]3+ 683.65 (calc. 684.71).
TMR was attached at the 3′ end of d(TC5) using a C7 linker employing thiol maleimide chemistry to give 3′-TMR-d(TC5). TMR was attached to the 5′-end of d(TC5) through a C6 linker using succinimidyl chemistry to give 5′-TMR-d(TC5).
Native gel electrophoresis
Polyacrylamide gel electrophoresis studies were performed using the TMR labeled DNA oligomers. Samples at 50 μM strand concentration were prepared by mixing equimolar 3′-TMR-d(TC5) and unlabeled p(C5T) following the protocol described earlier. A total of 20% of polyacrylamide (Sigma) gels were used to assess the mobility of different structures. Robinson–Britton buffer (pH 4.5) was used to make the gels and was also used as the running buffer. All gels were run at 70 V for 6 h at 4°C. Orange-G (Sigma) was used as a tracking dye. Fluorescently labeled bands were visualized using Ethidium Bromide filter.
Nano-electrospray ionization mass spectrometry (Nano ESI-MS)
All mass spectrometry were performed on Micromass ESI-MS Q-TOF Ultima Mass Spectrometer (Manchester, UK) with micro-channel plate detector. Nano ESI-MS spectra were collected in positive ion mode except where mentioned. An equimolar solution of 0.5 mM p(C5T) and 0.5 mM d(TC5) in 30 mM sodium acetate buffer were heated to 90°C, annealed to room temperature over 3 h and equilibrated at 4°C for 8 h. The parent DNA4 and PNA4 i-motifs were prepared similarly. Just prior to injection, the equilibrated sample was diluted 5-fold with Milli-Q water at pH 4.5 (pH adjusted with formic acid) and then loading into the nano-flow capillary. Source temperature of 70°C, capillary voltage of 1.5 kV, cone voltage of 60 V were the source parameters employed during acquisition of mass spectra.
UV-visible spectrophotometric studies
Concentration dependent thermal melts
All the UV measurements were done on a Varian Cary 300Bio UV-VIS spectrometer equipped with a Peltier temperature controller. Stock solutions of PNA and DNA were made in salt-free, Millipore water. Samples were prepared from a stock solution of 1 mM DNA d(TC5) and 2 mM PNA p(C5T) by diluting in 30 mM Na-acetate buffer (pH 4.5), to achieve the desired strand concentration. In the context of the hybrid i-motif, the term strand concentration refers to the total strand concentration corresponding to d(TC5) and p(C5T) in the sample. To form the hybrid i-motif of desired strand concentration, an equimolar mixture of d(TC5) and p(C5T) at appropriate concentrations were heated and maintained at 90°C for 10 min, cooled to 4°C over 3 h at a rate of 0.5°C/min and equilibrated at 4°C for 8 h. The parent PNA4 and DNA4 i-motifs were formed similarly. Melting experiments were performed by monitoring absorbance at 295 nm from 20 to 90°C using a heating rate of 0.5°C/min. Thermal denaturation experiments were performed on hybrid i-motifs at strand concentrations ranging from 1 to 100 μM.
pH dependent studies
The pH dependent thermal melts on the hybrid i-motif were carried out at a total strand concentration of 50 μM. Samples were formed as mentioned earlier in a buffer at the desired pH. The pH range 3–7 was investigated. A total of 30 mM NaH2PO4/H3PO4 of the buffer was used at pH 3, 30 mM CH3COONa/CH3COOH buffer was used for the pH range 3.5–5.5 and 30 mM NaH2PO4/Na2HPO4 buffer was used for the pH regime 6–7. The samples were also used for pH dependent CD studies (see below).
CD studies
CD spectra were recorded on a JASCO-J-720 spectrophotometer where temperature control was achieved using a water bath (Neslab RTE 100). Hybrid i-motifs at a strand concentration of 50 μM were prepared as described earlier in 30 mM Sodium acetate buffer at pH 4.5. This was then diluted with buffer to give a hybrid i-motif of the desired strand concentration for CD experiments. Spectra were recorded from 330 to 220 nm and are presented at an average of six successive scans. Finally all the spectra are subtracted from a baseline corresponding to buffer alone. Temperature dependent CD spectra (CD melts) were recorded on both hybrid i-motif and DNA i-motif at a total strand concentration of 100 μM in 30 mM sodium acetate buffer (pH 4.5), at a heating rate of 0.3°C/min.
NMR experiments
All NMR spectra were recorded on Bruker AV-700 spectrometer. A total of 1 mM strand concentration in 30 mM sodium acetate buffer at pH 4.5 was used to prepare samples for all 1-D and proton exchange experiments whereas, for 2-D experiments 30 mM sodium acetate-d3 was used to prepare samples. Water suppression was achieved using an Excitation Sculpting solvent suppression programme (28). For 1D experiment 132 scans were taken, the spectral bandwidth was maintained at 2 KHz, with an acquisition time of 1.6 s and a 2 s recycling delay. The methyl chemical shift at 1.91 p.p.m. of Na-acetate was used as the internal standard. For NOESY experiments, (900 × 4096) complex points were collected, a 2 KHz spectral width was employed in both dimensions with acquisition times of 0.1 s in t2 and 0.02 s in t1, using a 250 ms mixing time.
Fluorescence studies
Homo-FRET studies were carried out on hybrid i-motifs at a strand concentration of 25 μM, prepared by mixing 1:1 N-TMR-p(C5T) : d(TC5) in 30 mM Sodium acetate buffer at pH 4.5. Similarly 1:1 5′-TMR-d(TC5): p(C5T) as well as 3′-TMR-d(TC5): p(C5T) were also made using the same protocol. For fluorescence quenching, samples were made by mixing 1:1 N-Dabcyl-p(C5T) : 3′-TMR-d(TC5) or 1:1 N-Dabcyl-p(C5T): 5′-TMR-d(TC5) to achieve a strand concentration of 25 μM using the above mentioned protocol. All fluorescence studies were carried out on a FLUOROLOG-SPEX spectrometer. Temperature dependent melting experiments followed by Homo-FRET were done on a Corbett Rotor GENE RT–PCR instrument, over a temperature range of 30–90°C and a heating rate of 0.5°/min. All the samples were made at 25 μM strand concentration in 30 mM sodium acetate buffer (pH 4.5). Quenching was followed at 580 nm after exciting TMR at 530 nm. Prior to fluorescence measurement, uniformity in concentrations of the fluorescent samples were confirmed by UV spectrometry. In order to ensure the complete complexation of the N-TMR-PNA strands and rule out contributions from uncomplexed non-FRETting species, homo-FRET samples incorporated a 10% excess of unlabeled DNA strands. Similarly, 3′-TMR-DNA and 5′-TMR-DNA were mixed with a 10% excess of unlabelled PNA.
