Abstract
Trypanosomes represent an excellent model for the post-transcriptional regulation of gene expression because their genome is organized into polycistronic transcription units. However, few signals governing developmental stage-specific expression have been identified, with there being no compelling evidence for widespread conservation of regulatory motifs. As a tool to search for common regulatory sequences we have used the nuclear-encoded components of the cytochrome oxidase (COX) complex of the trypanosome respiratory chain. Components of this complex represent a form of post-transcriptional operon because trypanosome mitochondrial activity is unusual in being developmentally programmed. By genome analysis we identified the genes for seven components of the COX complex. Each mRNA exhibits bloodstream stage-specific instability, which is not mediated by the RNA silencing pathway but which is alleviated by cycloheximide. Reporter assays have identified regulatory regions within the 3′-untranslated regions of three COX mRNAs operating principally at the translational level, but also via mRNA stability. Interrogation of the mapped regions via oligonucleotide frequency scoring provides evidence for genome-wide conservation of regulatory sequences among a large cohort of procyclic-enriched transcripts. Analysis of the co-regulated subunits of a stage-specific enzyme is therefore a novel approach to uncover cryptic regulatory sequences controlling gene expression at the post-transcriptional level.
INTRODUCTION
The analysis of cohorts of mRNAs involved in functionally co-ordinated processes is providing a useful tool for the identification of regulatory signals controlling gene expression in eukaryotes (1). Of particular value has been the characterization of the regulatory signals and RNA binding proteins that control nuclear-encoded genes responsible for mitochondrial function. For example, in Saccharomyces cerevisiae, mitochondrial function is regulated dynamically in response to a shift between a fermentable and non-fermentable carbon source, oxygen and haem. This has enabled the experimental analysis (2) and computational prediction (3,4) of cryptic regulatory motifs governing expression.
The requirement to identify control elements in mRNAs which are subject to regulated expression is particularly acute in the parasite Trypanosoma brucei. These organisms, the causative agents of human and animal African trypanosomiasis, are evolutionarily divergent protozoa that are transmitted between mammalian hosts by the tsetse fly (Glossina spp) (5). The different environments that the parasite encounters during its complex life-cycle represent extreme challenges that the cell must overcome, this necessitating regulation of such fundamental processes as metabolism, cell morphology and cell cycle control (6). Unusually, however, these processes are not controlled by regulated transcription. This is because the genome is organized into polycistronic transcription units in which co-transcribed genes are not necessarily co-regulated (7). This genome organization requires resolution of polycistronic pre-mRNAs via the RNA processing reactions of trans-splicing and polyadenylation, generating mRNAs with a 39 nt 5′ spliced leader sequence and 3′ poly(A) tail. Interestingly, trans-splicing and polyadenylation appear to be mechanistically coupled, with the polyadenylation of the upstream gene in an array being co-ordinated with trans-splicing for the downstream gene (8,9). This linkage of RNA processing events for adjacent genes which can display differential mRNA abundance make it unlikely that RNA processing efficiency is a major contributor to the control of gene expression. Instead, the emphasis of trypanosome gene regulation is at the level of mRNA stability, although the importance of translational control is becoming increasingly clear.
In several cases the regulation of mRNA stability has been examined for genes that show differential expression during the life-cycle. The best-characterized genes are those of the major surface antigens of the bloodstream and procyclic form. In bloodstream forms, the variant surface glycoprotein (VSG) mRNA is specifically stabilized through signals in its 3′-untranslated region (3′-UTR), this also mediating destabilization upon transformation to procyclic forms in vitro (10,11). More detailed characterization has been carried out for the insect stage mRNAs encoding EP and GPEET procyclin. In both cases regulatory motifs can be identified in their 3′-UTR which regulate both the stability of each transcript and also their translational competence (12). For example, EP procyclin contains both 16mer and 26mer elements that contribute to mRNA stability and translation efficiency (13–16). For GPEET procyclin an element has also been identified in the 3′-UTR that confers regulation in response to glycerol and hypoxia (17). Although well characterized, VSG, EP and GPEET procyclin genes are all unusual in being transcribed by RNA polymerase I (pol I) (18) and their expression is very stringently regulated in response to life-cycle differentiation signals (19). This contrasts with the majority of regulated trypanosome genes, which are transcribed by RNA polymerase II (pol II). Although evidence for mRNA regulation of several stage-regulated pol II transcripts has been obtained, regulatory signals remain cryptic (7). This contrasts with at least some stage-regulated genes in the related kinetoplastids, Trypanosoma cruzi and Leishmania donovani where common regulatory elements can be identified (20,21).
In an attempt to decipher the complex regulatory information among stage-regulated genes in T.brucei, we have focussed on the cytochrome oxidase complex (COX; complex IV of the electron transport chain). Unusually, in trypanosomes the COX complex is developmentally regulated, being absent in bloodstream forms and induced upon transformation to the insect form (22). This reflects the differential biochemistry of bloodstream forms (where glycolysis driven by blood glucose provides sufficient ATP) and procyclic forms (where mitochondrial elaboration is required in the glucose-sparse tsetse midgut) (23). The COX complex is also a good model for the analysis of regulated gene expression because at least 10 components of the complex are nuclear-encoded (24), with several identified COX genes being dispersed throughout the T.brucei genome (25). In consequence each component must be co-incidentally up-regulated during differentiation in order to construct a functional multi-component enzyme complex. Therefore, analysis and comparison of the regulatory mechanisms governing each component has the potential to uncover hitherto cryptic signals not recognizable among larger groups of stage-regulated genes, or in individual genes.
Here we report the identification and verification of seven nuclear-encoded subunits of the T.brucei cytochrome oxidase complex. We demonstrate for each component that the regulation involves differential mRNA stability and, for at least three subunits, translational control mechanisms. Moreover, we show that differential mRNA abundance results from specific destabilization of each transcript in bloodstream forms. By reporter assays in stably transformed parasites we have mapped the signals governing differential gene expression in the 3′-UTR of three of the COX genes. This has uncovered the presence in the COX V mRNA of a regulatory element related to that found in procyclin transcripts, the so-called 26mer sequence. Moreover, genome-wide computational analysis of predicted 3′-UTRs demonstrates that the core of this motif is significantly overrepresented in procyclic-enriched transcripts. An element common in developmentally regulated transcripts encoding mitochondrial proteins is also identified. This reveals hitherto unexpected conservation of common regulatory signals between pol I and pol II transcribed protein coding genes during the life-cycle of T.brucei which may assist bioinformatic prediction of expression profile.
MATERIALS AND METHODS
Trypanosomes
Bloodstream and procyclic form trypanosomes were T.brucei brucei s427. For the analysis of Argonaute null mutants, bloodstream forms of T.brucei STIB 247 were used. Parasites were grown routinely in HMI-9 at 37°C in 5% CO2 (bloodstream forms) or at 27°C in SDM-79 (procyclic forms). For parasite transfection, 10–15 µg of linearized DNA was electroporated using a BTX ECM830 electroporator. Parasites were selected using 0.5 µg/ml phleomycin (CAT reporter constructs; into bloodstream forms).
Northern blotting and signal quantification; transcriptional and translational inhibition
RNA was extracted from ∼5 × 107 to 1 × 108 trypanosomes using an RNAeasy RNA purification kit (Qiagen). Approximately 3 µg of purified RNA was resolved on formaldehyde-agarose gels and transferred to nylon membrane by capillary blotting. For transcript detection, digoxygenin-labelled riboprobes were used, signal detection being achieved via a Bio-Rad Fluor-S imager. For each probe used for quantitation, the linearity of signal was verified over the detection range, generating R2 values between 0.94 and 0.99. To accurately quantify the respective levels of COX transcripts in bloodstream and procyclic forms, serial dilutions of RNA from each life-cycle stage were hybridized with each COX-specific riboprobe, thereby determining the relative expression level, and confirming linearity of detection at least over a log range of signal. Loading was normalized to rRNA levels in each lane, this being determined also via a Bio-Rad Fluor-S imager. Decay measurements for mRNAs were carried out after treatment of either bloodstream or procyclic form cultures with 5 µg/ml actinomycin D, samples being taken at 0, 30, 60 and 90 min (for bloodstream forms) or 0, 120, 240 and 480 min (for procyclic forms), these time points being established after preliminary analysis of the decay kinetics in each life-cycle stage. For cycloheximide treatment, samples were incubated with 50 µg/ml cycloheximide for 0, 60, 120 and 240 min. In both cases RNA from parallel untreated samples was also prepared and analysed.
Analysis of mRNA processing sites
To map sites of polyadenylation RT–PCR was used, with 3′ ends being amplified for each transcript via a gene-specific primer hybridizing to the 5′ end of the coding region and a generic oligo-dT ADAPT 3′ primer recognizing the poly(A) tail (5′ GGC CAC GCG TCG ACT AGT ACT TTT TTT TTT TTT TT 3′). Amplified products were then subjected to a second round of amplification using an oligonucleotide recognizing the 3′ end of each COX coding region and the primer AUAP (5′ GGC CAC GCG TCG ACT AGT AC 3′), which binds to the specific oligonucleotide sequence incorporated into the 5′ end of the ADAPT primer. The resulting products were gel purified and then sequenced to determine the site of polyadenylation in each life-cycle stage for each gene. For 5′ end mapping, a spliced leader specific primer was used in combination with a primer binding in the 5′ end of each COX gene.
