Abstract
Yeast cells expressing the Glu418Lys human topoisomerase I mutant display a camptothecin resistance that slowly decreases as a function of time. Molecular characterization of the single steps of the catalytic cycle of the purified mutant indicates that it has a relaxation activity identical to the wild-type protein but a different DNA sequence specificity for the cleavage sites when compared to the wild-type enzyme, as assayed on several substrates. In particular the mutant has a low specificity for CPT sensitive cleavable sites. In fact, the mutant has, at variance of the wild-type enzyme, a reduced preference for cleavage sites having a thymine base in position −1 of the scissile strand. This preference, together with the strict requirement for a thymine base in position −1 for an efficient camptothecin binding, explains the temporary camptothecin resistance of the yeast cell expressing the mutant and points out the importance of the DNA sequence in the binding of the camptothecin drug.
INTRODUCTION
Human topoisomerase I controls the superhelical structure of DNA playing a critical role in important biological processes, such as DNA replication, transcription and recombination (1–3). The enzyme is composed of 765 amino acids and has four distinct domains: the NH2-terminal domain (1–214), the core domain (215–635), the linker domain (636–712) and the COOH-terminal domain (713–765) (4–6).
The outline of the catalytic cycle has been unveiled in recent years. The active site tyrosine (Tyr-723) starts the catalytic cycle of the enzyme through a nucleophilic attack on the DNA backbone, resulting in the breakage of one DNA strand and formation of a covalent intermediate between the enzyme and the DNA 3′-phosphate. After changing the linking number a second nucleophilic attack, driven by the 5′-hydroxyl DNA end, restores an intact double-stranded DNA, after which the enzyme is released. The covalent intermediate between topoisomerase I and DNA is a transient step and when CPT is added the cleavage-religation equilibrium is shifted toward cleavage. Collision between the stabilized covalent complex intermediate and the replication fork results in double-strand DNA breaks and triggers a cascade of event leading to apoptosis (1,7). The X-ray diffraction study of the ternary complex formed by the human topoisomerase I, bound to the preferred sequence double-strand DNA with CPT or topotecan, a CPT derivative in clinical use as an anticancer agent (8,9), has provided details of the interaction of the drug with the catalytic region, proposing a molecular explanation for the resulting slowdown of the religation step.
Several topoisomerase I mutations, located either near or far from the active site, have been shown to render the enzyme resistant to the drug (8,10). The 3D structures of the ternary complexes with the native enzyme (5) and with some specific mutants (8,10,11) have clarified the molecular basis for the resistance for most of the mutations located near the active site, but have still left unexplained the effects of those located far from it, such as the Ala653Pro, the Glu418Lys and the Thr729Ala mutants (12–14). In the case of the Ala653Pro mutation, a combined experimental and simulative approach has demonstrated that the drug resistance is correlated to a difference in linker mobility that induces an altered control of the religation process (12), indicating the importance of the protein concerted motions in the catalytic cycle. The combined use of experimental and theoretical techniques has been adopted also for the characterization of functional and dynamical properties of the human topoisomerase I Thr718Ala mutant, which shows a lethal phenotype (15).
In this study, we have investigated at molecular level the Glu418Lys mutant, previously identified in a CPT-resistant clone of a human nasopharyngeal carcinoma treated for a long period with a CPT derivative (13). Our results indicate that the yeast cells expressing the mutant show a time-dependent resistance to CPT and that the purified mutant displays a different cleavage pattern compared to the wild-type enzyme. The Glu418Lys mutant, in particular, shows a larger preference for CPT unsensitive sites, as the ones having a thymine base in position −1 of the scissile strand (16), providing an explanation for the temporary drug resistance observed in yeast cells expressing the mutant.
MATERIALS AND METHODS
Yeast strains and plasmids
Camptothecin (Sigma) was dissolved in Me2SO to a final concentration of 4 mg/ml and stored at −20°C. Anti-FLAG M2 affinity gel, FLAG peptide and M2 monoclonal antibody were purchased from Sigma. Saccharomyces cerevisiae strains JN2-134 (MATα rad52::LEU2, trp1, ade2-1, his7, ura3-52, ise1, top1, leu2) and EKY3 (ura3-52, his3Δ200, leu2Δ1, trp1Δ63, top1::TRP1, MATα) were described previously (17,18). Plasmid YEpGAL1-wild-type and YCpGAL1-wild-type in which the human topoisomerase I is expressed under the galactose inducible promoter in a multi-copy plasmid was described previously (19). Glu418Lys was generated by oligonucleotide-directed mutagenesis of the YEpGAL1-wild-type and YCpGAL1-wild-type. The epitope-tagged construct YEpGAL1-e-wild-type contains the N-terminal sequence FLAG: DYKDDDY (indicated with ‘e’), recognized by the M2 monoclonal antibody. The epitope-tag was subcloned into YEpGAL1-Glu418Lys to produce YEpGAL1-e-Glu418Lys.
Drug sensitivity
Cultures of JN2-134 transformed with plasmids YEpGAL1-wild type, YEpGAL1-Glu418Lys and YEpGAL1 were grown at an OD A595 of 0.3, serially 10-fold diluted, and 5 µl spotted onto selective media containing dextrose or galactose. CPT sensitivity was assayed on dextrose plates containing 25 mM HEPES (pH 7.2) and 0.5 µg/ml CPT. Plates were incubated at 30°C for 3 days.
Cell viability assay
Overnight cultures of EKY3 transformed with plasmids YCpGAL1-wild-type, YCpGAL1-Glu418Lys, YCpGAL1-Ala653Pro (12) and YCpGAL1 were diluted 1:100 into selective media containing 2% raffinose. At an A595 of 0.3 the cultures were induced for 1 h with 2% galactose. After induction, cells were treated with 50 µM CPT or 1% Me2SO. At various time points aliquots were serially diluted and plated onto selective media containing 2% dextrose. The number of colonies was counted following incubation at 30°C and plotted relative to the number obtained at time 0 (drug addition).