The distance between the fluorophores was calculated, with the associated limitations of FRET, using the donor quenching method (29) employing the formula E = (1 − FD/FDA) = 1/[1 + R/R0]1/6 (for details see Supplementary Data). The Förster distance (R0) for TMR–TMR quenching due to homo-FRET was 44 Å and TMR-Dabcyl quenching was 26 Å consistent with the literature values (30). A coarse gained model of the hybrid tetramer, incorporating the linkers was constructed based on the NMR parameters of the d(TC5) DNA4 i-motif (1) to obtain theoretical estimates of the relevant distances. Anisotropy of all the fluorophores was less than 0.1 indicating that they were freely rotating (Supplementary Data).
RESULTS AND DISCUSSION
Design consideration
The oligonucleotide d(TC5) incorporated a stretch of cytosines at the 3′ end where tandem C residues are shown to potentiate tetramerization into i-motifs (31). The analogous PNA sequence has also been shown to form i-motifs (14). We synthesized p(C5T) by solid phase synthesis using Fmoc chemistry (See Materials and Methods section). A T residue was incorporated at the N-terminus in order to prevent possible higher order structure formation. A lysine residue was attached to C-terminus to enhance solubility of the PNA sequence in water. Furthermore, since tetramolecular DNA4 and PNA4 i-motifs formed from d(TC5) and p(C5T) are well characterized (14,31) they would prove good markers for comparison of any hybrid i-motif formed.
Gel electrophoresis shows formation of a hybrid complex
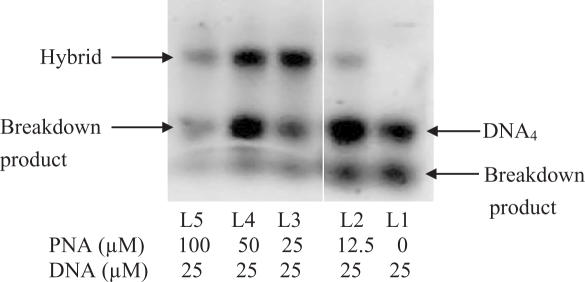
In order to see if a 1:1 mixture of C-rich DNA and PNA could form a complex, various concentrations of p(C5T) were annealed with a fixed concentration (25 μM) of 3′-TMR-d(TC5) in 30 mM Sodium acetate buffer (pH 4.5) as described in the experimental section and subjected to gel electrophoresis. Figure 1 shows the electrophoretic mobility shift of samples incorporating p(C5T) relative to 3′-TMR-d(TC5) alone. For a given DNA concentration, incremental increase in PNA concentration resulted in a complex with electrophoretic mobility slower relative to the DNA-only complex. In the case of both complexes, an extra band with higher mobility was observed that we assign to breakdown products of the slower-migrating parent complex. Thus, a mixture of 1:1 PNA : DNA forms a complex that is distinct from the parent DNA complex.
Figure 1.
20% Non-denaturing polyacrylamide gel at 25 μM 3′-TMR-d(TC5) with increasing p(C5T) concentration, running buffer: Robinsson–Britton (CH3COOH = H3PO4 = H3BO3 = 0.04 M in 100 mM Na+), pH 4.5 at 70 V for 6 h.
Nano-electrospray ionization mass spectrometry studies evidence a hybrid tetramer
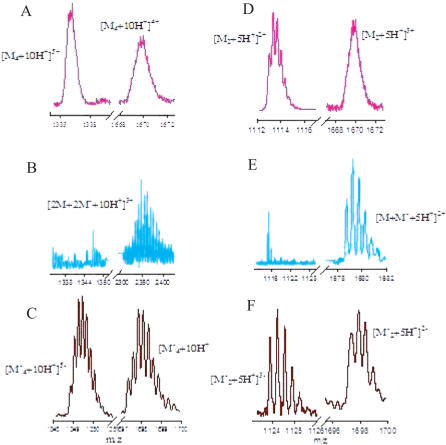
In order to check the molecularity of the PNA–DNA complex evidenced by gel electrophoresis, the complex was subjected to nano-electrospray ionization mass spectrometry (Nano ESI-MS). An equimolar solution of p(C5T) and d(TC5) at 0.5 mM each was prepared as described in the Materials and Methods section. An aliquot of this sample was diluted 5-fold and analyzed by non-covalent mass spectrometry to seek evidence for complex formation. The Nano ESI mass spectrum of this sample showed multiple peaks in m/z regime 2320 to 2400 (Figures 1B and 2). These peaks were equidistant, with a constant separation of 7.4 ± 0.1 m/z units indicating that these peaks corresponded to a triply charged species of m/z 2243.6, which was multiply sodiated. The associated molecular weight (6730.8 ± 0.9 Da) of this species corresponds to a four-stranded entity ([2M+2M'+10H+]3+) that is composed of two strands each of p(C5T) and d(TC5) with ten additional protons (calc. mol. wt. 6730.14 Da). Moreover, this molecular weight falls exactly in between the measured molecular weights of the respective parent DNA4 (6775.2 ± 0.6 Da) and PNA4 (6685.9 ± 1.06 Da) i-motifs that each incorporate an extra 10 additional protons, required for C-H+C base pairing. The consistent presence of these 10 extra protons in the observed 2[p(C5T)] : 2[d(TC5)] complex suggests that the strands in this tetramer may also be held together by C-H+C base pairs as seen in the parent i-motifs. Importantly, peaks corresponding to the DNA4 or PNA4 i-motifs (14) were completely absent suggesting that in a 1:1 mixture of p(C5T) and d(TC5), the formation of the homotetrameric complexes are suppressed.
Figure 2.
Nano ESI mass spectra of i-motif tetraplexes and duplexes: (A and C) are representative partial mass spectra showing quadruply and quintuply charged states of PNA4 and DNA4 i-motifs, (B) shows the triply charged state of a tetrameric PNA–DNA complex. (D and F) are representative partial mass spectra showing m/z regimes corresponding to doubly and triply charged states of PNA–PNA and DNA–DNA homoduplexes in the respective PNA4 and DNA4 i-motifs. (E) Shows the doubly charged state of a PNA–DNA heteroduplex in the tetrameric PNA–DNA complex. DNA4 i-motif mass spectra were recorded in negative ion mode.