Construct generation and CAT assays
Reporter constructs were based on the T.brucei expression vector pHD449, with the tetracycline resistance cassette being replaced by the coding region for choramphenicol acetyl transferase (CAT). This was achieved by digesting pHD449 with HindIII and BamHI to excise the TETR gene, this being replaced by a CAT gene PCR amplicon provided with a 5′HindIII site and 3′ BamHI site. To insert each COX gene 3′-UTR, each intact COX intergenic region was amplified using primers binding immediately after the COX gene stop coding and immediately upstream of the ATG codon of the downstream gene. An exception to this was COX V where the downstream (‘hypothetical unlikely’) gene was 1.4 kb downstream and the next open reading frame was over 2 kb from the COX V stop codon. In this case, a 3′ primer-binding site was chosen ∼30 bp downstream of the last of three prominent polypyrimidine tracts in the intergenic region. Each 5′ and 3′ primer had, respectively, BamHI and Bbs1 restriction sites incorporated to allow cloning into CAT449. The resulting PCR products were digested with BamHI and Bbs1 and these inserted into BamHI/Bbs1 digested CAT 449 to allow replacement of the endogenous truncated aldolase 3′-UTR in the vector. To insert regulatory regions in to intact CAT449 for analysis of function out of context, CAT 449 was digested with BamHI and each COX regulatory region, amplified with primers containing BamHI at each end, inserted. The resulting constructs were sequenced to ensure fidelity of insertion and to verify insertion copy number and orientation.
Levels of CAT protein derived from each reporter construct were determined by CAT ELISA assay (Roche) according to the manufacturer's instructions, values being determined on a Dynex technologies MRX II ELISA plate reader. In each case a CAT standard curve was constructed using known concentrations of CAT protein and these used to determine CAT levels in each lysate, this being verified to be in the linear range by lysate dilution over a 1000-fold range. CAT standard curves had a linear regression value typically of 0.998, providing accurate determination of CAT to as low as 0.001 ng/nl.
Oligonucleotide frequency analysis
Oligomer counting was performed using the oligo-analysis tool, a web-based tool forming part of the Regulatory Sequence Analysis Tools (RSAT) suite (http://rsat.scmbb.ulb.ac.be/rsat/). The tool identifies oligomers ranging in length from 3 to 8 nt that are more frequent within the UTR sequence of a group of co-regulated genes compared to a set of non-regulated genes. To identify possible regulatory elements within the 3′-UTR regions of the COX components from T.brucei and T.congolense, counts were performed on either the 3′-UTR sequence up to the experimentally mapped polyadenylation site or, if this site had not been mapped, the first 300 nt from the COX gene stop codon. Oligomer frequency counts were performed at various oligomer lengths from 3 to 8 and compared to ‘background counts’ performed on a database of 3′-UTR sequences from a mixed population of genes considered to be non-directionally regulated. For the background dataset, 300 nt of 3′-UTR sequence were retrieved from every annotated gene on T.brucei chromosomes 1 and 2, totalling 883 sequences. The same method was used for analysis of the microarray data with the genes shown to be upregulated in either the procyclic stage or bloodstream stage considered to be co-regulated. Again 300 nt of the 3′-UTR were retrieved for each gene and counted in the analysis. All statistics and significance values were determined according to (3).
RESULTS
Identification of the nuclear-encoded T.brucei COX genes
Components of the cytochrome oxidase complex of the kinetoplastid Crithidia fasciculata, have been biochemically purified and N-terminal sequences derived for six subunits (26). We used these peptide sequences to search the T.brucei genome database (http://www.genedb.org/genedb/tryp/) in order to identify genes encoding putative orthologues in that organism [(27) and this study]. The largest nuclear-encoded subunit (COX IV) has been characterized subsequently from Leishmania tarentolae and its homologue in T.brucei identified previously (28).
Table 1 shows each of the predicted T.brucei proteins aligned with the N-terminal sequence derived from C.fasciculata or L.tarentolae. Notably alignment of the derived open reading frame for subunits V–X with the determined N-terminal sequence of each from C.fasciculata predicts a cleaved leader sequence of between 1 [COX VI, VII; (27)] and 27 amino acids (COX VIII) whereas comparison of the T.brucei COX IV gene with the L.tarentolae COX IV protein predicts a 44 amino acid leader. The genes for these seven predicted nuclear-encoded cytochrome oxidase components are unlinked in T.brucei, being distributed over six distinct chromosomes and each has been annotated onto the T.brucei genome database (GeneDB co-ordinates for each gene are given in Table 1).
Table 1.
Identification of COX subunits in the genome of T.brucei
| Predicted leader sequence | Mature N-terminus (actual or predicted) | Organism | Protein | GeneDB identifier | Reference |
|---|---|---|---|---|---|
| MFTRRAVSSVVGVTGSAAVVTSSPLSVQRRY | DHDRWYGHALELDTHNYKFNGEP | L.tarentolae | COX IV | (28) | |
| MFARRSLIATVAAATATKPTSSAAQSNANGTAATQSTLLQQRRY | DHDRWYGHALELDSHNYKFTGEP | T.brucei | COX IV | Tb927.1.4100 | |
| FFGKGWDNASLDTIFSSML | C.fasciculata | COX V | (26) | ||
| MKRFVTPFLATVPLSRN | FFGKGWDNAALDTIFSCML | T.brucei | COX V | Tb09.160.1820 | |
| PHADHRKYKIQREEMP--PHFSDFNDP RF | C.fasciculata | COX VI | (26) | ||
| M | PFVDHNKYKIQREDLPALPHFTDFNDPRF | T.brucei | COX VI | Tb10.100.0160 | |
| PFVDHNKYKIQREDLPALPHFTDFNDPRF | T.brucei | COX VI-TY | (27) | ||
| PRPFGVWAPATTLAEYRARIPNPFAYSFKWVYSMKKEIFY | C.fasciculata | COX VII | (26) | ||
| M | PRPFGVWAPATTLAEYRARIPGMSNFKLRWVFGARREVYY | T.brucei | COX VII | Tb927.3.1410 | |
| GGDMHSSDRFKAAWDEIPLHM | C.fasciculata | COX VIII | (26) | ||
| MIRRTAPAVSFTTSHRALMLR TNRPLL | SADMHSLERFKVAWDEMPVH | T.brucei | COX VIII | Tb927.4.4620 | |
| YMLAFNSKAKARPNFGLRGVGYWH-EVY nKPGQsY | C.fasciculata | COX IX | (26) | ||
| MFSCALRTSRRT | YINAFNAKAKARPNFGLRGVGYWTSEVYHKPGQNY | T.brucei | COX IX | Tb10.6k15.2180 | |
| LHFPISAPPIEIDYLDNDPLEFAVRTEArKwGF | C.fasciculata | COX X | (26) | ||
| MLRRAGSRVACACSVPQARS | LHFPITPPPIEIEYLDNDPLEFAVRTEARKWRF | T.brucei | COX X | Tb11.01.4702 |
In each case the biochemically derived protein sequence of corresponding subunits from either C.fasciculata (26) or Leishmania tarantole (28) was used as a search tool to identify the T.brucei gene. Analysis of the determined N-terminal protein sequence and genomic analysis of the translated gene sequence allowed prediction of the N-terminal leader sequence on each subunit. For T.brucei COX VI this has been verified experimentally (27).
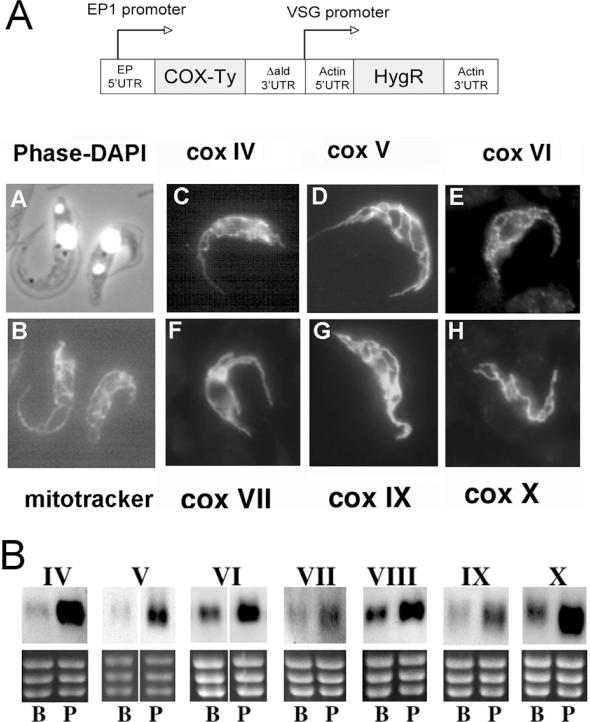
To verify the identity of each subunit we used an epitope-tagging approach to visualize their predicted mitochondrial location. Thus, each of the identified genes was amplified from T.brucei genomic DNA and inserted into the T.brucei expression vector pHD451 (29) modified to incorporate a Ty1 epitope tag (30) at the C-terminus of the encoded transgenic proteins (Figure 1A, schematic representation). Each construct was transiently transfected into procyclic form cells, which were subsequently analysed for expression and localization of the ectopically expressed protein after paraformaldehyde fixation and detergent permeabilization (27). Figure 1A shows cells transfected with tagged COX IV-COX X (panels C–H) and reveals, in each case, lattice-like staining characteristic of the procyclic form mitochondrion and matching the distribution of the mitochondrial vital dye, Mitotracker Red (Figure 1A, panel B). This analysis confirmed the predicted mitochondrial localization of each identified subunit, supporting their identification as components of the biochemically characterized kinetoplastid cytochrome oxidase complex.