Purification of DNA topoisomerase I
To purify the topoisomerase I epitope tagged EKY3 cells were transformed with YEpGAL1-e-wild-type and YEpGAL1-e-Glu418Lys, grown on SC-uracil plus 2% dextrose and diluted 1:100 in SC-uracil plus 2% raffinose. At an OD A595 of 1.0, the cells were induced with 2% galactose for 6 h. Cells were then harvested by centrifugation, washed with cold water and resuspended in 2 ml buffer/g cells using a buffer containing 50 mM Tris (pH 7.4), 1 mM EDTA, 1 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid, 10% (v/v) glycerol completed with protease inhibitors cocktail (Roche 1 836 153) and supplemented with 0.1 mg/ml sodium bisulfite and 0.8 mg/ml sodium fluoride. After addition of 0.5 vol of 425–600 µm diameter glass beads the cells were disrupted by vortexing for 30 s alternating with 30 s on ice. The lysate was centrifuged and KCl final concentration 0.15 M was added to the sample before loading onto 2 ml ANTI-FLAG M2 Affinity Gel column equilibrated as described in the technical bulletin (Sigma). The column was washed with 20 column volumes of TBS [50 mM Tris–HCl and 150 mM KCl (pH 7.4)]. Elution of FLAG-fusion topoisomerase I was performed by competition with five column volumes of a solution containing 100 µg/ml FLAG peptide in TBS. Fractions of 500 µl were collected and glycerol final concentration 40% was added; all preparations were stored at −20°C. The fractions were resolved by SDS–PAGE; protein concentration and integrity were measured through immunoblot assay, using the epitope-specific monoclonal antibody M2. After normalization to the same amount of protein, the activity of the wild-type and mutant DNA topoisomerase I, as assayed by relaxation of supercoiled DNA in 150 mM KCl, was found to be almost identical. In all the biochemical experiments the same amount of wild-type and mutated protein has been used.
DNA topoisomerase I activity in vitro
Topoisomerase I activity was assayed with a DNA relaxation assay (17–20). Topoisomerase I preparations were incubated in 30 µl reaction volume containing 0.5 µg of negatively supercoiled plasmid pHC624 and reaction buffer [20 mM Tris–HCl (pH 7.5), 0.1 mM Na2EDTA, 10 mM MgCl2, 50 µg/ml acetylated BSA and 150 mM KCl). Moreover, to assess the effects of ionic strength on enzyme activity KCl was added in the DNA relaxation assay. Reactions were stopped with a final concentration of 0.5% SDS after 1 h at 37°C and electrophoresis of the samples was carried out in a 1% agarose gel.
Kinetics of cleavage using oligonucleotide substrate
Oligonucleotide substrates CL14 (5′-GAAAAAAGACTTAG-3′) or CL14-A (5′-GAAAAAAGACATAG-3′) or CL14-G (5′-P-GAAAAAAGACGTAG-3′) were radiolabelled with [γ-32P]ATP at their 5′ end. The CP25 complementary strand (5′-TAAAAATTTTTCTAAGTCTTTTTTC-3′) or the CP25-T (5′-TAAAAATTTTTCTATGTCTTTTTTC-3′) or the CP25-C (5′-TAAAAATTTTTCTACGTCTTTTTTC-3′) were phosphorylated at their 5′ end with unlabelled ATP. CL14/CP25 or CL14-A/CP25-T or CL14G/CP25C were annealed as described previously (12). The suicide cleavage reactions were carried out by incubating 20 nM of the duplex with an excess of enzyme in 20 mM Tris (pH 7.5), 0.1 mM Na2EDTA, 10 mM MgCl2, 50 µg/ml acetylated BSA and 150 mM KCl at 23°C in a final volume of 50 µl as described by Yang and Champoux (21). A 5 µl sample of the reaction mixture was removed before addition of the protein and used as the zero time point. At various time points 5 µl aliquots were removed and the reaction stopped with 0.5% SDS. After ethanol precipitation samples were resuspended in 5 µl of 1 mg/ml trypsin and incubated at 37°C for 30 min. Samples were analyzed by denaturing urea/PAGE. The percentage of CL1 or CL2 were determined by PhosphorImager and ImageQuant software and normalized on the total amount of radioactivity in each lane.
Kinetics of religation using oligonucleotide substrate
CL14/CP25 (20 nM), prepared as described above was incubated with an excess of enzyme for 60 min at 23°C followed by 30 min at 37°C in 20 mM Tris (pH 7.5), 0.1 mM Na2EDTA, 10 mM MgCl2, 50 µg/ml acetylated BSA and 150 mM KCl. Religation reactions were initiated by adding a 200-fold molar excess of R11 oligonucleotide (5′-AGAAAAATTTT-3′) or R13 (5′-TTAGAAAAATTTT-3′) over the duplex CL14/CP25 in the presence or absence of 100 µM CPT. At 37°C a various time point 5 µl aliquots were removed and the reaction stopped with 0.5% SDS. After ethanol precipitation samples were resuspended in 5 µl of 1 mg/ml trypsin and incubated at 37°C for 30 min. Samples were analyzed by denaturing urea/PAGE. The percentage of the remaining covalent complex (CL1 or CL2) was determined by PhosphorImager and ImageQuant software and normalized on the total amount of radioactivity in each lane.
Cleavage/religation equilibrium
Oligonucleotide CL25 (5′-GAAAAAAGACTTAGAAAAATTTTTA-3′) was radiolabelled with [γ-32P]ATP at its 5′ end. The CP25 complementary strand (5′-TAAAAATTTTTCTAAGTCTTTTTTC-3′) was phosphorylated at its 5′ end with unlabelled ATP. The two strands were annealed at a 2-fold molar excess of CP25 over CL25. A final concentration of 20 nM duplex CL25/CP25 was incubated with an excess of enzyme at 25°C in 20 mM Tris (pH 7.5), 0.1 mM Na2EDTA, 10 mM MgCl2, 50 µg/ml acetylated BSA and 150 mM KCl, in the presence or absence of 1, 2.5, 5, 10, 30 µM CPT. After 30 min, the reaction was stopped by adding 0.5% SDS and digested with trypsin after ethanol precipitation. Reaction products were resolved in 16% acrylamide-7 M urea gels and the percentage of cleavage was determined by PhosphorImager and ImageQuant software.