Fragmentation reveals that the tetramer is comprised of PNA–DNA duplexes. PNA is capable of forming parallel stranded duplexes held together by C-C+ base pairing (14). PNA has also been shown to form Watson–Crick base paired PNA–PNA (16) and PNA–DNA duplexes (18). Given that the stoichiometry of PNA: DNA in the tetrameric complex is 2:2, two possible molecular arrangements exist for the sub-units in the proposed hybrid i-motif. The first is that of a PNA–PNA homoduplex and a DNA–DNA homoduplex that could intercalate. The second is one where two PNA–DNA heteroduplexes could intercalate. This could be resolved by dissociating the hybrid complex by Nano ESI-MS/MS. Because the net charge on the complex is zero, it exhibited poor flying characteristics thereby limiting detailed ESI-MS/MS studies. However, with a higher cone voltage, it was possible to dissociate the tetrameric complex into smaller fragments. This resulted in high intensity peaks at m/z 1680 ± 0.02 (Figure 1E) where the isotopic separation indicated that the peaks were doubly charged, with an associated molecular weight of 3360.9 ± 0.04 Da. This corresponded to a p(C5T):d(TC5) heteroduplex with five additional protons (calc. mol. wt. 3361.1 Da). Furthermore, the complex did not show any other peaks corresponding to DNA–DNA homoduplexes (in negative ion mode) or PNA–PNA homoduplexes (in positive ion mode) despite the net charge and good flight characteristics of these homoduplexes (14). These results support a model where the hybrid complex is composed of two identical sub-units, namely the PNA–DNA heteroduplex. Taken together, our Nano ESI-MS studies suggest that at pH 4.5, a 1:1 ratio of p(C5T):d(TC5), forms a tetrameric complex that is held together by 10 extra protons. Furthermore this complex is composed of two PNA–DNA heteroduplexes each of which is held together by five additional protons.
Hybrid complex shows a thermal transition characteristic of C-H+C base pairs
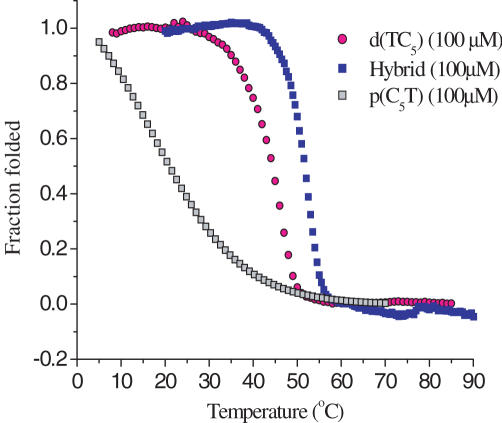
Stability as a function of concentration: DNA duplexes and triplexes show positively sloped sigmoidal curves when the UV absorbance at 260 nm is monitored as a function of temperature (T) whereas the melting transition of the i-motif, which is held together by C.H+-C base pairs, is characterized by a distinctive inverse sigmoidal melting curve at 295 nm (32–35). UV melting studies were conducted on complexes that were formed as mentioned in the Materials and Methods section from an equimolar solution of d(TC5) and p(C5T) at the desired strand concentration. UV melting profiles of a 1:1 d(TC5): p(C5T) complex at 100 μM strand concentration showed an inverse sigmoidal curve at 295 nm with an associated T1/2 of 51.8 ± 0.2°C (Figure 3). This is about 6–7°C higher than that of either the PNA4 or DNA4 i-motifs, indicating that the hybrid is more stable than the parent structures (14). This increased stability could be possibly because the DNA4 i-motif is net negatively charged, the PNA4 i-motif is net positively charged, but the hybrid is electrically neutral. Interestingly, melting temperatures of the hybrid complex at strand concentrations from 2–200 μM showed negligible variation with concentration (See Supplementary Table 1). A striking feature of the hybrid complex was that the melting transitions were quite prominent even at concentrations as low as 2 and 4 μM even though neither d(TC5) nor p(C5T) showed well-behaved melting transitions below 25 μM (see Supplementary Data). These characteristics of the hybrid complex suggest that it is much more efficiently associated with either of the parent complexes at lower concentrations. Furthermore, the hybrid showed a more cooperative melting transition than either of the parent i-motifs.
Figure 3.
UV melting profiles at 295 nm of [p(C5T)]4, [d(TC5)]4 and hybrid at comparable strand concentrations in 30 mM Acetate buffer (pH 4.5).
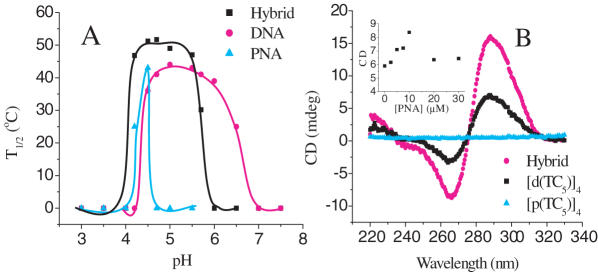
Stability as a function of pH: The thermal denaturation of i-motifs is strongly pH dependent as the formation of C-H.C+ base pairs require hemi-protonation of the cytosine nucleobases. Melting studies on the hybrid, p(C5T) alone and its DNA analog, d(TC5), were performed at different pH values and the corresponding melting temperatures were plotted as a function of pH (Figure 4A). At a strand concentration of 25 μM, T1/2 of the hybrid as a function of pH showed a bell-shaped curve (Figure 4A) (31). In our hands d(TC5) showed melting transitions only in the pH regime 4.2–6.8 whereas p(C5T) showed melting transitions over a much narrower pH range (4.2–4.5) (14). The hybrid i-motif, on the other hand, showed thermal transitions over an intermediate pH regime of 4.2–5.7, probably reflecting the contribution of DNA and PNA strands. Interestingly, the hybrid complex showed maximum stability at pH 4.3–4.5 with an associated T1/2 of 51.5 ± 0.2°C. Given that the pKa value of N3 on cytidine mono phosphate is 4.4–4.5 (36), the maximum number of C-C+ base pairs would be expected to form at a pH equal to the pKa of N3 on cytosine. This strongly indicates that the structure is held together by C-C+ base pairs, such as those found in DNA i-motifs. Increasing or decreasing the pH from the optimal pH value (4.3–4.5) had a marked effect on the stability of the hybrid i-motif, which showed abrupt lowering of T1/2 values and less cooperative melting profiles (See Supplementary Data). For pH regimes outside of 4.2–5.7 there was no detectable i-motif formation by the hybrid.