Figure 1.
(A) Mitochondrial localization of identified COX subunits. For each subunit, the coding region was inserted into a trypanosome expression vector such that the expressed protein included a C-terminal Ty1 epitope tag. Transient transfection of each construct into procyclic forms resulted in expression of an epitope-tagged protein which, in each case, localized to the mitochondrion of the procyclic form (panels C–H). This localization also matched the staining observed with the mitochondrial vital dye, Mitotracker red (panel B). A phase contrast image of the cells in panel B stained with DAPI is shown in panel A, this visualizing the position of the cell nucleus and the mitochondrial genome (kinetoplast). (B) Northern blots demonstrating the relative expression of the identified COX subunit transcripts in bloodstream (B) or procyclic (P) form trypanosomes. In each case, a section of the ethidium stained gel is shown (containing the rRNAs) to demonstrate relative loading.
The COX subunit mRNAs are regulated at the level of stage-specific differential stability
Activity of the cytochrome oxidase complex is developmentally regulated in T.brucei, and the nuclear-encoded subunit COX VI has been shown previously to be stage-regulated, its mRNA being enriched in procyclic forms (31). In order to determine if regulation at the RNA level was a general feature of the nuclear-encoded COX genes, northern blots of bloodstream and procyclic RNA were hybridized with riboprobes specific for each. Figure 1B demonstrates that although low levels of mRNA are detectable for each subunit in bloodstream forms, there is increased abundance in procyclic forms. Quantitative analysis of the level of several of the COX transcripts in bloodstream and procyclic forms revealed a mean differential expression of ∼3-fold between bloodstream and procyclic forms (COX V, 4-fold; COX VI, 2.5-fold; COX IX, 2.6-fold).
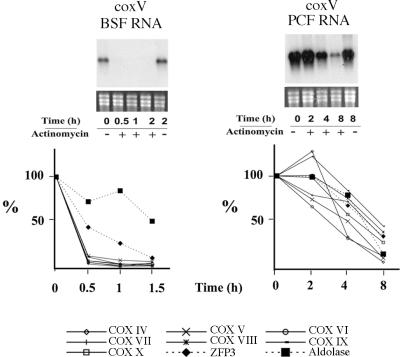
Because of the importance of post-transcriptional regulation in trypanosome gene expression, the most likely mechanism for regulated mRNA abundance is via developmental changes in mRNA stability. In order to investigate the stability of each COX transcript in bloodstream and procyclic forms we used actinomycin D to inhibit transcription and, thereafter, followed the decay of each mRNA in each life-cycle stage by northern blotting. In each case transcript levels were quantitated with respect to the amount of rRNA loaded and after verifying the linearity of detection with each probe. Figure 2 (upper panel) shows a representative northern blot of COX V mRNA decay in bloodstream (upper left hand panel) and procyclic forms (upper right hand panel) at time points after transcriptional inhibition with actinomycin D. Analysis of the decay of transcripts in procyclic cells revealed that even after 4 h COX V transcripts were still abundant (50% of COX V mRNA remained with respect to its abundance before the addition of actinomycin D; Figure 2 lower right panel). This contrasts with bloodstream forms where after only 30 min, COX V transcripts were barely detectable (Figure 2, lower left panel; COX V mRNA is <10% of the initial abundance after 30 min). The same analysis for each of the other identified COX transcripts is shown in Figure 2 (lower panels) with the abundance of each being quantified at time points after actinomycin D addition to either bloodstream or procyclic forms. In each case the COX transcripts were relatively unstable in bloodstream forms with respect to procyclic forms, with a mean half-life of ∼4 h in procyclic forms whereas in bloodstream cells the half-life was <15 min. Contrasting with this, analysis of a constitutively expressed control transcript, TbZFP3 (A. Paterou, P. Walrad, P. Craddy, K. Fenn and K. R. Matthews, manuscript submitted), or bloodstream-specific transcripts such as aldolase, revealed far greater stability in bloodstream forms (t1/2 = 40 min or 1.5 h, respectively). Although these transcripts were less stable in bloodstream forms than in procyclic forms, consistent with previous observations (10,16), this instability was far less than the >16-fold difference in half-life observed with each COX transcript. Together this analysis demonstrated that each of the COX mRNAs is differentially regulated between bloodstream and procyclic forms, with differences in mRNA stability being a significant contributor to this.
Figure 2.
Stability of COX subunit mRNAs in bloodstream and procyclic form trypanosomes. The upper panels show representative northern blots of COX V transcripts from cells untreated (−) or treated (+) with 5 µg/ml actinomycin D. RNA was isolated at the times shown. The lower panels show a quantitation of all COX subunit mRNAs in each life-cycle stage in the presence of actinomycin D, with each data point representing the level of mRNA with respect to its abundance before the addition of actinomycin D. Control transcripts of TbZFP3 (constitutively expressed) and aldolase (bloodstream enriched) are also shown.
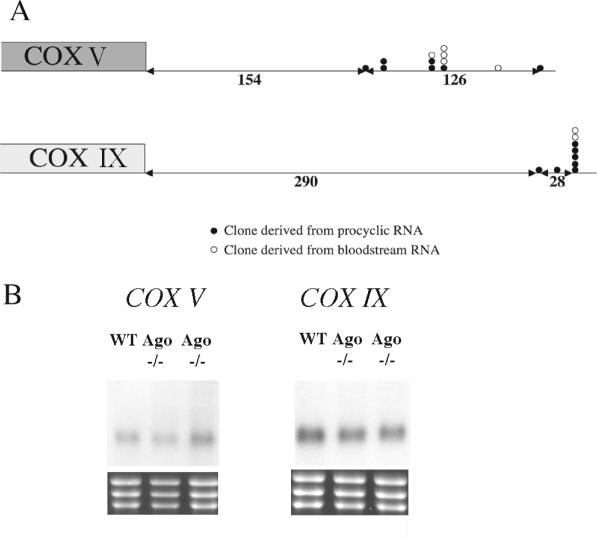
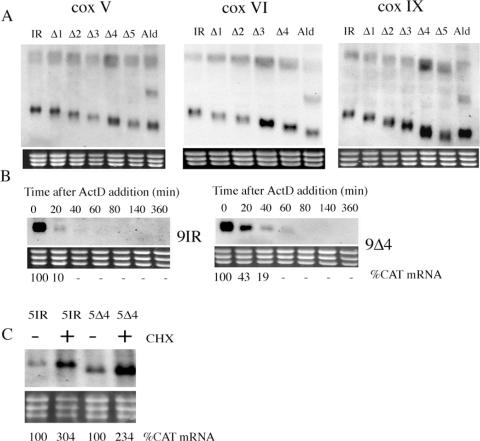
To investigate the basis of the differential mRNA stability of COX transcripts between bloodstream and procyclic forms we assayed three potential regulatory mechanisms: (i) the use of distinct mRNA processing sites in each life-cycle stage (32,33); (ii) the destabilization of regulated transcripts via the RNA silencing pathway (34); and (iii) control via labile protein regulators. With respect to the differential usage of RNA processing sites in distinct life-cycle stages, RT–PCR was performed on bloodstream and procyclic RNA using primers specific for the 3′ end or 5′ end of the COX V and IX mRNA. This revealed that although several sites of polyadenylation are used for each gene, these did not differ between different life-cycle stages (Figure 3A). Similarly spliced leader addition sites were identical in bloodstream and procyclic forms (data not shown). To investigate potential involvement of the RNA silencing machinery in regulated control of the COX transcripts, the relative abundance of COX V and COX IX mRNA was assayed in an Argonaute null mutant bloodstream form line and the corresponding wild-type parent (35). Argonaute is a functional homologue of slicer in the trypanosome RNA silencing machinery (36), with null mutants being incapable of RNAi. Figure 3B reveals that no difference in the abundance of either transcript was detected in the Argonaute null mutant when compared to wild-type parents, eliminating a role for the RNAi machinery in stage-regulation of these transcripts.
Figure 3.
(A) Mapping of the site of polyadenylation for two COX subunit mRNAs (COX V and IX) in bloodstream (open circles) or procyclic forms (closed circles). The sites of polyadenylation are distributed, but there is no consistent difference between the sites used in each life-cycle stage. (B) mRNA abundance of COX V and COX IX in wild-type bloodstream forms of T.brucei brucei STIB 247 and in two independently derived Ago null mutants of the same strain (35).