The cleavage religation equilibrium was also assayed incubating enzyme preparations with a 900 bp double-stranded DNA fragment, radiolabelled at one unique 3′ end, in the presence or absence of CPT. The 900 bp DNA fragment, containing a high affinity DNA topoisomerase I cleavage site, was single 32P-end-labelled and purified from plasmid pBlueAK3-1 as described (20). Enzyme preparations were incubated in 20 µl reactions with DNA, reaction buffer and, where indicated, 1 µM CPT. Dimethyl sulfoxide (DMSO) was added to the no-drug controls. Following 8 min of incubation at 37°C in the case of no drug experiment, and 0.25, 1, 4 or 8 min, for kinetic assay in the presence of CPT, reactions were stopped by the addition of 1% SDS. The cleaved DNA fragments were resolved in 7 M urea/8% polyacrylamide gels and visualized by phosphorimager.
RESULTS
The Glu418Lys mutant is active and partially resistant to CPT in yeast cells
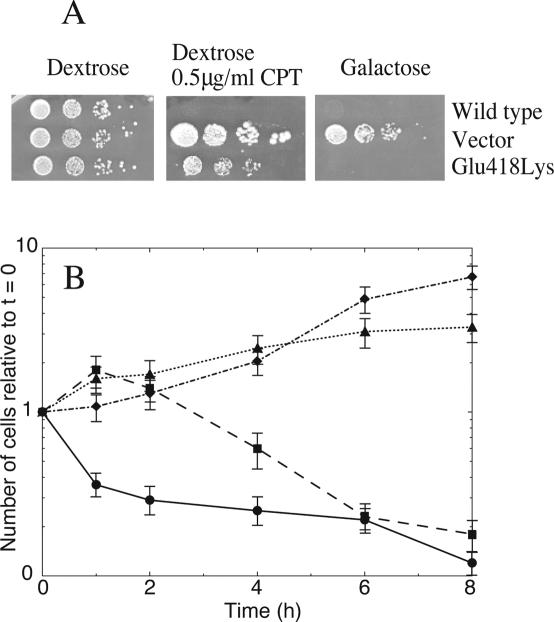
The resistance and the in vivo activity of Glu418Lys topoisomerase I mutant has been tested in a yeast drug sensitivity assay which makes use of the JN2-134 (rad52Δ) strain that does not tolerate galactose induced topoisomerase I expression from a multicopy vector and that, due to the leaky expression of the same vector, is sensitive to CPT (22). Figure 1A, showing the result for the wild-type and the Glu418Lys mutant, indicates that substitution of the glutamic 418 residue with a lysine confers slight resistance, and also demonstrates that the mutant is active in vivo because the yeast strain dies when the enzyme is overexpressed on galactose.
Figure 1.
Glu418Lys is partially resistant to CPT in vivo. (A) Exponentially growing cells in dextrose, transformed with YEpGAL1-wild-type, YEpGAL1-Glu418Lys or YEpGAL1 (vector), serially 10-fold diluted starting from an A595 of 0.3; 5 µl, and spotted onto selective media in the presence of dextrose (left) or dextrose plus 0.5 µg/ml CPT (middle), or induced with galactose (right). (B) Number of colonies relative to that obtained at time 0 plotted against time for wild-type, (circle), Glu418Lys mutant (square), Ala653Pro mutant (diamond) and vector (triangle). Exponentially growing cells in dextrose transformed with YCpGAL1-wild-type, YCpGAL1-Glu418Lys, YCpGAL1-Ala653Pro or YCpGAL1 (Vector) were diluited 1:100 into selective medium containing 2% raffinose. After induction with 2% galactose cells were treated with 50 µM CPT or 1% Me2SO. At various time point aliquots were serially diluted and plated onto selective media containing 2% dextrose. Three different plates were averaged for each data point, and the error bars indicate the standard deviation of the individual values from the mean.
To better elucidate the partial drug resistance, a cell viability assay in liquid culture has been carried out. EKY3 cells transformed with a single copy plasmid (YCp) expressing Glu418Lys or wild-type or Ala653Pro (that is known to be CPT resistant) (12), or the vector were induced with galactose and treated with 50 µM CPT. At various time intervals, aliquots were removed and plated onto selective medium to score for viable cells. Figure 1B shows that the cells expressing the Glu418Lys mutant display a time-dependent CPT resistance. In fact the Glu418Lys expressing cells in the first 2 h are increasing in numbers just like the Ala653Pro expressing cells. However, at longer times, the Ala653Pro transformed cells continue their growth, while the Glu418Lys transformed ones gradually stop growing, indicating the occurrence of a time dependent process that renders them sensitive to CPT. After 6–8 h of treatment with CPT, the Glu418Lys viable cells reach the same level of drug sensitivity as the cells expressing the wild-type protein. This experiment confirms that the Glu418Lys mutant is active in vivo and indicates the existence of a molecular mechanism that permits the survival of the cells expressing the mutated enzyme only during the first hours from drug addition, slowly switching them toward CPT sensitivity. It can be excluded that the time-dependent survival of the yeast cells expressing the Glu418Lys mutant is due to a slow uptake of CPT since the yeast cells expressing topoisomerase I wild-type or the Ala653Pro mutant display a fast, not time-dependent, response to the drug, positive or negative, respectively.
The catalytic activity of the Glu418Lys mutant has also been tested in vitro, serially diluting and incubating identical wild-type and mutant protein concentrations with the same amount of supercoiled plasmid DNA. The Glu418Lys mutant shows a relaxation efficiency similar to the wild-type protein, as well as the same ionic strength dependence (data not show). In fact, relaxation experiments carried out at salt concentration ranging between 50 and 300 mM show that the mutant Glu418Lys and the wild-type enzymes have an identical salt dependent relaxation efficiency, indicating that the single mutation apparently does not perturb the electrostatic interaction between the enzyme and the DNA, despite the fact that the mutation involves a net charge difference.
