Abstract
We present MultiGO, a web-enabled tool for the identification of biologically relevant gene sets from hierarchically clustered gene expression trees (http://ekhidna.biocenter.helsinki.fi/poxo/multigo). High-throughput gene expression measuring techniques, such as microarrays, are nowadays often used to monitor the expression of thousands of genes. Since these experiments can produce overwhelming amounts of data, computational methods that assist the data analysis and interpretation are essential. MultiGO is a tool that automatically extracts the biological information for multiple clusters and determines their biological relevance, and hence facilitates the interpretation of the data. Since the entire expression tree is analysed, MultiGO is guaranteed to report all clusters that share a common enriched biological function, as defined by Gene Ontology annotations. The tool also identifies a plausible cluster set, which represents the key biological functions affected by the experiment. The performance is demonstrated by analysing drought-, cold- and abscisic acid-related expression data sets from Arabidopsis thaliana. The analysis not only identified known biological functions, but also brought into focus the less established connections to defense-related gene clusters. Thus, in comparison to analyses of manually selected gene lists, the systematic analysis of every cluster can reveal unexpected biological phenomena and produce much more comprehensive biological insights to the experiment of interest.
INTRODUCTION
The first computational steps of gene expression analysis encompass the pre-processing of the data and the use of statistical tests to detect genes with altered expression (1–3). Different clustering approaches can then be used to group together genes with similar expression profiles, in order to present the data in a more comprehensible and interpretable form (2,3). A common clustering approach is hierarchical clustering (HC). In HC, genes are classified into a series of nested clusters by iteratively joining, or disjoining, two elements (3,4). Alternatively, genes can be groupedinto a predetermined number of clusters using partitional clustering approaches, as in e.g. k-means clustering (3,5). Regardless of the chosen clustering approach, however, numerous alternative clusters, and even result series, may be generated.
The next step after performing the clustering is to extract the biological information and to interpret the biological relevance of the generated clusters. This is often done for the clusters by calculating the statistical significance of the functions that the genes in different clusters are involved in (6–15). The biological information for the genes can be extracted, e.g. using text mining tools for biological and medical literature (9–11) or controlled vocabularies, such as Gene Ontology (GO) (16). The difference between these information sources is that in the first the same function may be described using numerous alternative wordings, whereas in the latter this is eliminated. After gene functions have been extracted, their significance is typically evaluated using standard statistical methods, such as the binomial distribution (BD) or hypergeometric distribution (HD) or chi-squaretest. In this evaluation, the frequency of the term within a set of query genes, i.e. the genes within the cluster, is compared against the frequency of the same term in a background gene set, e.g. in the entire transcriptome of the organism. The resulting P-value illustrates the chance to randomly find a GO-term that is at least as significant as the tested one.
Various computational tools do exist for the determination of the biological relevance of a single gene set, or cluster, with respect to the statistical significance of the GO-terms of its genes (12–15). Although these tools have improved the interpretation of expression data by enabling the creation of rapid overviews of affected functions, they suffer from a deficiency of being mainly designed to analyse only a single gene set at a time. Thus, existing tools unnecessarily complicate the systematic analysis of the clustering results of large expression data sets and make their interpretation a laborious task, where numerous alternative clusters must be analysed one by one. This in turn complicates the understanding of the different affected biological functions of the experiment and of their various connections. For example, looking at only one gene set ignores the possibility that multiple cellular processes can be stimulated and affected by the experiment. Also, since all clusters are not necessarily analysed, there is a possibility of overlooking a specific cluster and a GO-term, e.g. the affected key function. There are some tools that estimate the optimal set of clusters using gene function annotations (7,8). Although these tools enumerate all clusters in the analysis, they still suffer from the deficiency of not reporting all significant GO-terms.
To address the above problems, we developed a web-enabled tool, MultiGO. Our tool analyses every gene cluster, assigns a representative function for each, and reports all functionally enriched clusters with respect to the GO classification. The biggest advantage of the tool is gained when MultiGO is used to analyse expression trees created using HC. These analyses may discover unexpected connections between different clusters, e.g. two clusters that are located in distinct branches of the tree but share similar functions. MultiGO can also highlight the key biological functions that are affected by the experiment by determining the most optimal set of clusters from the trees using Fisher's combined P-value test (17).
We demonstrate the functionality of MultiGO by analysing a set of abscisic acid (ABA) experiments combined with selected abiotic stress experiments, namely drought and cold, on Arabidopsis thaliana. ABA is a plant hormone, which is produced as a consequence of several abiotic stresses (18). It has also been shown that ABA is one of the key regulators of abiotic stress responses and the correct expression of, e.g. a subset of genes involved in drought and cold responses is dependent on ABA (19,20). Although in recent years the transcriptional regulatory circuit involved in ABA signalling has become more evident, the complete signalling network and its relation to abiotic stress signals is still not entirely understood (21,22). Therefore, the aim of the analysis was to investigate connections between abiotic stress and ABA affected functions. The analysis recovered known connections between ABA and the stresses and established a less well-characterized relationship to defense-related gene clusters.