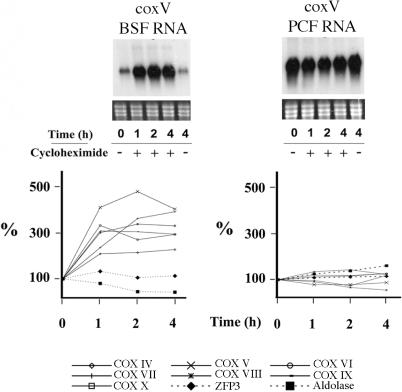
To determine if COX transcripts are actively destabilized in bloodstream cells or actively stabilized in procyclic cells via labile protein factors, bloodstream and procyclic cells were incubated in 50 µg/ml cycloheximide to inhibit de novo translation. Under these conditions any labile regulatory proteins controlling COX transcripts would be rapidly depleted leading either to a decrease in the abundance of COX transcripts in procyclic cells or an increase in bloodstream cells (37–40). The upper panels in Figure 4 show a northern blot of RNA extracted from bloodstream and procyclic cells at various time points after translational inhibition and hybridized to detect COX V mRNA. Figure 4 (lower panels) show quantification of corresponding northern blots probed for each of the remaining nuclear-encoded COX transcripts. This analysis revealed a marked increase in the abundance of all COX transcripts tested in bloodstream cells when treated with cycloheximide (3.7-fold, ±0.4 SE), but not in procyclic cells (1.2-fold, ±0.1 SE). In contrast to this, the constitutively expressed transcript TbZFP3 and bloodstream-specific aldolase transcript showed no significantly enhanced abundance in the presence of translational inhibition either in bloodstream or procyclic forms (Figure 4). These experiments establish that COX mRNAs are specifically destabilized in bloodstream forms, this being alleviated by translational inhibition.
Figure 4.
mRNA abundance of COX subunit mRNAs after treatment of bloodstream or procyclic form trypanosomes with cycloheximide. The upper panels show representative northern blots of COX V transcripts from bloodstream (BSF) or procyclic forms (PCF) untreated (−) or treated (+) with 50 µg/ml cycloheximide. RNA was isolated at the times shown. The lower panels show a quantification of all COX subunit mRNAs in each life-cycle stage in the presence of cycloheximide, with each data point representing the level of mRNA with respect to its abundance before the addition of cycloheximide. Control transcripts of TbZFP3 (constitutively expressed) and aldolase (bloodstream-enriched) are also shown.
CAT reporter assays identify elements that repress COX gene expression
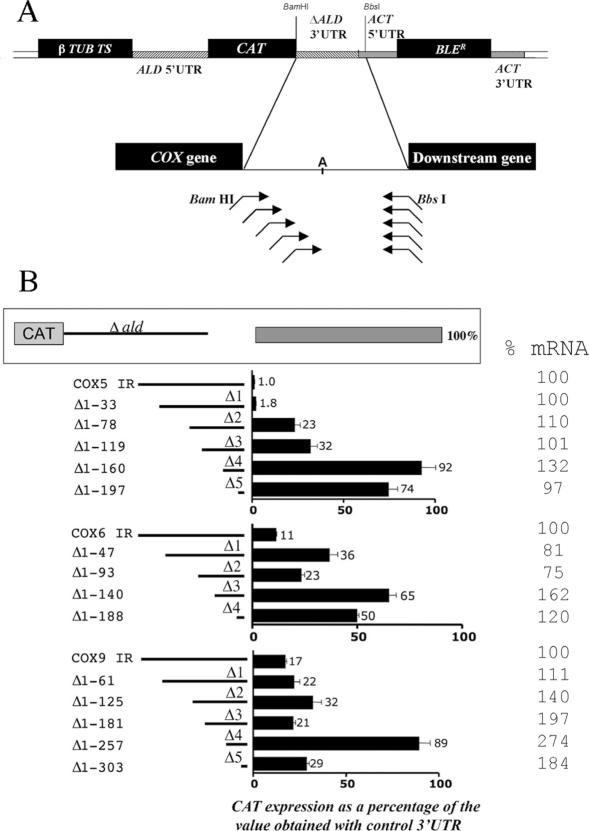
In order to dissect the sequences contributing to the bloodstream stage-specific repression of COX gene expression, three distinct and arbitrarily chosen COX genes were analysed for their regulatory signals. Thus, we created a series of CAT reporter constructs each bearing sequential deletions of the COX V, VI and IX 3′-UTRs (Figure 5A). These constructs were integrated into the T.brucei tubulin gene cluster, enabling read-through transcription of the reporter gene via pol II and accessory factors. This ensured that each operated in a transcription unit matching that of the endogenous gene. In each case the CAT reporter gene was provided with either a deleted 3′-UTR of the aldolase gene [which results in constitutive expression; (29)] or with ∼50 nt sequential deletions of the COX V, VI or IX downstream intergenic regions (Supplementary Figure 1 shows the sequence and deletion limits for each 3′-UTR). The deletions were progressively made from the 5′ end of each 3′-UTR toward the mapped region of polyadenylation for each gene, such that all constructs retained the endogenous 3′ end processing site and downstream intergenic sequences extending either to the next gene in the polycistron, or in the case of COX V (where the next gene is over 1.5 kb downstream) 164 nt downstream of the major site of polyadenylation (this region contains at least three prominent polypyrimidine tracts). Our strategy was to transfect these constructs into bloodstream cells and assay for increases in CAT production as the 3′-UTRs were progressively deleted, thereby mapping regions responsible for mRNA instability or translational repression in bloodstream forms. Figure 5B shows CAT activity assays from a minimum of two assays performed on protein lysates extracted from at least two independently derived stably transformed clonal cell lines for each construct. These assays revealed specific regions that when deleted cause a marked increase in CAT expression. For COX V, an overall 92-fold increase in CAT expression was observed during progressive truncation of the intergenic region, this comprising a 13-fold increase in CAT protein when nt 33–78 from the stop codon (i.e. the COX VΔ2 region) were deleted (compare Δ1–33 with Δ1–78) and a further 2.9-fold increase with deletion of nt 119–160 (COX VΔ4 region). For COX IX, a single deletion positioned between nt 181 and 257 from the stop codon (COX IX Δ4) caused a 5-fold increase in CAT activity with respect to the intact intergenic region, although this increase was abolished by further deletion close to the major site of polyadenylation (COX IXΔ5). In contrast to COX V and COX IX 3′-UTRs, deletions of the COX VI 3′-UTR sequence did not clearly define particular regions having a major impact on CAT gene expression, although truncation of nt 93–140 increased CAT expression 2.8-fold with respect to the intact intergenic region.
Figure 5.
(A) Investigation of regulatory signals within COX V, VI and IX 3′-UTRs. The intergenic region for each COX subunit was positioned downstream of the CAT gene, this being encompassed within a construct comprising a phleomycin resistance gene and targeted to integrate into the tubulin gene array. The site of polyadenylation is indicated within the intergenic region (denoted ‘A’). At least two stable cell lines were generated for each construct and two independent CAT assays performed on cell extracts from each cell line. (B) Resulting CAT expression for each COX intergenic region deletion are shown as a percentage of the CAT expression from an identical construct comprising a deleted aldolase gene 3′-UTR, which provides constitutive expression in bloodstream and procyclic forms. The CAT mRNA abundance for each construct is shown at the extreme right, as a percentage of the CAT mRNA abundance derived from constructs comprising each intact COX subunit intergenic region (IR). These values were derived from northern blots, representatives of which are shown in Figure 6.
mRNA abundance is not a major regulator of CAT production for COX V and VI
In order to distinguish the relative contribution of mRNA abundance to the observed CAT activity, we used northern blotting to determine the level of CAT mRNA derived from each truncation of the COX V, VI and IX intergenic regions (Figures 5B and 6A). This revealed that the CAT mRNA abundance derived from those constructs furnished with the COX V intergenic region differed little, exhibiting only a maximal 32% increase over the intact intergenic region despite a nearly 100-fold increase in CAT protein upon truncation of the 3′-UTR. This indicates that this 3′-UTR conferred regulation almost exclusively at the translational/post-translational level. Similarly, for COX VI, the CAT mRNA abundance exhibited a maximal 62% increase over the intact intergenic region (contrasting with a 6-fold protein difference), whereas for COX IX CAT mRNA abundance increased by 274% in the COX IXΔ4 truncation. This approximates to the differential mRNA abundance for the endogenous COX IX transcript between bloodstream and procyclic forms (2.6-fold; Figure 1B). Analysis of the COX IX Δ4 deletion further by actinomycin D treatment confirmed that the mRNA stability of the CAT transcript was increased over that of the intact intergenic region, although the rapid turnover of the latter prevented accurate quantitation of the relative increase in stability (Figure 6B). Importantly, RT–PCR analysis (for COX V) or CAT transcript sizing (for COX V, COX VI and COX IX) for mRNAs derived from all truncations revealed that no alteration of the polyadenylation site occurred with respect to the intact intergenic region for each gene (Figure 6A and data not shown). This eliminated perturbance in the site of 3′ end formation being responsible for the observed changes in CAT expression.
Figure 6.