Effect of CPT on the cleavage-religation equilibrium
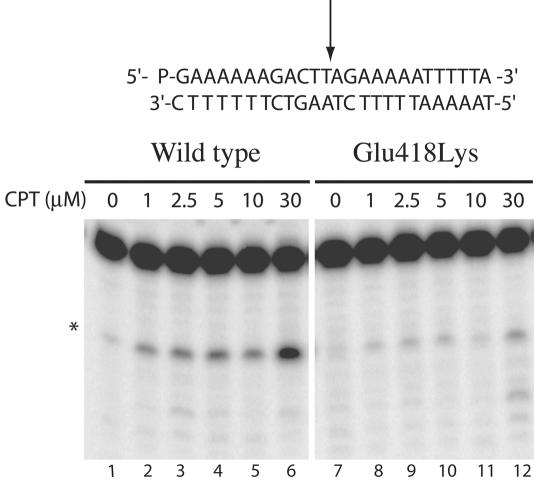
To investigate the mechanism of time dependence of the Glu418Lys mutant camptothecin resistance, the stability of the covalent DNA–enzyme complex has been analyzed using the 25mer full duplex oligonucleotides substrate CL25/CP25 in the absence or presence of increasing amount of CPT (from 1 to 30 µM). After 30 min of incubation, the reactions were stopped with SDS, the samples were treated with trypsin and the products analyzed by polyacrylamide–urea gel electrophoresis (Figure 2). The results, in the absence of the drug, are consistent with the finding that the cleavage-religation equilibrium is shifted toward religation for the wild-type protein, not being detectable any trapped cleavable complex (Figure 2, lane 1). When the wild-type protein is exposed to increasing amount of CPT the cleavage-religation equilibrium is shifted toward the cleavage as already reported (23,24), since the band corresponding to the substrate cleaved at the preferred CL1 site is clearly detectable (Figure 2, lanes 2–6). The Glu418Lys mutant in the absence of CPT, shows an equilibrium shifted toward religation as that of the wild-type protein (Figure 2, lane 7). In the presence of increasing amount of CPT the equilibrium is partially shifted toward cleavage as revealed by the substrate band appearing on the gel (Figure 2, lanes 8–12), i.e. less intense than for the wild-type, indicating that the Glu418Lys mutant is less sensitive to the drug, although it is still able to bind CPT and stabilize the cleavable complex. To better clarify this point and since the equilibrium constant Keq is given by the ratio between the cleavage and the religation rate (kcl/kr), the single steps of the enzyme catalytic cycle have been analyzed.
Figure 2.
Cleavage/religation equilibrium of the CL25/CP25 fully duplex DNA substrate. Gel electrophoresis of the products coming from the incubation of the wild-type topoisomerase I with the [γ-32P] end-labelled duplex DNA, shown at the top of the figure in the absence (lane 1) and presence (lanes 2–6) of increasing amount of CPT. The arrow at the DNA sequence indicates the CL1 site preferred by the wild-type protein. Lanes 7 and 8–12 show the same experiment with the Glu418Lys mutant. The asterisk indicates the band corresponding to the CL1 site.
Cleavage of the wild-type and Glu418Lys mutant with the preferred CL14/CP25 substrate
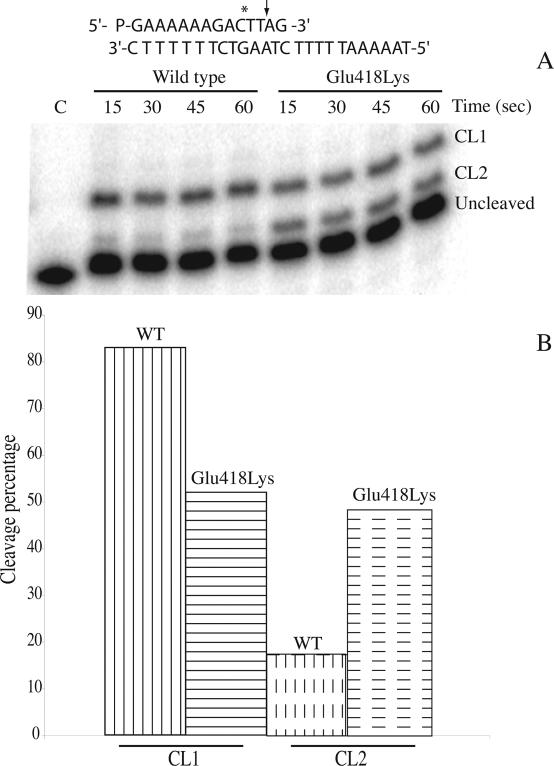
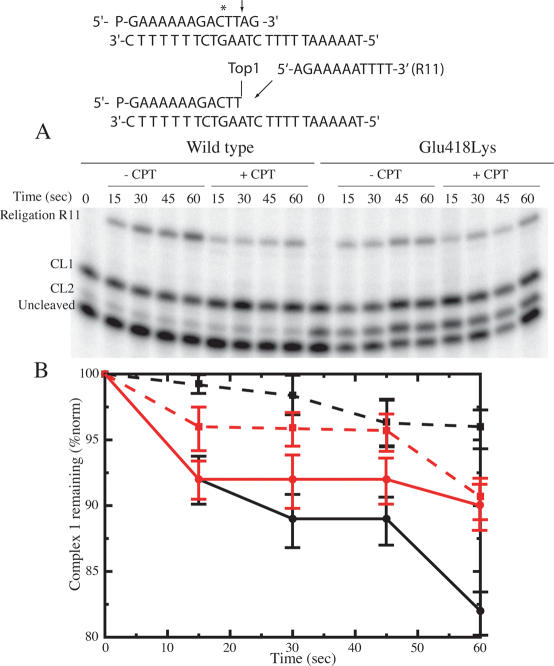
The time course of the cleavage of the wild-type and Glu418Lys mutant has been followed using a suicide cleavage substrate. In detail, a 5′ end radiolabelled oligonucleotide CL14 (5′-GAAAAAAGAC*TT↓AG-3′) has been annealed to the CP25 (5′-TAAAAATTTTTCTAAGTCTTTTTTC-3′) complementary strand, to produce a duplex with an 11 base 5′ single-strand extension. The arrow identifies the preferred cleavage (CL1) site for the enzyme, but the wild-type enzyme has been shown to cleave the oligonucleotide also at the site indicated by the asterisk (CL2), although to a lower extent (23). The religation step is precluded because the AG-3′ or TTAG-3′ oligonucleotides are too short to be religated, leaving the enzyme covalently attached to the 12 or 10 oligonucleotide 3′ end (23).