Figure 4.
(A) Plot of the melting temperatures of [p(C5T)]4, [d(TC5)]4 and hybrid tetraplexes, at 25 μM strand concentration, as a function of pH. The lines merely serve as a visual aid. (B) CD profiles of the hybrid, [p(C5T)]4, [d(TC5)]4 tetraplexes at 12.5 μM strand concentration at pH 4.5. Inset shows CD at 288 nm as a function of varying PNA concentration with DNA concentration were fixed at 10 μM.
CD spectroscopy of hybrid i-motif
In order to further characterize the hybrid i-motif we investigated secondary structure formation of the complex by CD spectroscopy. DNA4 i-motifs show a characteristic CD profile with a positive maximum near 285 nm, which is followed by a negative trough with a minimum centered near 265 nm (37). A sample of 12.5 μM p(C5T) alone in acetate buffer (pH 4.5), showed comparatively negligible CD as the only source of chirality is the C-terminal lysine (Figure 4B). A 12.5 μM solution of d(TC5) alone in 30 mM acetate buffer (pH 4.5), showed a characteristic, albeit modest, CD trace corresponding to the small fraction of DNA4 i-motif formed at this concentration. However, a 1:1 mixture of p(C5T):d(TC5) at 25 μM total strand concentration, showed a CD signal that is amplified nearly 2-fold, Figure 4B, reaffirming the formation of a completely different species in line with gel electrophoresis and UV studies. The amplification of the signal could be due to the greater degree of complexation of DNA strands at 12.5 μM by PNA, or the imposition of the chirality of the duplexed DNA backbone onto a larger number of nucleobases. Since the basic features of the hybrid CD profile preserved the CD characteristics of the DNA4 i-motif, it is likely that the nucleobases in the hybrid tetraplex experience a similar chiral environment.
In order to confirm the fraction of hybrid complex formed, complexes of d(TC5) at 10 μM incorporating incrementally increasing p(C5T) concentrations were made in 30 mM buffer, and CD at 288 nm of the resulting complexes were plotted as a function of p(C5T) concentration. It was observed that the CD value increased along with increase in PNA concentration till a ratio of 1:1 PNA:DNA was reached. The CD signal decreased thereafter, probably indicating that excess PNA strands interfered with hybrid complex formation due to the skewed strand ratios. Importantly, the maximum signal is attained only when an equimolar amount of PNA is added indicating that the DNA in the solution is completely saturated, confirming that the hybrid is indeed predominant.
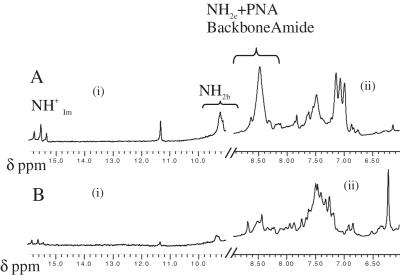
1D proton NMR and D2O exchange studies
In order to obtain more detailed structural information on the hybrid complex we performed high resolution 1H NMR studies in 30 mM sodium acetate buffer pH 4.5 at 1 mM strand concentration. The 1D NMR spectrum shows three distinct peaks between 15–16 δp.p.m. [Figure 5A (i)] characteristic of the imino protons in C-H+C base pairs in DNA4 i-motifs (1,38,39). Also present was a single thymine imino peak at 11.4 δp.p.m. indicating a single population of hybrid (38). The added presence of a single peak corresponding to the thymine CH3 at 1.8 δp.p.m. further supports the formation of a unique population that is C2-symmetric. Contributions from single-stranded DNA and PNA were negligible indicating that these species are below the NMR detection limit. This confirms the findings from CD experiments which indicated that the hybrid was the predominant species in solution. In order to gauge the extent of protection of various H-bonded protons in the complex we carried out D2O exchange experiments on the complex. The sample was lyophilized and resuspended in an equal volume of D2O and NMR spectra were collected at different time points (Figure 5A and B). Figure 5B shows the NMR spectrum after 6 h of D2O exchange. Interestingly a set of signals centered on 8.5 δp.p.m. disappeared, which corresponds to the non-hydrogen bonded amino protons (NH2e) and the PNA amide backbone protons. Importantly, the N+HIm protons exchanged more slowly (See Supplementary Data than the backbone amide protons, suggesting that the former are present in a more protected environment. This may be explained by possible intercalation of PNA–DNA duplexes, as in DNA4 i-motifs, to form an inner core that is solvent shielded, accounting for the slow exchange rate.
Figure 5.
(A) One dimensional proton NMR experiments in 95/5 H2O / D2O at 5°C recorded on a 700 MHz Bruker NMR spectrometer of 1 mM hybrid complex in 30 mM sodium acetate d3 at pH 4.5 showing (i) Imino proton () region C-H+-C base pairs, thymine imino and internally hydrogen bonded Cytosine amino protons (NH2b). (ii) External, non-hydrogen bonded Cytosine amino (NH2e), PNA backbone amide protons, sugar H1′ protons and Aromatic H6 proton region. (B) Corresponding regions of the 1D-NMR spectrum in D2O at t = 6 h after D2O addition.