(A) Northern blots of CAT mRNA provided with successive deletions of the COX V, COX VI or COX IX intergenic regions. In each case the rRNA region of the ethidium stained gel is provided to demonstrate relative loading. (B) Northern blots of CAT mRNA derived from cell lines in which the CAT gene is provided with the COX IX intergenic region (9IR) or a deletion (COX IX Δ4, ‘9Δ4’) which results in altered mRNA abundance. All cell lines were exposed to actinomycin D at t = 0 and RNA isolated at time points thereafter in order to follow transcript decay. The relative abundance of CAT mRNA is denoted beneath each lane. (C) Northern blot of CAT mRNA derived from cells transfected with constructs comprising either the intact intergenic region for COX V or a deleted derivative (COX VΔ4; ‘5Δ4’). In each case cells were either untreated (−) or treated (+) with cycloheximide, with mRNA being isolated 4 h after the addition of drug. CAT mRNA abundance is denoted beneath each lane.
Since little or no increase in the level of CAT mRNA was observed upon deletion of the COX V 3′-UTR, we examined whether this intergenic region exhibited elevation of CAT mRNA after exposure to cycloheximide, thereby matching the response of the endogenous COX V transcript. Figure 6C demonstrates that this was the case: the intact intergenic region for COX V showed a 300% elevation in CAT mRNA after cycloheximide treatment, contrasting with the response of the same construct bearing the aldolase intergenic region (which showed a 0–30% increase after cycloheximide treatment; data not shown). Moreover the response of the COX V 3′-UTR to cycloheximide was retained even in the COX VΔ4 truncation which lacks all but 50 nt of the COX V 3′-UTR, this construct exhibiting a 230% elevation (Figure 6C). These observations implicate sequences in the intergenic region downstream of the polyadenylation site as contributing to the response to cycloheximide. This matches previous observations with constructs containing the procyclin intergenic region, where deletion of the complete 3′-UTR also did not ablate the super-induction of mRNA in response to cycloheximide (41).
To conclude this section, we showed enhanced expression of CAT upon deletion of each COX 3′-UTR implicating the presence of negative control elements. For COX IX mRNA stability is an important component, whereas for COX V and COX VI control operates almost exclusively at the translational/post-translational level. This is consistent with analysis of endogenous COX VI expression where protein levels are stringently stage-regulated in a mechanism dominant to differential mRNA regulation (27). Combined, this highlights the complexity of regulation among components of even the same enzyme complex.
Identification of shared control elements with the EP procyclin genes
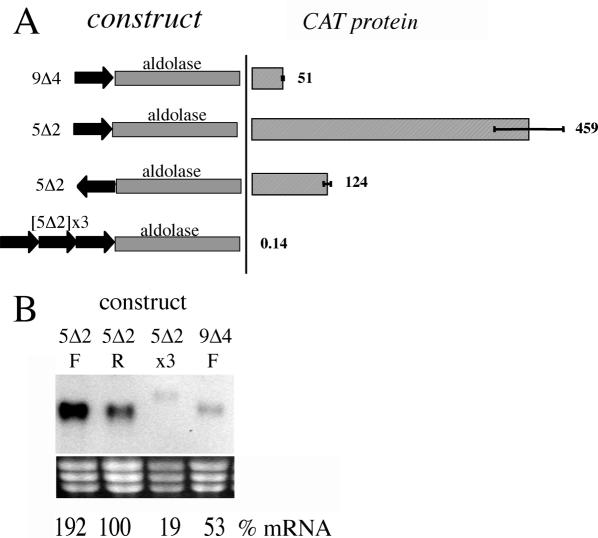
To analyse in more detail those regions identified as contributing to COX regulation we focused on the COX IX Δ4 and COX V Δ2 regions, each of which had a distinct effect on CAT gene expression. Initially we examined the COX IXΔ 4 region, which affected both CAT mRNA abundance and stability. To determine if this element was able to operate in isolation, this domain was inserted into the aldolase 3′-UTR immediately after the CAT gene stop codon. In this context, CAT protein expression was observed to be reduced to 50% of the aldolase control, with CAT mRNA being decreased to the same extent (Figure 7A and B). This established that this region could operate to down-regulate mRNA abundance in a context independent manner.
Figure 7.
(A) CAT activity generated by constructs in which either the COX IXΔ4 (‘9Δ4’) or COX VΔ2 (‘5Δ2’) regions are placed in front of the aldolase 3′-UTR, with 5Δ2 being either in forward or reverse orientation, or in multiple copies. Values are expressed as a percentage of the protein derived from a construct with the aldolase 3′-UTR alone. (B) Northern blot of CAT mRNA derived from the cell lines in which either the COX IXΔ4 (‘9Δ4’) or COX VΔ2 (‘5Δ2’) regions are placed in front of the aldolase 3′-UTR, with 5Δ2 being either in forward (F) or reverse (R) orientation, or in multiple copies (x3). mRNA quantification is expressed as a percentage of a cell line with the aldolase 3′-UTR alone, these being normalized to relative loading as determined by rRNA levels.
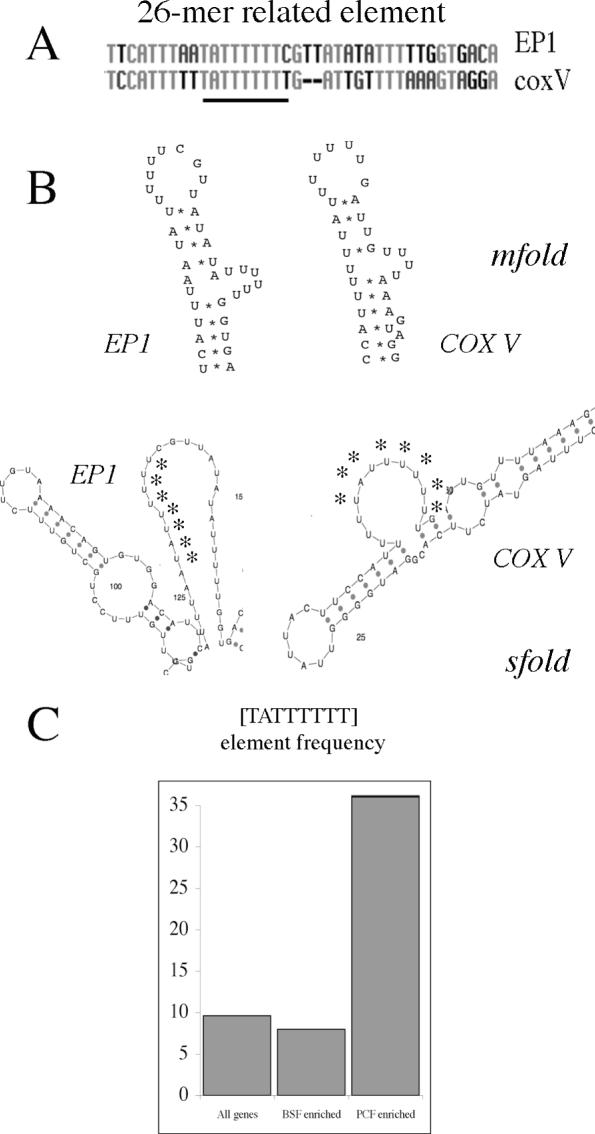
In contrast to the COX IX Δ4 region, our reporter assays demonstrated that the COX V Δ2 element operated exclusively at the level of translation. Moreover it gave the largest single effect of any of the mapped regions in COX V, VI or IX demonstrating the presence of a strong regulatory element within that domain. This prompted us to analyse this region for recognizable elements that may contribute to this regulatory role. Significantly, this revealed that this region contained a sequence motif similar to that of the 26mer regulatory element identified previously in the EP procyclin 3′-UTR (14,15) (Figure 8A and Supplementary Figure 2). The 26mer element is a region comprising oligoU sequence interrupted by a spacer region which has the potential to form a stem–loop structure containing a U-rich bulge (14). Interestingly, the element in COX V Δ2 could be folded into a very similar structure to that predicted for the 26mer in the EP procyclin 3′-UTR, with changes in one side of the stem from the 26mer sequence being matched by compensatory changes in the corresponding base pair partners on the other side of the stem (Figure 8A). Since structural mapping does not predict extensive stable base pair interactions in the EP procyclin 26mer element in vitro (42), we also analysed the predicted S-fold structures for both EP procyclin and COX V 3′-UTRs (43). This algorithm predicted that each element was in a single stranded bulge. Thus, although different methods assign different predicted structures, the folding of the EP procyclin 26mer and the related COX V element were similar in each case.
Figure 8.
(A) Alignment of the EP procyclin 26mer element with the corresponding region in the COX V 3′-UTR (the [TATTTTTT] element is underlined). (B) Structure prediction for the 26mer core element in EP procyclin and COX V 3′-UTRs. In the upper structures, an m-fold prediction (47) for the 26mer element in the procyclin 3′-UTR (14) is used to model the related region in the COX V 3′-UTR. The lower structures represent the centroid consensus derived by s-fold structure prediction (43) covering the same region. Asterisks indicate the UAUUUUUU RNA sequence of the core element. (C) Frequency (expressed as a percentage of genes within each group) of the 26mer core element [TATTTTTT] in the 300 nt downstream from all genes in the trypanosome genome, transcripts enriched in bloodstream forms (BSF-enriched), or transcripts enriched in procyclic forms (PCF-enriched). Expression data were derived from microarray data kindly provided by Christine Clayton, University of Heidelberg.