The suicide substrate was incubated with an excess of wild-type or mutant enzyme in a time course experiment. The cleaved DNA fragments resolved in a denaturing polyacrylamide gel are reported in Figure 3A. The cleavage reaction is very fast for both the wild-type and the mutant enzyme, with all the substrate already cleaved within 15 s, making it impossible to calculate the cleavage reaction rate (kcl). However, evaluation of the cleavage products, namely CL1 and CL2, indicates that the wild-type and mutated enzymes have different substrate distributions (Figure 3B). In fact, both enzymes give an identical percentage of total cleavage products but the wild-type enzyme, as shown in Figure 3B, preferentially cuts (∼85%) at the CL1 site (full vertical lines) and only moderately (∼15%) at the CL2 site (dashed vertical lines), whilst the mutated enzyme equally cuts at the CL1 (full horizontal lines) and the CL2 (dashed horizontal lines) sites. So a clear effect of the Glu418Lys mutation is to induce different substrate specificity. The DNA base cleavage preferences, studied for the wild-type topoisomerase I in the SV40 genome (16), has indicated that the enzyme shows a strong preference for T in position −1 in the scissile strand, as in the CL1 but not in the CL2 site, that has a C base in this position. The mutant then reduces the enzyme cleavage preference for a thymine base in position −1 in the scissile strand.
Figure 3.
Suicide cleavage experiment with the CL14/CP25 substrate for the wild-type and Glu418Lys mutant. (A) Time course of the suicide cleavage reaction carried out with the substrate described on the top of the figure. CL1 and CL2 identify the cleaved complexes at the site indicated by the arrow and the asterisk, respectively. Lane C, no protein added. (B) Percentage of the DNA substrate cleaved at the CL1 or CL2 site for the wild-type and the Glu418Lys mutant.
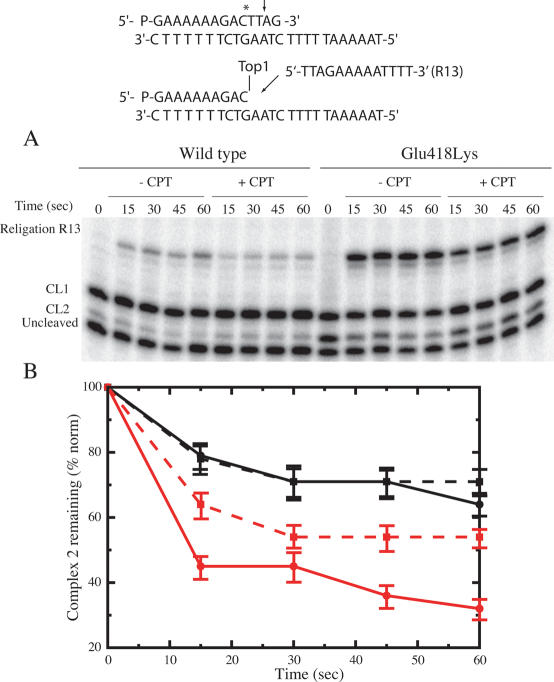
Religation of the wild-type and Glu418Lys mutant
Because of the different substrate specificities displayed by the wild-type and mutated enzyme, the DNA religation step has been studied for both enzyme by adding R11 (5′-AGAAAAATTTT-3′) or R13 (5′-TTAGAAAAATTTT-3′) oligonucleotides, that can bind to the CL1 or the CL2, respectively. The wild-type and mutant enzyme have been incubated for 60 min with the suicide substrate in order to generate the cleavage complexes. Once cleavage has occurred, R11 or R13 oligonucleotide are added to start the religation process in the absence or presence of 100 µM CPT. Aliquots have then been removed as function of time, the reaction stopped by SDS, trypsinized, analyzed by PAGE and the percentage of the remaining cleaved 1 or 2 complexes has been plotted. Figure 4A shows the religation kinetics upon addition of the R13 oligonucleotide. The CL1 band maintains the same intensity at different times, indicating that R13 is religated by the enzyme only at the CL2, whose band decreases in intensity as a function of time. As indicated in Figure 4B, the percentage of the remaining cleaved complex 2 as a function of time indicates that, in the absence of CPT, the religation rate of the Glu418Lys mutant (full red line) is faster than the wild-type enzyme (full black line). The presence of the drug does not affect the religation rate of the wild-type protein (black dashed lines) and only slightly that of the Glu418Lys mutant (dashed red line), as quantified and normalized on the total amount of radioactivity in each lane (Figure 4B). This result indicates that the binary enzyme–DNA CL2 oligonucleotide complex, that has a cytosine base in the −1 position, is not stabilized by CPT for both proteins. In line, previous experimental data (16), partially confirmed by a theoretical study (25) have shown that a thymine base in position −1 of the scissile strand is a strict requirement for CPT activity.
Figure 4.
Religation experiment with the R13 oligonucleotide for the wild-type and Glu418Lys topoisomerase I. (A) Time course of the religation experiment between the R13 substrate and the wild-type or Glu418Lys covalent complexes, in the absence or presence of 100 µM CPT. The R13 oligonucleotide is selectively religated to the CL2 site but not to the CL1 site. (B) Percentage of the remaining covalent complex 2 plotted at different time for the wild-type and Glu418Lys (black and red lines, respectively), in absence (full line) and presence (dashed line) of CPT. Three different religation experiments were averaged for each data point, and the error bars indicate the standard deviation of the individual values from the mean.
The religation experiment carried out upon addition of the R11 oligonucleotide is shown in Figure 5A. The intensity of the CL2 band is fairly constant at any time indicating that the R11 is religated only at the CL1 site. The plot of the percentage of the remaining CL1 (Figure 5B) indicates that in the absence of CPT the rate of religation is almost identical for both enzymes (full black and red lines). The addition of CPT abolishes the religation process in the first 30 s for the wild-type protein (dashed black line) and strongly reduces that of the Glu418Lys mutant (dashed red line), indicating that CPT is able to bind and inhibit the religation process for both enzymes.
Figure 5.