Homo-FRET reveals DNA–DNA and PNA–PNA strand alignments
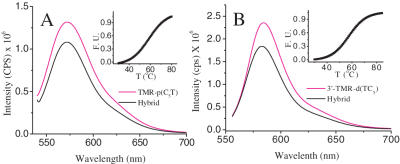
In order to elucidate strand polarity in this structure, hybrid i-motifs were formed where only the PNA strands were labeled with TMR. This was achieved by forming an i-motif using a 1:1 mixture of d(TC5) and N-TMR-p(C5T). In order to check that the incorporation of the fluorescent label did not drastically alter the hybrid structure, thermal melting of the complex was monitored by both UV and fluorescence (Figure 5A and B insets). These showed that the corresponding i-motifs were in fact marginally stabilized compared to their unlabelled counterparts. The introduction of the fluorescent label has been shown to alter i-motif stabilities and has also been used to follow i-motif formation (40). TMR self-quenches with an associated Förster's distance of 44 Å (30). As shown in Figure 6A, the fluorescence of TMR was quenched by ∼18% in the hybrid i-motif. This corresponds to an interfluorophore distance of 52 ± 5 Å, which is consistent with a theoretical estimate (50 Å, See Supplementary Data) of head-to-tail (or antiparallel) arrangement of both the PNA strands in the hybrid i-motif, taking into account (i) lengths of the linkers connected the fluorophores to strands and (ii) assuming complete intercalation of base pairs. A similar homo-FRET study was performed to check the DNA strand polarities in the hybrid i-motif using 1:1 3′-TMR-d(TC5) : p(C5T). This gave an interfluorophore distance of 51.7 ± 5 Å revealing that the DNA strands are also arranged in an antiparallel configuration in the hybrid i-motif. A complementary study on the same distance at a differently labeled site i.e. 5′-TMR-d(TC5) gave an interfluorophore distance of 53 ± 5 Å (see Supplementary Data) that reaffirms DNA strand polarity in the hybrid i-motif.
Figure 6.
(A) Homo-FRET experiments on hybrid i-motif of 1:1 TMR-p(C5T) : d(TC5) at 25 μM total strand concentration. The transfer efficiency was measured with respect to single-stranded 25 μM TMR-p(C5T) only (black). Inset: thermal denaturation of fluorescently labeled hybrid i-motif. Fluorescence intensity at 580 nm was followed at a function of temperature. (B) Analogous Homo-FRET measurements on hybrid i-motif with 3′ labeled DNA strands, 3′-TMR-d(TC5) : p(C5T). Transfer efficiency was measured with respect to single-stranded 25 μM 3′-TMR-d(TC5) only. Inset: UV melting of fluorescently labeled hybrid i-motif followed at 295 nm as a function of temperature.
Fluorescence quenching establishes relative strand polarities in the hybrid i-motif
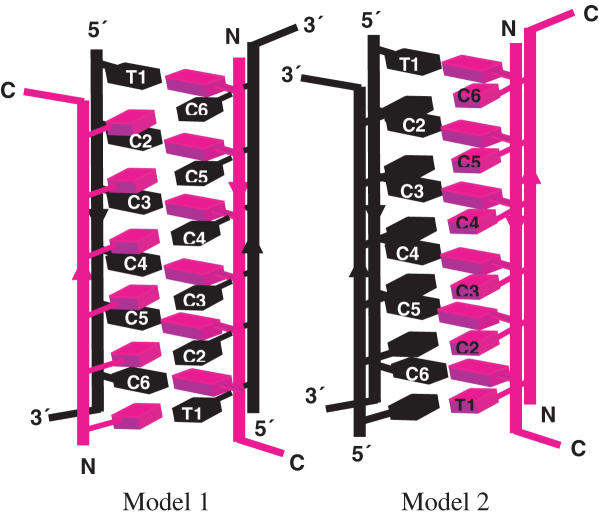
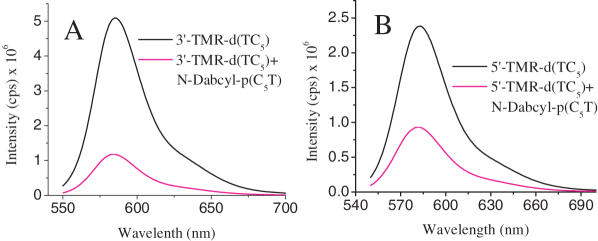
The two PNA–DNA duplexes can intercalate in two different configurations that are C2-symmetric to form the hybrid i-motif. Although, the hybrid need not have the same groove dimensions as the DNA4 i-motif, as the chiral environment of the latter is preserved in the former, we performed fluorescence quenching based distance measurements to ascertain this issue. If one assumes that the groove dimensions of the DNA4 i-motif are preserved in the hybrid, there are two possible configurations as shown in Figure 7. One, where each narrow groove has one DNA and one PNA backbone each (Model 1) and the other where one narrow groove has both the DNA strands and the second has both the PNA strands (Model 2). To address this, we formed an i-motif using 3′ TMR-d(TC5) and N-Dabcyl-p(C5T). Theoretical estimates of the interfluorophore distances, taking into account linker lengths and assuming groove dimensions commensurate with DNA4 i-motifs, were found to be ∼9 Å in the case of Model 1 and ∼20 Å in the case of Model 2. As shown in Figure 8A, a quenching efficiency of ∼80% was obtained, indicating a separation of 21.7 ± 5 Å consistent with Model 2. Fluorescence quenching measurements on a complex of 1:1 5′ TMR-d(TC5) and N-Dabcyl-p(C5T) yielded a distance of 24.5 ± 5 Å (calculated distance as per Model 2 was ∼23 Å). The two distances obtained are comparable with each other and rule out a situation as in Model 1, where there is a much larger difference between these two distances.
Figure 7.
Two possible models of the hybrid i-motif. Model 1: One DNA and one PNA strand in narrow groove. Model 2: Both DNA strands are in one of the narrow grooves.
Figure 8.
Fluorescence quenching experiments on the dual labeled hybrid i-motif of (A) 1:1 3′-TMR-d(TC5) and Dabcyl-p(C5T) and (B) 1:1 5′-TMR-d(TC5) and Dabcyl-p(C5T) at 25 μM strand concentration. The transfer efficiency was measured with respect to 25 μM 3′-TMR-d(TC5) only (black). Samples were excited at 530 nm and the emission was followed at 580 nm.