MATERIALS AND METHODS
In MultiGO, the preferred input is a hierarchically clustered gene expression tree. The most common HC approach used to process expression data is agglomerative clustering (4). In agglomerative HC, a distance matrix of all pairs of genes is first calculated. After this, the matrix is iteratively updated by combining the two closest elements, i.e. a gene or cluster, together and by recalculating the distances of the remaining elements (3,4). The result of the HC is then represented as an expression tree, which illustrates the combination events made at each successive stage of analysis and where the similarity of the combined elements is illustrated by the length of the branch connecting the elements, i.e. shorter branch lengths indicate higher similarity than longer ones. Since genes are buried in the nested structure of the tree, a gene can simultaneously belong to several clusters at different levels of hierarchy (3,4). In the context of gene expression analysis, HC can be used to group the genes and to visualize the expression data. From the resulting tree, groups of genes with correlated expression profiles are typically identified visually. For example, the tree is cut at a desired hierarchy level beyond which the user has defined that the genes are no longer co-expressed.
GO-terms
GO describes the biological process, molecular function or cellular component of a gene using a systematic classification that guarantees each function being described only once (16). GO-terms are associated with the submitted genes and clusters using the ontologies and annotations offered by the GO consortium (http://www.geneontology.org/). GO ontologies are structured as a directed acyclic graph (DAG) where terms, i.e. the biological description of the gene product, can have one or more parent and child terms (16). In the DAG, the parent term is a less specialized description than its child term (16). This means that a term is always less specifically described by its parents, which in turn are less specifically described by their parents, etc. all the way to very first parent term of DAG. When mapping the GO-terms this also means that if a gene is associated with a certain term, then it is indirectly associated with all parent terms of the term, and with their ancestral terms.
Statistical tests to evaluate individual clusters
In MultiGO, the enrichment of the GO-terms within each cluster is tested and the most significant GO-term is assigned to the cluster that satisfies the parameters. If the analysed cluster is removed or if the cluster does not contain any GO-terms above the P-value threshold, no GO-term is assigned to the cluster (see Supplementary Data for more detailed description of the parameters). The assigned GO-term is then assumed to be the representative function of the genes within the cluster.
The enrichment of the GO-terms within each cluster is tested using the HD or BD (Equations 1 and 2). In Equations 1 and 2, n is the number of genes in the cluster, k is the number of genes in the cluster with a given GO-term, N is the number of genes in the background distribution and D is the number of genes in the background distribution with the same GO-term. In Equation 2, p is the probability of finding a gene from the background distribution with the same GO-term (D/N).
| 1 |
| 2 |
Both statistical tests are widely used in similar tasks (12–15). HD calculates the probability (P-value) of randomly finding the same or higher number of genes having the same GO-term from the background gene list, e.g. the transcriptome of the organism. BD offers the same information and is slightly faster to calculate. However, since in BD the probability of finding a gene from the background distribution (p in Equation 2) is approximated and kept constant throughout the calculus, it is less exact than HD.
When many statistical tests are performed simultaneously, the chances of declaring results erroneously as statistically significant increase. This happens, because the given significance level may be appropriate for each individual test, but the probability of observing at least one significant P-value by chance increases when the number of tests increases. Therefore, the significance level of the individual comparisons should be modified to account for the number of comparisons performed. One approach for multiple hypothesis correction is to control the family-wise error rate (FWER) that is the probability that any reported GO-term is a false positive. In MultiGO, the FWER can be controlled using Bonferroni or Holm's step-down methods. An alternative approach is to calculate the false discovery rate (FDR) that is the expected proportion of false positive GO-terms in the results. In MultiGO, the FDR can be calculated using Benjamini-Hochberg's FDR method (23). While the FWER can erroneously discard more statistically significant GO-terms, this risk is minimized in FDR at the cost of reporting a few more false positives, i.e. GO-terms that are not significant.
The above correction methods assume that the performed comparisons are independent, which is not the case with the GO-terms. Due to the structure of DAG, GO-terms and their P-values are dependent and correlated (15). For example, if a term is significant then it is likely that its parents are also significant. Despite this caveat, correction methods can correct, at least, the worst errors and increase the reliability of the results when compared to uncorrected results.