Religation experiment with the R11 oligonucleotide for the wild-type and Glu418Lys topoisomerase I. (A) Time course of the religation experiment between the R11 substrate and the wild-type or Glu418Lys covalent complexes, in the absence or presence of 100 µM CPT. The R11 oligonucleotide is selectively religated to the CL1 site but not to the CL2 site. (B) Percentage of the remaining covalent complex 1 plotted at different time for the wild-type and Glu418Lys (black and red lines, respectively), in absence (full line) and presence (dashed line) of CPT. Note the difference in scale between this figure and Figure 4. Three different religation experiments were averaged for each data point, and the error bars indicate the SD of the individual values from the mean.
Confirmation of the different cleavage specificity of the Glu418Lys mutant
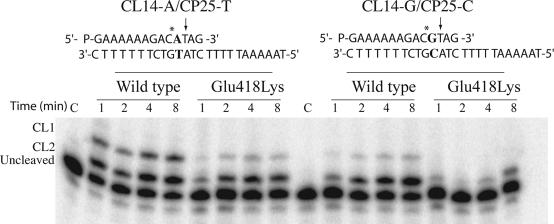
Different oligonucleotide substrates have been used in order to verify that the different sequence specificity of the mutant in respect of the wild-type is observed not only with the preferred CL14/CP25 substrate. Figure 6 shows the cleavage reaction with two different suicide substrates CL14-A/CP25-T and CL14-G/CP25-C, where a thymine in position +1 in respect of the CL2 site has been substituted with an adenine or a guanine, respectively. The experiment confirms the different sequence specificity of the mutant, and most important it shows that for both substrates the cleavage at the CL1 site, i.e. at a site having a thymine base in position −1 in the scissile strand, is much reduced in the mutant when compared to the wild-type enzyme. Actually in the case of the CL14-G/CP25-C substrate the mutant never cleaves at the CL1 site. It is interesting to notice that the cleavage reaction is slower, when compared with the one carried out with the CL14/CP25 substrate for both the wild-type and the mutated enzymes. In fact, Figures 3 and 6 have a time scale of seconds and minutes, respectively.
Figure 6.
Suicide cleavage experiment with the CL14-A/CP25-T or the CL14-G/CP25-C substrates for the wild-type and Glu418Lys mutant. Left side: time course of the suicide cleavage reaction carried out with the CL14-A/CP25-T substrate, described on the top of the figure. CL1 and CL2 identify the cleaved complexes at the site indicated by the arrow and the asterisk, respectively. Lane C, no protein added. Right side: time course of the suicide cleavage reaction carried out with the CL14-G/CP25-C substrate, described on the top of the figure.
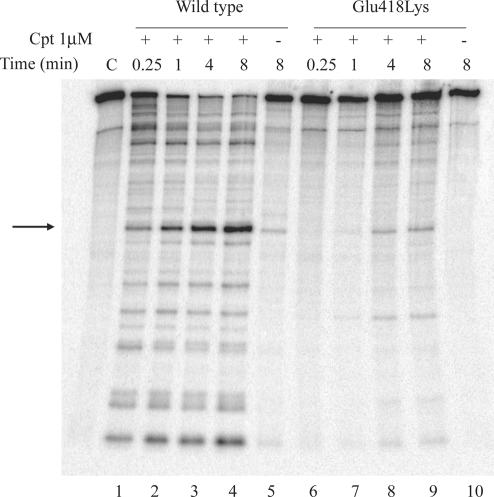
Different sequence specificity is also shown by the cleavage-religation experiments carried out as a function of time for both enzymes with a linear 900 bp oligonucleotide, in absence or in presence of 1 µM of CPT (Figure 7). The experiment shows in the case of the wild-type the presence of a large number of bands diagnostic of several CPT trapped cleavable complexes, much higher than in the case of the mutant. Actually at short times, in the case of the mutant only few bands are barely detectable, that partially increase as a function of time, indicating that the mutant is able to cleave only at few CPT sensitive sites.
Figure 7.
Cleavage/religation equilibrium of a 900 bp fully duplex DNA substrate. Gel electrophoresis of the products coming from the incubation of a 900 bp 32P-end-labelled DNA duplex with the wild-type topoisomerase I (lanes 1–5) or Glu418Lys mutant (lanes 6–10), as a function of time, in the absence (lanes 5 and 10) or in the presence of 1 µM CPT (lanes 1–4 and 6–9). Control, no enzyme added (C). Arrow indicates the preferred cleavage site.
DISCUSSION
The results presented here provide a molecular explanation for the slight CPT resistance of the Glu418Lys mutant of topoisomerase I, first reported by Chang et al. (13) and confirmed by the assay shown in Figure 1A. Interestingly the mutation doesn't affect enzyme activity in vivo and in vitro, as shown by the death of the yeast strain when the mutant is overexpressed on galactose (Figure 1A) and by in vitro plasmid relaxation assay (data not shown). Yeast EKY3 cells, expressing the Glu418Lys mutant, show a CPT resistance in the first 2 h, but after this time the resistance starts to decrease, reaching the same level of sensitivity of the cells expressing the wild-type protein after 6–8 h of treatment with CPT (Figure 1B). In a similar assay, carried out by Chang and co-authors, the time dependence of the CPT resistance was not observed, likely because a different yeast strain (JN2134) (17) was used (13).
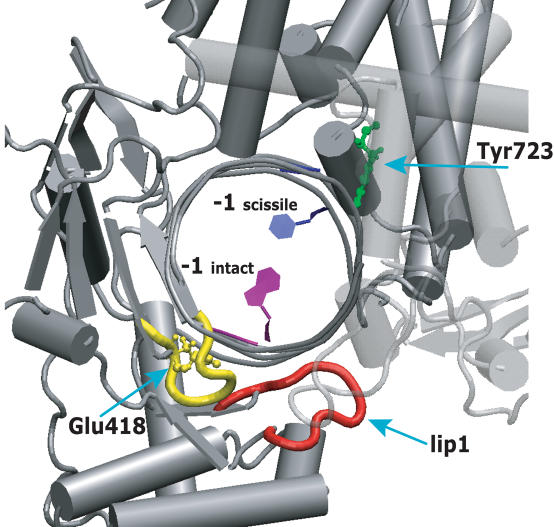
The mutated amino acid is located in the highly conserved subdomain I of the core domain, on the loop formed by residues Thr417-Ser423 that directly contacts Lip 1 (see Figure 8). The loop, located between β strands 10 and 11, also contacts the −1 and −2 DNA bases in the intact strand by means of both direct and water mediated hydrogen bonds (5,26). Based on structural consideration, it has been proposed that the Glu418Lys mutation would disrupt the order of the 417–423 loop and then the relative positioning of the Lip 1 region, likely altering a protein-drug contact and so explaining its resistance to CPT (11). However the equilibrium results indicate that CPT is able to interact, altough with less efficiency, with the Glu418Lys mutant, stabilizing the cleavable complex (Figure 2).