In the EP procyclin 3′-UTR, the 26mer operates in a context-specific manner, with distinct effects depending upon the adjacent 3′-UTR or coding sequence (16). To analyse whether the COX V Δ2 region exhibited the same characteristics, this region was placed immediately after the CAT stop codon in the aldolase 3′-UTR (Figure 7A and B). In this context the element conferred a 450% increase in CAT protein and 192% increase in CAT mRNA whereas the same sequence in inverted orientation had no significant effect on CAT expression. In contrast to this, when three copies of the COX V Δ2 region were inserted, CAT protein was reduced to 0.14% of the intact aldolase 3′-UTR and the CAT mRNA was barely detectable. This was not due to the activation of an alternative, perhaps less efficient, polyadenylation site because the transcript size detected on northern blots matched that of the intact aldolase 3′-UTR taking into account the additional size provided by the inserted element (Figure 7B).
We conclude that the major regulatory element in the COX V intergenic region is related structurally and functionally to the 26mer regulatory element mapped in the EP procyclic 3′-UTR. However, it effects on CAT gene expression appear to be strongly context dependent.
Oligonucleotide counting as a tool to identify regulatory sequences in developmentally expressed genes
We sought a bioinformatics approach to search for conserved regulatory sequences either among many procyclic-specific genes, or the more restricted COX gene subset. In order to achieve this we employed an approach that searches for oligonucleotides that are statistically overrepresented in a co-regulated gene set when compared to their frequency in the intergenic region of other genes (3). This approach has been used successfully to identify nuclear-encoded mitochondrial genes in yeast that are regulated in response to catabolite repression (4). In order to derive oligonucleotide frequency tables for a large cohort of trypanosome genes that do not show co-regulation, we downloaded and analysed the first 300 nt after the stop codon of genes predicted on chromosome I and II of the T.brucei genome to provide a training set comprising 883 genes. This length of 3′-UTR was chosen as representative of the average length of 3′-UTRs for experimentally characterized trypanosome mRNAs and closely matches the 348 nt median 3′-UTR length determined recently by a predictive algorithm (44). The resulting frequency tables were then screened with the 300 nt downstream of those genes up-regulated in procyclic forms as determined by microarray analysis (kindly provided by Stefanie Brems, Joerg Hoheisel and Christine Clayton, University of Heildelberg and publicly available at http://www.zmbh.uni-heidelberg.de/Clayton/default.shtml) in order to identify sequence elements overrepresented in this group. This identified an overrepresented octamer oligonucleotide set, of which the most statistically significant was the sequence TATTTTTT. Interestingly, this oligonucleotide sequence comprises the core of the 26mer element identified in EP and GPEET procyclin, as well as in PGKB, PPDE (45) and COX V (this study). Scanning of the transcripts up-regulated in procyclic forms found that 56/179 (31%) of procyclic-enriched transcripts contained precisely this element (Figure 8C and Supplementary Table 1), whereas only 8% of bloodstream-enriched transcripts (23/287 transcripts) contained the sequence. Moreover, analysis of the frequency of the TATTTTTT element in the 3′-UTR of all genes predicted in the genome (10 765 genes analysed) revealed that only 9.6% of all putative genes contained this sequence (Figure 8C). This emphasizes that when transcripts known to be up-regulated in procyclic forms are considered, there is significant overrepresentation of the 26mer core element.
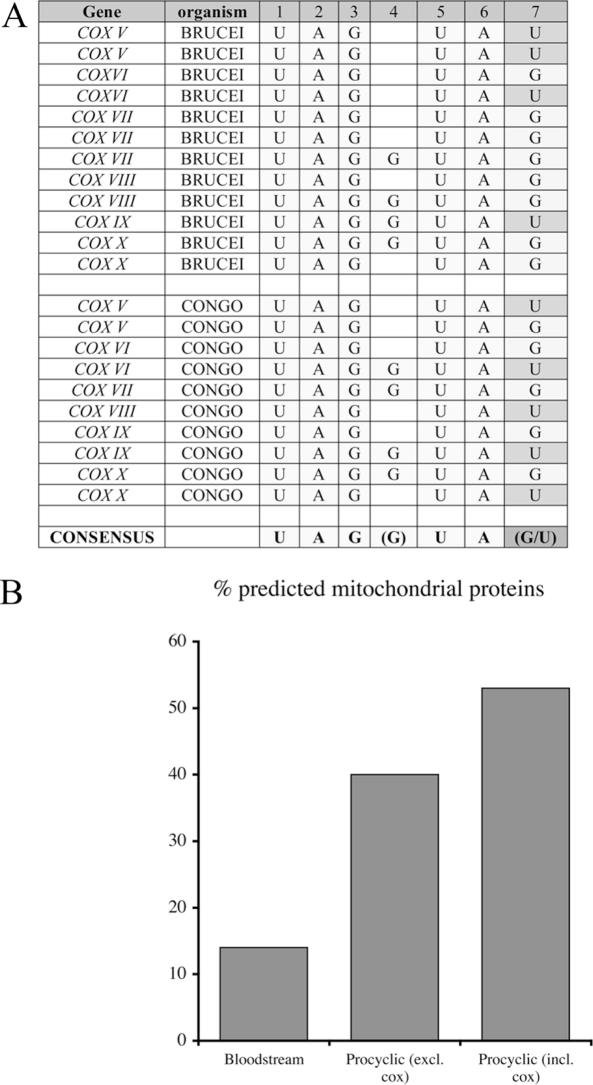
Not all COX 3′-UTRs analysed contained a U-rich element in regions mapped as contributing to gene expression. Moreover, not all deletions of regions containing U-rich elements resulted in changes to gene expression. Therefore, we also applied the oligonucleotide counting approach to search for motifs specifically overrepresented in the COX subunit 3′-UTRs. In this case, the trypanosome oligonucleotide frequency dataset was screened using the sequence of the seven 3′-UTRs of the nuclear-encoded COX subunits. In addition, the search set was expanded by inclusion of cytochrome oxidase subunits identified in T.congolense, a closely related trypanosomatid where respiratory activity is also stage-regulated. All seven cytochrome oxidase subunits were identified by TBLASTX searching of that genome with the T.brucei protein sequences isolated in this study. The resulting search identified a consensus sequence UAG (G) UA (G/U) which was present in 6/7 COX genes analysed whether derived from T.brucei or T.congolense (Figure 9A). Interestingly, a copy of this element is in nt 1–47 of the COX VI intergenic region (UAGUAGUAG) where deletion results in a 3-fold increase in CAT activity in bloodstream forms and a related sequence UAaGUAUAUA is in the COX IXΔ4 nt 181–257 region, whose deletion results in a 4-fold increase in CAT protein and 2.6-fold increase in CAT mRNA abundance. Analysis of transcripts enriched in bloodstream or procyclic forms also indicated that this element was overrepresented in procyclic forms, with 18% (32/179) of transcripts containing the element compared to 9.7% of bloodstream-enriched transcripts (28/287). Moreover, even excluding the COX subunits, 40% of transcripts with the element were predicted to be mitochondrial on the basis of PSORTII or manual analysis, this including the respiratory chain components rieske iron–sulphur protein, cytochrome C1 and a subunit of the F1 ATPase. This contrasts with the bloodstream-enriched transcripts of which only 14% were predicted to encode proteins with a mitochondrial location (Figure 9B).
Figure 9.
(A) The 3′-UTR of COX transcripts conserve the oligonucleotide consensus UAG (G) UA (G/U). The 3′-UTR of COX subunits identified in T.brucei and by bioinformatic interrogation of the incomplete T.congolense genome were analysed for overrepresented oligonucleotide sequences. The occurrence of variants of the identified consensus is shown for each transcript, some genes containing multiple representatives. (B) The frequency of predicted mitochondrially located proteins containing the conserved oligonucleotide in either bloodstream or procyclic-enriched transcripts. Mitochondrial location was determined by a combination of PSORT II and manual analysis.
We conclude that in addition to the 26mer element identified in COX V, additional elements can be recognized among the COX gene family which may be conserved among a wider subset of procyclic-enriched transcripts, particularly those associated with mitochondrial function.
DISCUSSION
To date, widely conserved regulatory signals governing stage-specific gene expression in trypanosomes have eluded detection, preventing the prediction of gene expression by bioinformatic approaches. Here we have characterized the signals that contribute to the expression of a developmentally regulated multi-subunit enzyme complex in order to recognize conserved sequence elements likely to control stage-specific gene expression. Consistent with previous analyses, this has revealed the importance of both mRNA stability and translational efficiency in the control of developmentally regulated nuclear gene expression. However, one identified element in the cytochrome oxidase subunit V mRNA was closely related in sequence, predicted secondary structure and function to the 26mer element identified previously as important in the control of the procyclic form surface antigens, EP and GPEET procyclin. Moreover, the core of this conserved element was found by an oligonucleotide frequency analysis approach to be conserved in a large cohort of transcripts that are up-regulated in the procyclic form of the life cycle. This predicts that the 26mer core element is an important regulatory element for a large set of stage-regulated pol I and pol II transcribed genes. This facilitates bioinformatic prediction of gene expression profiles in T.brucei.