NOESY shows sugar–sugar contacts between DNA backbones of fully intercalated PNA–DNA duplexes
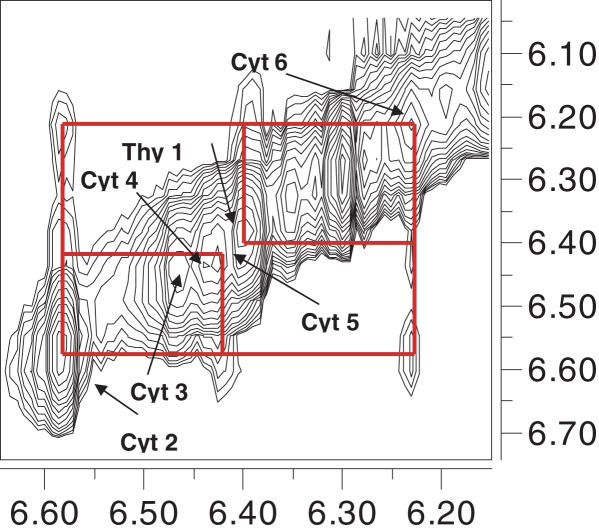
In order to address the validity of assumptions made in the FRET and quenching experiments which were (i) complete intercalation of the base pairs and that (ii) the groove dimensions in the hybrid and the DNA4 i-motif could be comparable, NOESY experiments were recorded in H2O. NOESY on the hybrid showed several cross peaks, a few of which are indicated in Figure 9. Cross peaks were observed in the NH2b–NHIm, NH2e–NHIm, NHIm–NHIm, H1′–H1′, H2′–H2′′ regions as seen in analogous spectra of i-motifs (1). Intense cross peaks between NH2e and sugar protons as well as inter-sugar H1′–H1′ protons clearly indicate that the two DNA strands are in close proximity and are present in a narrow groove with a separation comparable to DNA4 i-motifs (1,41). This finding is in line with our fluorescence quenching distance measurements. Figure 9 shows NOE's corresponding to the H1′ region indicating several short sugar–sugar H1′–H1′ contacts between the DNA backbones. The assignments of NOE's are consistent with the previously reported DNA4 i-motif structure showing a base stacking order of Thy-C6-C2-C5-C3-C4 revealing that the PNA–DNA duplexes are indeed completely intercalated, reaffirming the results from the fluorescence-based distance measurements. Focused cross peaks between the glycine protons (3.4–4 δp.p.m.) and the NH2e protons (8.2–9 δp.p.m.) signify that the PNA strands are rigidified into a unique conformation by virtue of it forming the hybrid i-motif (Supplementary Data). Thus NOESY on the hybrid complex reveals that it is indeed a structure formed by the complete intercalation of C–C+ base paired PNA–DNA duplexes where the DNA backbones are close enough to invoke sugar–sugar interactions as seen in DNA4 i-motifs.
Figure 9.
Partial NOESY spectrum showing sugar H1′–H1′ contacts of 1 mM p(C5T) and d(TC5) in 30 mM Na-acetate-d3, pH 4.5 recorded on a 700 MHz Bruker spectrometer with a mixing time of 250 ms.
CONCLUSIONS
This paper describes the first four-stranded i-motif composed of DNA and PNA. Nano-electrospray ionization mass spectroscopy revealed that a 1:1 mixture of PNA and DNA strands with tandem C residues resulted in the formation of a tetrameric species. Fragmentation studies revealed that the tetramer was formed from two PNA–DNA heteroduplexes. Temperature dependant UV studies on the complex revealed a thermal transition characteristic of C–C+ base pairs. Furthermore, the thermal stability of the hybrid complex as a function of pH revealed that it exists over a pH regime 4.2–5.7 with maximum stability at pH 4.5, the pKa of cytosine. This reaffirms that the hybrid complex was formed as a consequence of hemi-protonated C–C+ base pairs as seen in i-motifs. NMR spectroscopy revealed the existence of a single population of a C2-symmetric complex and five fully intercalated C–C+ base pairs. The central imino protons exchanged slowly with D2O, reaffirming their existence in a relatively solvent shielded environment, consistent with hybrid i-motif formation. Homo-FRET experiments revealed that in the hybrid i-motif, the PNA strands are antiparallel with respect to each other, as are the DNA strands. This is consistent with the CD data, which indicated that the hybrid possessed similar topology to the DNA4 i-motif (37). Fluorescence quenching studies revealed that the PNA–DNA heteroduplexes were arranged, such that both DNA strands occupied one narrow groove and the PNA strands occupied the other groove. This was confirmed by NMR experiments that showed sugar–sugar contacts in the form of NOEs between the H1′ protons of both the DNA backbones as in DNA4 i-motifs.
Because of the altered affinities for duplex formation in the hybrid i-motif, it can form at concentrations much lower than the parent DNA4 or PNA4 i-motifs. The hybrid i-motif was also more thermally stable than either of the parent DNA4 and parent PNA4 i-motifs. This is likely to be because two of negatively charged backbones in the DNA4 i-motif are replaced by neutral polyamide PNA backbones thereby reducing the electrostatic repulsion associated with multi-stranded structures. Furthermore, the DNA4 i-motif is net negatively charged, the PNA4 i-motif is net positively charged and the hybrid is an electrically neutral complex. The PNA4 i-motif exists over a narrow pH regime, while the DNA4 i-motif exists over a much wider pH window. The hybrid i-motif, however, exists over a pH regime intermediate between the parent DNA4 and PNA4 i-motifs. This is probably due to the lower number of DNA backbones that confers on the complex a limited ability to tolerate pH change. CD revealed that the hybrid has chiral characteristics similar to that of the parent DNA4 i-motif. The hybrid i-motif is clearly a robust structural element with attractive physico–chemical properties that may be used as a nanoscale structural element. We are currently exploiting the fast kinetics of hybrid i-motif formation and abolition to develop nucleic acid-based molecular switches in our laboratory.