Figure 8.
Human topoisomerase I structure in complex with duplex DNA. The main chain of the 417–423 loop, contacting the intact strand of the lip 1 (represented in red), is shown in yellow. The mutated Glu418 and the catalytic Tyr723 residues are represented in ball and stick, in green and yellow colours, respectively. The −1 base of the scissile strand, strictly required for CPT binding, is coloured in blue; the −1 base of the intact strand, that contacts the 417–423 loop, is represented in purple. All other enzyme residues and DNA bases are represented in grey. Core subdomain III is partial transparent.
The time-dependent CPT resistance cannot then be simply explained by the inability of the mutant to interact with CPT, but can actually be rationalized by the different cleavage specificity shown by the Glu418Lys mutant. The first important result is, in fact, that the mutant has a substrate specificity very different from that of the wild-type enzyme. In fact, as shown in Figures 3 and 6, the reaction of the wild-type and the mutant with three different suicide substrates (CL14/CP25, CL14-A/CP25-T and CL14-G/CP25-C) produces different pattern distributions. In detail, as shown in Figure 3 and reported in the literature (27), addition of the wild-type to a CL14/CP25 substrate gives rise to a suicide cleavage that is mainly (∼85%) localized on the site indicated by the arrow (CL1) and only in minimal part (∼15%) on the site indicated by the asterisk (CL2). Pommier and coauthors have actually demonstrated that a T base in position −1 in the scissile strand is strongly preferred in the cleavage process (16). The Glu418Lys mutant loses this preference being the cleavage distribution over the CL1 and CL2 sites almost identical (Figure 3). This result is confirmed by the cleavage experiment carried out with the CL14-A/CP25-T and CL14-G/CP25-C substrates. In this case the CL1 band is almost absent in the gel referring to the mutant (Figure 6) despite the fact that the base mutation is in proximity of the CL2 site. A general feature of the Glu418Lys mutation is then to reduce the enzyme cleavage preference for a thymine base in position −1 in the scissile strand.
The religation experiment, carried out on the cleavage products of the CL14/CP25 substrate, indicates that CPT strongly inhibits the R11 religation process for both the native and the mutated enzyme but it is inefficient in inhibiting the religation of R13 (Figure 4). In fact, the R13 religation occurs selectively with the oligonucleotide cleaved at the CL2 site, indicating that CPT is unable to bind this complex. In line in vivo and in vitro experiments have shown the strict requirement of a thymine base in position −1 of the scissile strand in order to permit CPT binding and stabilization of the binary complex (16,28). All these findings provide an explanation for the time dependent CPT resistance displayed by the cell expressing the mutant (Figure 1B). In fact, the increased preference of the mutant for DNA substrates not having a thymine in position −1 of the scissile strand permits to a large population of molecules to be cleaved in vivo in a position that does not permit CPT binding.
The different sequence specificity of the mutant is also confirmed by the cleavage/religation equilibrium experiment of Figure 7, that indicates for the wild-type enzyme the presence of a large number of CPT sensitive cleavage sites. In the case of the mutant, at short times, just few bands, diagnostic of the stabilization of a ternary complex, are detected. Only after some minutes, the mutant cleaves at sites able to be stabilized by CPT. So we propose that the number of sites cleaved by the enzyme in CPT sensitive sequences increase as a function of time, in a way sufficient to provoke cell death, as shown by the cell viability assay (Figure 1B). The CPT time dependent resistance of the Glu418Lys mutant is then due to its different DNA substrate preference likely induced by the perturbation of the direct and water mediated interactions of the Thr417-Ser423 loop with the DNA intact strand (5,26). The indirect effect caused on the drug binding pocket by the repositioning of Lip 1 should be trascurable, because CPT efficiently binds to the cleavage 1 complex of both the native and the mutated enzyme, strongly reducing the religation rate of the R11 oligonucleotide in both cases (Figure 5).
In conclusion, we have shown for the first time the occurrence of a different cleavage substrate specificity for the single Glu418Lys human topoisomerase I mutant. The different specificity points out the important role of core subdomain I in the substrate recognition and confirms the strict requirement of T base at the −1 position of the scissile strand for CPT binding, independently from the 418 residue nature.