The 26mer element was identified in the EP procyclin gene 3′-UTR regions and demonstrated to have a role in mRNA stability and translational control (14,15). Similarly the PGKB and PPDE 3′-UTRs, which contain the U-rich core of the 26mer sequence contribute to gene expression via stage-regulated mRNA stability (45). In the COX V 3′-UTR, however, the 26mer core element demonstrated a role only at the translational level, at least when in the context of its complete 3′-UTR. This is not necessarily surprising: mutational analysis of the 26mer element derived from the EP procyclin gene revealed that very limited mutation resulted in the loss of effect on mRNA stability and yet retained the effects on translation (16). Thus, subtle differences in sequence appear to have quite important consequences in the action of the element. This is further reinforced by analysis of the function of the element when inserted into an unrelated intergenic region downstream from the aldolase gene. In this case, incorporation of the element resulted in a surprising increase in CAT expression, although three copies of the element almost completely abolished CAT mRNA and protein. Similar to this, the EP procyclin 26mer element has also been observed to generate both increases and decreases of CAT gene expression when placed in to an unrelated 3′-UTR. Thus, the 26mer element exhibits context-specific effects on gene expression.
We exploited an oligonucleotide counting approach to identify overrepresented motifs among procyclic-enriched transcripts. This revealed that the core element of the 26mer sequence (UAUUUUUU at the RNA level) was significantly overrepresented among the 3′-UTRs of regulated genes, being present in the 3′-UTR of one-third of procyclic form enriched transcripts, contrasting with 9.6% of all 3′-UTRs and 8% of 3′-UTRs for bloodstream-enriched transcripts. In fact, the frequency of such U-rich elements is even higher since the top five overrepresented octamer oligonucleotide sequences identified in the procyclic-enriched mRNA population were entirely comprised of AU-rich sequence, these being recognizable in 44% of procyclic-enriched transcripts. Interestingly, when the overall AU content of intergenic regions derived from all trypanosome genes, bloodstream-enriched or procyclic-enriched transcripts is compared it is remarkably consistent (59, 60 and 58%, respectively) reinforcing the overrepresentation of the AU-rich core sequence as a discrete element regulating developmental gene expression. In contrast to procyclic-enriched transcripts, the 3′-UTR of bloodstream-enriched transcripts did not contain any clearly overrepresented oligonucleotide sequence. These genes may, therefore, have more cryptic signals or be recognized by their absence of the 26mer element (and, possibly, other contributing sequences). Indeed the presence of the 26mer core element in 8% of these transcripts may represent short mRNAs erroneously included by the arbitrary 300 nt 3′-UTR cut-off used in our analysis.
The regulation of AU-rich sequences in mammalian cells can be governed by a number of RNA binding proteins including HuR. Consistent with this, ectopic expression of human HuR results in the stabilization of AU-rich containing transcripts in T.brucei bloodstream forms (45). More recently, AU-rich mRNA destabilization has been found to be effected in mammalian cells by the activity of the RNA silencing pathway, involving microRNAs and components of the RNAi machinery (34). Although there is to date no evidence for microRNA regulation of gene expression in trypanosomes, we investigated whether regulation via RNAi could contribute to the regulated expression of COX transcripts. Our analysis revealed that COX transcripts were of equivalent abundance in wild-type cells and in cells ablated for the Argonaute component of the RNAi machinery in trypanosomes. This demonstrates that regulated expression of these genes was not effected by the RNA silencing machinery, matching the capacity of Argonaute null mutants to complete differentiation events with normal kinetics (35).
Although the RNAi machinery had no effect on COX gene expression, cycloheximide treatment generated a significant and stage-specific increase in the abundance of COX mRNA but not constitutively expressed transcripts. As has been proposed previously in the analysis of several regulated genes in T.brucei, this may reflect the expression of labile negative regulatory factors which destabilize developmentally regulated mRNAs (39,40). More recently, however it has been recognized that cycloheximide can prevent the trafficking of mRNAs to cytoplasmic ribonuclear granules, which act as sites of mRNA degradation (46). Inhibition of this pathway may therefore provide an alternative explanation for the enhanced abundance of COX transcripts in bloodstream forms upon cycloheximide treatment. Interestingly, and consistent with observations for EP procyclin (41), the cycloheximide mediated enhancement of CAT transcripts harbouring the COX V intergenic region was not mediated by sequences contained within the 3′-UTR. This invokes, therefore, either a role for sequence elements in the intergenic region downstream of the polyadenylation site, or a more complex response linked to translation of the gene.
Oligonucleotide counting was originally applied to the identification of signals controlling catabolite repression in yeast, resulting in the recognition of regulated nuclear-encoded mitochondrial mRNAs (4). This identified the consensus CYUGUAA—UA, which was subsequently confirmed to be a generic sequence involved in regulation of nuclear-encoded mitochondrial transcripts by the yeast Puf3 mRNA regulator (recognition sequence UGUR—UA) (2). Our analysis of a subset of nuclear-encoded cytochrome oxidase genes in both T.brucei and T.congolense revealed that these contained a significantly overrepresented oligonucleotide sequence (UAG [G] UA [G/T]) that is similar to (but distinct from) the yeast element. Interestingly, deletion of some regions containing this element, or close relatives of it, also caused an increase in CAT expression in bloodstream forms suggesting that this element may be a contributor to COX gene regulation. Other similar sequences were also present in regions whose deletions did not effect CAT gene expression, however, suggesting context may also be important. Nonetheless, analysis of regulated transcripts annotated via microarray indicated significant overrepresentation of the element in procyclic-enriched mRNAs when compared to bloodstream-enriched transcripts. Moreover, 40% of the non-COX transcripts (50% of all transcripts) containing the element were predicted mitochondrial proteins, several of which contained multiple copies of the element (see supplementary Table 2). This indicates this element is likely to be a contributor to stage-regulation of a subset of mRNAs in trypanosomes, particularly those encoding mitochondrial components.
We initiated our study as a route to search for common sequences among a tightly co-regulated gene family, the developmentally regulated components of the cytochrome oxidase complex. The co-regulation of the cohort of stage-regulated nuclear-encoded mitochondrial proteins is an excellent example of a post-transcriptional operon. Our findings illustrate that even within a clearly related and co-ordinately regulated gene set, the signals controlling expression are complex. Nonetheless, by a combination of bioinformatic analysis and detailed experimental validation we have uncovered the general importance of one element, the 26mer and related sequences, among procyclic-specific genes, as well as a more gene-specific motif which may contribute to the regulation of a much more restricted set of mRNAs, including those of the respiratory chain. The identification of these elements provides the necessary tools to isolate the regulatory protein factors that control the expression of this mRNA cohort in a procyclic-specific manner.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We are indebted to Professor Christine Clayton, Stephanie Brems and Joerg Hoheisel for permission to use microarray data. We also thank Professor Stephen Hajduk and members of his laboratory for discussions throughout this work. M.M. was supported by a doctoral training account from the BBSRC. Work in Keith Matthews' laboratory is supported by a programme grant from the Wellcome Trust. Funding to pay the Open Access publication charges for this article was provided by a programme grant to KM by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Moore M.J. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 2.Gerber A.P., Herschlag D., Brown P.O. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Helden J., Andre B., Collado-Vides J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 1998;281:827–842. doi: 10.1006/jmbi.1998.1947. [DOI] [PubMed] [Google Scholar]
- 4.Jacobs Anderson J.S., Parker R. Computational identification of cis-acting elements affecting post-transcriptional control of gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 2000;28:1604–1617. doi: 10.1093/nar/28.7.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vickerman K. Developmental cycles and biology of pathogenic trypanosomes. Br. Med. Bull. 1985;41:105–114. doi: 10.1093/oxfordjournals.bmb.a072036. [DOI] [PubMed] [Google Scholar]