Importantly, the hybrid i-motif presents a structural variant of the DNA4 i-motif which helps assess relatively open-ended issues regarding duplex intercalation and stability in the DNA4 i-motif. Here, the intercalation of two DNA–DNA duplexes gives rise to a remarkably narrow groove positioning two negatively charged DNA backbones extremely close resulting in greater electrostatic repulsion. Thus, one of the prevailing issues regarding DNA i-motif formation has been in understanding why the two DNA–DNA duplexes should intercalate at all. It has been widely suggested that the sugar–sugar contacts all along the backbones of the two strands located in the narrow grooves of the i-motif favors tetramer formation (42,43) although evidence for the contrary exists as well (14,15,25,44). The hybrid i-motif is a model system that is well-placed to address this issue, being a structural variant of the DNA4 i-motif, where the DNA–DNA duplex is replaced by a PNA–DNA duplex, which can intercalate in two different configurations, see Models 1 and 2, Figure 7. In one, Model 1, electrostatic repulsion may be minimized by positioning one DNA and one PNA backbone in each of the narrow grooves. In the other, Model 2, the sugar–sugar contacts may be maximized by positioning both DNA backbones in one of the narrow grooves. The strand arrangements in the hybrid i-motif is exclusively one where both DNA strands occupy a single narrow groove, invoking sugar–sugar interactions, in spite of the associated electrostatic repulsion. Thus the hybrid i-motif offers a platform to evaluate the factors governing tetramer formation in i-motifs by allowing the system to prioritize electrostatic repulsion versus sugar–sugar contacts along the DNA backbones. We observe that in the hybrid i-motif, the stability gained from sugar–sugar contacts outweighs the effects of electrostatic repulsion. In this regard, the structure of the hybrid i-motif suggests that sugar-sugar contacts in i-motifs are not probably a consequence of, but possibly drive i-motif tetramerization.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Drs Shankar Balasubramanian and G. V. Shivashankar for critical input, Ms V. Rangaraju and Mr K. Raghunathan for technical assistance and NCBS for funding. The authors thank Dr Siddhartha Sarma and the Biological NMR facility, NMR Research Centre, IISc. S.M. and A.H.W thank the CSIR for Fellowships. Dedicated to Prof. S. Chandrasekaran on his sixtieth birthday. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gehring K., Leroy J.L., Guéron M. A tetrameric DNA structure with protonated cytidine–cytidine base pairs. Nature. 1993;363:561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- 2.Esmaili N., Leroy J.L. i-motif solution structure and dynamics of the d(AACCCC) and d(CCCCAA) tetrahymena telomeric repeats. Nucleic Acids Res. 2005;33:213–224. doi: 10.1093/nar/gki160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang C.H., Berger I., Lockshin C., Ratliff R., Moyzis R., Rich A. Stable loop in the crystal structure of the intercalated four-stranded cytosine-rich metazoan telomere. Proc. Natl Acad. Sci. USA. 1995;92:3874–3878. doi: 10.1073/pnas.92.9.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phan A.T., Gueron M., Leroy J.L. The solution structure and internal motions of a fragment of the cytidine-rich strand of the human telomere. J. Mol. Biol. 2000;299:123–144. doi: 10.1006/jmbi.2000.3613. [DOI] [PubMed] [Google Scholar]
- 5.Leroy J.L., Gueron M., Mergny J.L., Helene C. Intramolecular folding of a fragment of the cytosine rich strand of telomeric DNA into an i-motif. Nucleic Acids Res. 1994;22:1600–1606. doi: 10.1093/nar/22.9.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmed S., Kintanar A., Henderson E. Human telomeric C-strand tetraplexes. Nature Struct. Biol. 1994;1:83–88. doi: 10.1038/nsb0294-83. [DOI] [PubMed] [Google Scholar]
- 7.Miyoshi D., Matsumura S., Li W., Sugimoto N. Structural polymorphism of telomeric DNA regulated by pH and divalent cation. Nucleosides Nucleotides Nucleic Acids. 2003;22:203–221. doi: 10.1081/NCN-120019528. [DOI] [PubMed] [Google Scholar]
- 8.Gallego J., Chou S.H., Reid B.R. Centromeric pyrimidine strands fold into an intercalated motif by forming a double hairpin with a Novel T:G:G:T tetrad: solution structure of the d(TCCCGTTTCCA) dimmer. J. Mol. Biol. 1997;273:840–856. doi: 10.1006/jmbi.1997.1361. [DOI] [PubMed] [Google Scholar]
- 9.Catasti P., Chen X., Deaven L.L., Moyzis R.K., Bradbury E.M., Gupta G. Cystosine-rich strands of the insulin minisatellite adopt hairpins with intercalated Cytosine+·Cytosine pairs. J. Mol. Biol. 1997;272:369–382. doi: 10.1006/jmbi.1997.1248. [DOI] [PubMed] [Google Scholar]
- 10.Nonin-Lecomte S., Leroy J.L. Structure of a C-rich strand fragment of the human centromeric satellite III: a pH-dependent intercalation topology. J. Mol. Biol. 2001;309:491–506. doi: 10.1006/jmbi.2001.4679. [DOI] [PubMed] [Google Scholar]
- 11.Fojtik P., Vorlickova M. The fragile X chromosome (GCC) repeat folds into a DNA tetraplex at neutral pH. Nucleic Acids Res. 2001;29:4684–4690. doi: 10.1093/nar/29.22.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mergny J.L., Lacroix L. Kinetics and thermodynamics of i-DNA formation: phosphodiester versus modified oligodeoxynucleotides. Nucleic Acids Res. 1998;26:4797–5803. doi: 10.1093/nar/26.21.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snoussi K., Nonin-Lecomte S., Leroy J.L. The RNA I-motif. J. Mol. Biol. 2001;309:139–153. doi: 10.1006/jmbi.2001.4618. [DOI] [PubMed] [Google Scholar]
- 14.Krishnan-Ghosh Y., Stephens E., Balasubramanian S. PNA forms i-motif. Chem. Commun. 2005:5278–5280. doi: 10.1039/b510405j. [DOI] [PubMed] [Google Scholar]
- 15.Diederichsen U. Oligomers with intercalating cytosine-cytosine+ base pairs and peptide backbone: DNA i-motif analogues. Angew. Chem. Int. Ed. 1998;37:2273–2276. doi: 10.1002/(SICI)1521-3773(19980904)37:16<2273::AID-ANIE2273>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 16.Rasmussen H., Kastrup J.S., Nielsen J.N., Nielsen J.M., Nielsen P.E. Crystal structure of a peptide nucleic acid (PNA) duplex at 1.7 Å resolution. Nature Struct. Biol. 1997;4:98–101. doi: 10.1038/nsb0297-98. [DOI] [PubMed] [Google Scholar]