Acknowledgments
The authors thank S. Z. Pedersen for critical reading, P. Benedetti for useful discussion, and M.-A. Bjornsti for kindly providing the oligos used for the religation experiment. The experimental contribution of E. Ghidini, O. Sarra and C. Tesauro in the laboratory is also acknowledged. This work was partly supported by grants from COFIN 2005 and a fellowship from the FIRC to S. Castelli. Funding to pay the Open Access publication charges for this article was provided by MIUR project COFIN 2005.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chen A., Liu L.F. DNA topoisomerase: essential enzymes and lethal targets. Ann. Rev. Pharmacol. Toxicol. 1994;34:191–218. doi: 10.1146/annurev.pa.34.040194.001203. [DOI] [PubMed] [Google Scholar]
- 2.Nitiss J. Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim. Biophys. Acta. 1998;1400:63–82. doi: 10.1016/s0167-4781(98)00128-6. [DOI] [PubMed] [Google Scholar]
- 3.Wang J.C. DNA topoisomerases. Ann. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 4.Stewart L., Ireton G.C., Champoux J.J. The domain organization of human topoisomerase I. J. Biol. Chem. 1996;271:7602–7608. doi: 10.1074/jbc.271.13.7602. [DOI] [PubMed] [Google Scholar]
- 5.Redinbo M.R., Stewart L., Kuhn P., Champoux J.J., Hol W.G.J. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 6.Stewart L., Redinbo M.R., Qiu X., Hol W.G.J., Champoux J.J. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 7.Pommier Y., Pourquier P., Fan Y., Strumberg D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta. 1998;1400:83–106. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 8.Staker B.L., Hjerrild K., Feese M.D., Behnke C.A., Burgin A.B., Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl Acad. Sci. USA. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Staker B.L., Feese M.D., Cushman M., Pommier Y., Zembower D., Stewart L., Burgin A.B. Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex. J. Med. Chem. 2005;48:2336–2345. doi: 10.1021/jm049146p. [DOI] [PubMed] [Google Scholar]
- 10.Pommier Y., Pourquier P., Yoshimasa U., WU J., Laco G.S. Topoisomerase I inhibitors: selectivity and cellular resistance. Drug Resistance Updates. 1999;2:307–318. doi: 10.1054/drup.1999.0102. [DOI] [PubMed] [Google Scholar]
- 11.Chrencik J.E., Staker B.L., Burgin A.B., Pourquier P., Pommier Y., Stewart L., Redinbo M.R. Mechanisms of camptothecin resistance by human topoisomerase I mutations. J. Mol. Biol. 2004;339:773–784. doi: 10.1016/j.jmb.2004.03.077. [DOI] [PubMed] [Google Scholar]
- 12.Fiorani P., Bruselles A., Falconi M., Chillemi G., Desideri A., Benedetti P. Single mutation in the linker domain confers protein flexibility and camptothecin resistance to human topoisomerase I. J. Biol. Chem. 2003;278:43268–43275. doi: 10.1074/jbc.M303899200. [DOI] [PubMed] [Google Scholar]
- 13.Chang J.Y., Liu J.F., Juang S.H., Liu T.W., Chen L.T. Novel mutation of topoisomerase I in rendering cells resistant to camptothecin. Cancer Res. 2002;62:3716–3712. [PubMed] [Google Scholar]
- 14.Kubota N., Kanazawa F., Nishio K., Takeda Y., Ohmori T., Fujiwara T., Terashiman Y., Saijo N. Detection of topoisomerase I gene point mutation in CPT-11 resistant lung cancer cell line. Biochem. Biophys. Res. Comm. 1992;188:571–577. doi: 10.1016/0006-291x(92)91094-7. [DOI] [PubMed] [Google Scholar]
- 15.Chillemi G., Fiorani P., Castelli S., Bruselles A., Benedetti P., Desideri A. Effect on DNA relaxation of the single Thr718Ala mutation in human topoisomerase I: a functional and molecular dynamics study. Nucleic Acids Res. 2005;33:3339–3350. doi: 10.1093/nar/gki642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaxel C., Capranico G., Kerrigan D., Kohn K.W., Pommier Y. Effect of local DNA sequence on topoisomerase I cleavage in the presence or absence of camptothecin. J. Biol. Chem. 1991;266:20418–20423. [PubMed] [Google Scholar]
- 17.Bjornsti M.-A., Benedetti P., Viglianti G.A., Wang J.C. Expression of human DNA topoisomerase I in yeast cells lacking yeast DNA topoisomerase I: restoration of sensitivity of the cells to the antitumor drug camptothecin. Cancer Res. 1989;49:6318–6323. [PubMed] [Google Scholar]
- 18.Kauh E.A., Bjornsti M.-A. CT1 mutants suppress the camptothecin sensitivity of yeast cells expressing wild-type DNA topoisomerase I. Proc. Natl Acad. Sci. USA. 1995;92:6299–6303. doi: 10.1073/pnas.92.14.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bjornsti M.-A., Wang J.C. Expression of yeast DNA topoisomerase I can complement a conditional-lethal DNA topoisomerase I mutation in Esherichia coli. Proc. Natl Acad. Sci. USA. 1987;84:8971–8975. doi: 10.1073/pnas.84.24.8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benedetti P., Fiorani P., Capuani L., Wang J.C. Camptothecin resistance from a single mutation changing glycine 363 of human DNA topoisomerase I to cysteine. Cancer Res. 1993;53:4343–4348. [PubMed] [Google Scholar]
- 21.Yang Z., Champoux J.J. Reconstitution of enzymatic activity by the association of the cap and catalytic domains of human topoisomerase I. J. Biol. Chem. 2002;277:30815–30823. doi: 10.1074/jbc.M205302200. [DOI] [PubMed] [Google Scholar]
- 22.Fiorani P., Amatruda J.F., Silvestri A., Butler R.H., Bjornsti M.A., Benedetti P. Domain interactions affecting human DNA topoisomerase I catalysis and camptothecin sensitivity. Mol. Pharmacol. 1999;56:1105–1115. doi: 10.1124/mol.56.6.1105. [DOI] [PubMed] [Google Scholar]
- 23.Stewart L., Ireton G.C., Champoux J.J. A functional linker in human topoisomerase I is required for maximum sensitivity to camptothecin in a DNA relaxation assay. J. Biol. Chem. 1999;274:32950–3296. doi: 10.1074/jbc.274.46.32950. [DOI] [PubMed] [Google Scholar]
- 24.Hsiang Y.H., Hertzberg R., Hecht S., Liu L.F. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 25.Xiao X., Cushman M. An ab initio quantum mechanics calculation that correlates with ligand orientation and DNA cleavage site selectivity in camptothecin-DNA-topoisomerase I ternary cleavage complexes. J. Am. Chem. Soc. 2005;127:9960–9961. doi: 10.1021/ja042485n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chillemi G., Castrignano T., Desideri A. Structure and hydration of the DNA-human topoisomerase I covalent complex. Biophys J. 2001;81:490–500. doi: 10.1016/S0006-3495(01)75716-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen A.H., Gocke E., Bonven B.J., Nielsen O.F., Westergaard O. Topoisomerase I has a strong binding preference for a conserved hexadecameric sequence in the promoter region of the rRNA gene from Tetrahymena pyriformis. Nucleic Acids Res. 1985;13:1543–1557. doi: 10.1093/nar/13.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pondarre C., Strumberg D., Fujimori A., Torres-Leon R., Pommier Y. In vivo sequencing of camptothecin-induced topoisomerase I cleavage sites in human colon carcinoma cells. Nucleic Acids Res. 1997;25:4111–4116. doi: 10.1093/nar/25.20.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]