- 6.Matthews K.R. The developmental cell biology of Trypanosoma brucei. J. Cell Sci. 2005;118:283–290. doi: 10.1242/jcs.01649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clayton C.E. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LeBowitz J.H., Smith H.Q., Rusche L., Beverley S.M. Coupling of poly (A) site selection and trans-splicing in Leishmania. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 9.Matthews K.R., Tschudi C., Ullu E. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- 10.Ehlers B., Czichos J., Overath P. RNA turnover in Trypanosoma brucei. Mol. Cell. Biol. 1987;7:1242–1249. doi: 10.1128/mcb.7.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jefferies D., Tebabi P., Pays E. Transient activity assays of the Trypanosoma brucei variant surface glycoprotein gene promoter: control of gene expression at the posttranscriptional level. Mol. Cell. Biol. 1991;11:338–343. doi: 10.1128/mcb.11.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roditi I., Furger A., Ruepp S., Schurch N., Butikofer P. Unravelling the procyclin coat of Trypanosoma brucei. Mol. Biochem. Parasitol. 1998;91:117–130. doi: 10.1016/s0166-6851(97)00195-3. [DOI] [PubMed] [Google Scholar]
- 13.Hehl A., Vassella E., Braun R., Roditi I. A conserved stem–loop structure in the 3′ untranslated region of procyclin mRNAs regulates expression in Trypanosoma brucei. Proc. Natl Acad. Sci. USA. 1994;91:370–374. doi: 10.1073/pnas.91.1.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furger A., Schurch N., Kurath U., Roditi I. Elements in the 3′ untranslated region of procyclin mRNA regulate expression in insect forms of Trypanosoma brucei by modulating RNA stability and translation. Mol. Cell. Biol. 1997;17:4372–4380. doi: 10.1128/mcb.17.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hotz H.R., Hartmann C., Huober K., Hug M., Clayton C. Mechanisms of developmental regulation in Trypanosoma brucei: a polypyrimidine tract in the 3′-untranslated region of a surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res. 1997;25:3017–3025. doi: 10.1093/nar/25.15.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hotz H.R., Biebinger S., Flaspohler J., Clayton C. PARP gene expression: control at many levels. Mol. Biochem. Parasitol. 1998;91:131–143. doi: 10.1016/s0166-6851(97)00196-5. [DOI] [PubMed] [Google Scholar]
- 17.Vassella E., Den Abbeele J.V., Butikofer P., Renggli C.K., Furger A., Brun R., Roditi I. A major surface glycoprotein of Trypanosoma brucei is expressed transiently during development and can be regulated post-transcriptionally by glycerol or hypoxia. Genes Dev. 2000;14:615–626. [PMC free article] [PubMed] [Google Scholar]
- 18.Gunzl A., Bruderer T., Laufer G., Schimanski B., Tu L.C., Chung H.M., Lee P.T., Lee M.G. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot Cell. 2003;2:542–551. doi: 10.1128/EC.2.3.542-551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pays E. Regulation of antigen gene expression in Trypanosoma brucei. Trends Parasitol. 2005;21:517–520. doi: 10.1016/j.pt.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 20.Boucher N., Wu Y., Dumas C., Dube M., Sereno D., Breton M., Papadopoulou B. A common mechanism of stage-regulated gene expression in Leishmania mediated by a conserved 3′-untranslated region element. J. Biol. Chem. 2002;277:19511–19520. doi: 10.1074/jbc.M200500200. [DOI] [PubMed] [Google Scholar]
- 21.Di Noia J.M., D'Orso I., Sanchez D.O., Frasch A.C. AU-rich elements in the 3′-untranslated region of a new mucin-type gene family of Trypanosoma cruzi confers mRNA instability and modulates translation efficiency. J. Biol. Chem. 2000;275:10218–10227. doi: 10.1074/jbc.275.14.10218. [DOI] [PubMed] [Google Scholar]
- 22.Priest J.W., Hajduk S.L. Developmental regulation of mitochondrial biogenesis in Trypanosoma brucei. J. Bioenerg. Biomembr. 1994;26:179–191. doi: 10.1007/BF00763067. [DOI] [PubMed] [Google Scholar]
- 23.Bringaud F., Riviere L., Coustou V. Energy metabolism of trypanosomatids: adaptation to available carbon sources. Mol. Biochem. Parasitol. 2006;149:1–9. doi: 10.1016/j.molbiopara.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 24.Grossman L.I., Lomax M.I. Nuclear genes for cytochrome c oxidase. Biochim. Biophys. Acta. 1997;1352:174–192. doi: 10.1016/s0167-4781(97)00025-0. [DOI] [PubMed] [Google Scholar]
- 25.Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 26.Speijer D., Muijsers A.O., Dekker H., Dehaan A., Breek C.K.D., Albracht S.P.J., Benne R. Purification and characterization of cytochrome-c-oxidase from the insect Trypanosomatid Crithidia fasciculata. Mol. Biochem. Parasitol. 1996;79:47–59. doi: 10.1016/0166-6851(96)02648-5. [DOI] [PubMed] [Google Scholar]
- 27.Tasker M., Timms M., Hendriks E., Matthews K. Cytochrome oxidase subunit VI of Trypanosoma brucei is imported without a cleaved presequence and is developmentally regulated at both RNA and protein levels. Mol. Microbiol. 2001;39:272–285. doi: 10.1046/j.1365-2958.2001.02252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maslov D.A., Zikova A., Kyselova I., Lukes J. A putative novel nuclear-encoded subunit of the cytochrome c oxidase complex in trypanosomatids. Mol. Biochem. Parasitol. 2002;125:113–125. doi: 10.1016/s0166-6851(02)00235-9. [DOI] [PubMed] [Google Scholar]
- 29.Biebinger S., Wirtz L.E., Lorenz P., Clayton C. Vectors for inducible expression of toxic gene products in bloodstream and procyclic Trypanosoma brucei. Mol. Biochem. Parasitol. 1997;85:99–112. doi: 10.1016/s0166-6851(96)02815-0. [DOI] [PubMed] [Google Scholar]
- 30.Bastin P., Bagherzadeh Z., Matthews K.R., Gull K. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 1996;77:235–239. doi: 10.1016/0166-6851(96)02598-4. [DOI] [PubMed] [Google Scholar]
- 31.Matthews K., Gull K. Identification of stage specific and differentiation enriched transcripts during transformation of the African trypanosomes from its bloodstream to procyclic form. Mol. Biochem. Parasitol. 1998;95:81–95. doi: 10.1016/s0166-6851(98)00100-5. [DOI] [PubMed] [Google Scholar]
- 32.Erondu N.E., Donelson J.E. Differential expression of two mRNAs from a single gene encoding an HMG1-like DNA binding protein of African trypanosomes. Mol. Biochem. Parasitol. 1992;51:111–118. doi: 10.1016/0166-6851(92)90206-y. [DOI] [PubMed] [Google Scholar]
- 33.Clement S.L., Koslowsky D.J. Unusual organization of a developmentally regulated mitochondrial RNA polymerase (TBMTRNAP) gene in Trypanosoma brucei. Gene. 2001;272:209–218. doi: 10.1016/s0378-1119(01)00538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jing Q., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 35.Janzen C.J., van Deursen F., Shi H., Cross G.A., Matthews K.R., Ullu E. Expression site silencing and life-cycle progression appear normal in Argonaute1-deficient Trypanosoma brucei. Mol. Biochem. Parasitol. 2006;149:102–107. doi: 10.1016/j.molbiopara.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi H., Tschudi C., Ullu E. Functional replacement of Trypanosoma brucei Argonaute by the human slicer Argonaute 2. RNA. 2006;12:943–947. doi: 10.1261/rna.20806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graham S.V., Barry J.D. Polysomal, procyclin mRNAs accumulate in bloodstream forms of monomorphic and pleomorphic trypanosomes treated with protein synthesis inhibitors. Mol. Biochem. Parasitol. 1996;80:179–191. doi: 10.1016/0166-6851(96)02674-6. [DOI] [PubMed] [Google Scholar]
- 38.Schurch N., Furger A., Kurath U., Roditi I. Contributions of the procyclin 3′ untranslated region and coding region to the regulation of expression in bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 1997;89:109–121. doi: 10.1016/s0166-6851(97)00107-2. [DOI] [PubMed] [Google Scholar]
- 39.Webb H., Burns R., Ellis L., Kimblin N., Carrington M. Developmentally regulated instability of the GPI-PLC mRNA is dependent on a short-lived protein factor. Nucleic Acids Res. 2005;33:1503–1512. doi: 10.1093/nar/gki298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gruszynski A.E., van Deursen F.J., Albareda M.C., Best A., Chaudhary K., Cliffe L.J., Del Rio L., Dunn J.D., Ellis L., Evans K.J., et al. Regulation of surface coat exchange by differentiating African trypanosomes. Mol. Biochem. Parasitol. 2006;147:211–223. doi: 10.1016/j.molbiopara.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 41.Fluck C., Salomone J.Y., Kurath U., Roditi I. Cycloheximide-mediated accumulation of transcripts from a procyclin expression site depends on the intergenic region. Mol. Biochem. Parasitol. 2003;127:93–97. doi: 10.1016/s0166-6851(02)00310-9. [DOI] [PubMed] [Google Scholar]
- 42.Drozdz M., Clayton C. Structure of a regulatory 3′ untranslated region from Trypanosoma brucei. RNA. 1999;5:1632–1644. doi: 10.1017/s1355838299990623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan C.Y., Lawrence C.E., Ding Y. Structure clustering features on the Sfold Web server. Bioinformatics. 2005;21:3926–3928. doi: 10.1093/bioinformatics/bti632. [DOI] [PubMed] [Google Scholar]
- 44.Benz C., Nilsson D., Andersson B., Clayton C., Guilbride D.L. Messenger RNA processing sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 2005;143:125–134. doi: 10.1016/j.molbiopara.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 45.Quijada L., Guerra-Giraldez C., Drozdz M., Hartmann C., Irmer H., Ben-Dov C., Cristodero M., Ding M., Clayton C. Expression of the human RNA-binding protein HuR in Trypanosoma brucei increases the abundance of mRNAs containing AU-rich regulatory elements. Nucleic Acids Res. 2002;30:4414–4424. doi: 10.1093/nar/gkf577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brengues M., Teixeira D., Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuker M., Jacobson A.B. Using reliability information to annotate RNA secondary structures. RNA. 1998;4:669–679. doi: 10.1017/s1355838298980116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.