- 17.Brown S.C., Thomson S.A., Veal J.M., Davis D.G. NMR solution structure of a peptide nucleic acid complexed with RNA. Science. 1994;265:777–780. doi: 10.1126/science.7519361. [DOI] [PubMed] [Google Scholar]
- 18.Eriksson M., Nielsen P.E. Solution structure of a peptide nucleic acid–DNA duplex. Nature Struct. Biol. 1996;3:410–413. doi: 10.1038/nsb0596-410. [DOI] [PubMed] [Google Scholar]
- 19.Petersson B., Nielsen B.B., Rasmussen H., Larsen I.K., Gajhede M., Nielsen P.E., Kastrup J.S. Crystal structure of a partly self-complementary peptide nucleic acid (PNA) oligomer showing a duplex-triplex network. J. Am. Chem. Soc. 2005;127:1424–1430. doi: 10.1021/ja0458726. [DOI] [PubMed] [Google Scholar]
- 20.Betts L., Losey J.A., Veal J.M., Jordan S.R. A nucleic acid triple helix formed by a peptide nucleic acid–DNA complex. Science. 1995;270:1838–1841. doi: 10.1126/science.270.5243.1838. [DOI] [PubMed] [Google Scholar]
- 21.Krishnan-Ghosh Y., Stephens E., Balasubramanian S. A PNA4 Quadruplex. J. Am. Chem. Soc. 2004;126:5944–5945. doi: 10.1021/ja031508f. [DOI] [PubMed] [Google Scholar]
- 22.Krishnan-Ghosh Y., Whitney A.M., Balasubramanian S. Dynamic covalent chemistry on self-templating PNA oligomers: formation of a bimolecular PNA quadruplex. Chem. Commun. 2005:3068–3070. doi: 10.1039/b503578c. [DOI] [PubMed] [Google Scholar]
- 23.Datta B., Bier M.E., Roy S., Armitage B.A. Quadruplex formation by a guanine-rich PNA oligomer. J. Am. Chem. Soc. 2005;127:4199–4207. doi: 10.1021/ja0446202. [DOI] [PubMed] [Google Scholar]
- 24.Datta B., Schmitt C., Armitage B.A. Formation of a PNA2-DNA2 hybrid quadruplex. J. Am. Chem. Soc. 2003;125:4111–4118. doi: 10.1021/ja028323d. [DOI] [PubMed] [Google Scholar]
- 25.Sharma N.K., Ganesh K.N. PNA C-C+ i-motif: superior stability of PNA TC8 tetraplexes. Chem. Commun. 2005:4330–4332. doi: 10.1039/b506870c. [DOI] [PubMed] [Google Scholar]
- 26.Liu D., Balasubramanian S. A proton-fuelled DNA nanomachine S. Angew. Chem. Int. Ed. 2003;42:5734–5736. doi: 10.1002/anie.200352402. [DOI] [PubMed] [Google Scholar]
- 27.Liedl T., Simmel F. Switching the conformation of a DNA molecule with a chemical oscillator. Nano Lett. 2005;5:1894–1898. doi: 10.1021/nl051180j. [DOI] [PubMed] [Google Scholar]
- 28.Hwang T.L., Shaka A.J. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magnetic Res. Series A. 1995;112:139–282. [Google Scholar]
- 29.Berney C., Danuser G. FRET or No FRET: a quantitative comparison. Biophys. J. 2003;84:3992–4010. doi: 10.1016/S0006-3495(03)75126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernacchi S., Mély Y. Exciton interaction in molecular beacons: a sensitive sensor for short range modifications of the nucleic acid structure. Nucleic Acids Res. 2001;29:e62. doi: 10.1093/nar/29.13.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leroy J.L., Gehring K., Kettani A., Guéron M. Acid multimers of oligo-cytidine strands: stoichiometry, base pair characterization and proton exchange properties. Biochemistry. 1993;32:6019–6031. doi: 10.1021/bi00074a013. [DOI] [PubMed] [Google Scholar]
- 32.Phan A.T., Mergny J.L. Human telomeric DNA: G-quadruplex, i-motif and Watson–Crick double helix. Nucleic Acids Res. 2002;30:4618–4625. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mergny J.L., Lacroix L., Han X., Leroy J.L., Helene C. Intramolecular folding of a pyrimidine oligodeoxynucleotides into an i-DNA motif. J. Am. Chem. Soc. 1995;117:8887–8898. [Google Scholar]
- 34.Lacroix L., Mergny J.L., Leroy J.L., Helene C. Inability of RNA to form the i-motif: implications for triplex formation. Biochemistry. 1996;35:8715–8722. doi: 10.1021/bi960107s. [DOI] [PubMed] [Google Scholar]
- 35.Mergny J.-L., Jing L., Lacroix L., Amrane S., Chaires J.B. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Research. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blackburn G.M., Gait M. Nucleic Acids in Chemistry and Biology. 2nd edn. Oxford: Oxford University Press; 1996. [Google Scholar]
- 37.Edwards E.L., Patrick M.H., Ratliff R.L., Gray D.M. A.T and C.C+ base pairs can form simultaneously in a novel multistranded DNA complex. Biochemistry. 1990;29:828–836. doi: 10.1021/bi00455a033. [DOI] [PubMed] [Google Scholar]
- 38.Collin D., Gehring K. Stability of chimeric DNA/RNA cytosine tetrads: implications for i-motif formation by RNA. J. Am. Chem. Soc. 1998;120:4069–4072. [Google Scholar]
- 39.Brazier A.J., Fisher J., Cosstick R. Stabilization of the DNA I-motif structure by incorporation of 3′-S-phosphorothiolate linkages. Angew. Chem. Int. Ed. 2006;45:114–117. doi: 10.1002/anie.200503110. [DOI] [PubMed] [Google Scholar]
- 40.Mergny J.L. Fluorescence energy transfer as a probe for tetraplex formaition: the i-motif. Biochemistry. 1999;38:1573–1581. doi: 10.1021/bi982208r. [DOI] [PubMed] [Google Scholar]
- 41.Leroy J.L., Guéron M. Solution structure of the i-motif tetramers of d(TCC), d(5methylCCT) and d(T5methylCC): novel NOE connection between amino protons and sugar protons. Structure. 1995;3:101–120. doi: 10.1016/s0969-2126(01)00138-1. [DOI] [PubMed] [Google Scholar]
- 42.Malliavin T.E., Gau J., Snoussi K., Leroy J.L. Stability of the I-motif structure is related to the interactions between phosphodiester backbones. Biophys. J. 2003;84:3838–3847. doi: 10.1016/S0006-3495(03)75111-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berger E., Egli M., Rich A. Inter-strand C-H···O hydrogen bonds stabilizing four-stranded intercalated molecules: stereoelectronic effects of O4′ in cytosine-rich DNA. Proc. Natl Acad. Sci. USA. 1996;93:12116–12121. doi: 10.1073/pnas.93.22.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leroy J.L., Snoussi K., Guéron M. Investigation of the energetics of C-H...O hydrogen bonds in the DNA i-motif via the equilibrium between alternative intercalation topologies. Magnetic Res. Chem. 2001;39:S171–S176. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.