Statistical tests to analyse the optimal cluster set
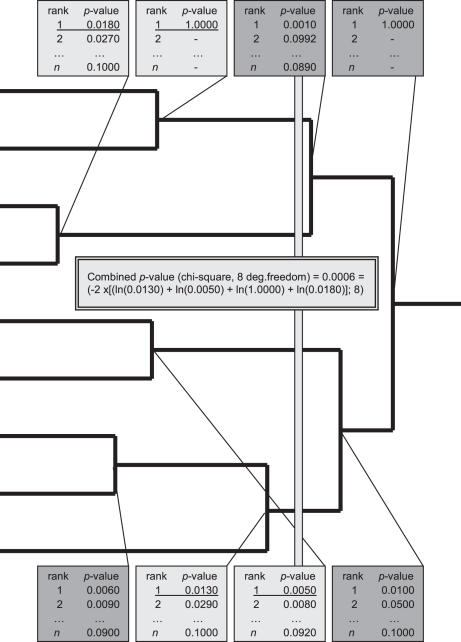
The most optimal cluster set, i.e. the best cutting level of the expression tree, and the corresponding biological key functions are searched in MultiGO by calculating the overall statistical significance of the GO-terms of the clusters located at a given level of similarity (Figure 1). This calculus is repeated at every level of similarity in the tree, starting from the single gene level and ending at the level of one cluster containing all the genes of the experiment. The overall statistical significance is calculated using Fisher's combined probability test (Equation 3, Figure 1) (17). In Equation 3, Pi is the corrected P-value of the most significant GO-term of the ith cluster and k is the number of clusters created at the position or passing it, i.e. clusters created farther from the root and bypassing the position. The overall P-value for the given clusters is then calculated from the test score using chi-square distribution with 2k degrees of freedom.
| 3 |
When calculating the significance, Pi is set to one for clusters that do not contain significant GO-terms and for clusters that have been filtered, i.e. clusters that contain an improper number of genes according to the corresponding parameters (see Supplementary Data for more detailed description of the parameters). In Fisher's combined probability test, P-values to be combined (Pi) are assumed to be independent (17). In MultiGO, independence is aspired using non-overlapping clusters and using the P-values of the best GO-terms. The use of non-overlapping clusters guarantee that the GO-terms combined in Equation 3 are not influenced by shared genes, whereas only choosing the GO-term with the most significant P-value avoids the correlations of GO-terms due to the structure of the DAG.
Figure 1.
Fisher's combined probability test is used to calculate the overall P-values for the clusters and to estimate the cutting point of the expression tree. Light gray line illustrates the height that is calculated in the example. Light gray boxes show the selected clusters where the P-values used in the calculus are underlined (the P-value of the best GO-term of the selected cluster).
Acquisition and analysis of experimental data
A.thaliana was grown in 1:1 peat:vermiculite (Finnpeat B2; Kekkilä Oyj, Tuusula, Finland) with a 12-h light period at 22°C. Three week-old Col-0 wild type and plants overexpressing a gene early responsive to dehydration (ERD15, 24) under control of the cauliflower mosaic virus 35S promoter were used for experiments. Samples were collected to liquid nitrogen 90 min after spraying with 100 μM ABA or water as a control. Total RNA was isolated with Qiagen Plant RNA extraction kit. cRNA synthesis, hybridization to Affymetrix ATH1 arrays and chip-scans were performed at the NASC's Affymetrix Service (25). The data set of the experiment has been published under NASCARRAYS-321. The experimental data was combined with data of three public experiments retrieved from NASC (http://affymetrix.arabidopsis.info) and TAIR (http://www.arabidopsis.org) (25,26). The public data included expression data sets of drought stress time course in shoots (ME00338), drought and cold stresses in mutant and wild-type leaves (NASCARRAYS-70) and ABA time course in seedlings (ME00333) (Supplementary Table 1). All expression sets contained arrays for both the controls and the treatments, which provided the removal of the biological variability coming from the experimental tissues used.
All expression sets were pre-processed as one entity using Robust Multichip Average (RMA) and Mas5Calls (27–29). RMA computes the log2 scale expression values using background-corrected probe-specific correction of the perfect match probes, quantile normalization and median polish summarizing (28). Low expression genes were detected from the unprocessed data using Mas5Calls (29), and genes flagged as absent on every array were removed. After pre-processing, genes that had in the public experiments a significantly altered expression between the sample and the corresponding control (Supplementary Table 1) were pooled together by taking their union. The significance of the genes was detected using regularized t-test with 0.001 as the P-value threshold for statistical significance (30). These processes resulted in 11 875 genes whose expression values were averaged between the replicate arrays of the experiments. (Using 0.05 or 0.01 as the P-value threshold would have yielded respectively 17 982 and 15 344 genes.) The mean expression values of the controls were then subtracted from the corresponding mean values of the samples to eliminate any bias deriving from the different monitored tissues. Genes were clustered using HC with different linkage methods (complete, single and average) and with different distance metrics (Euclidean and Pearson correlation coefficient) (4). RMA, mas5calls and the regularized t-test (Cyber-T) were performed in Bioconductor (31) extension to R computing environment (http://www.r-project.org/). Clustering was done using TIGR MultiExperiment Viewer (version 3.1) (32).
METHODOLOGICAL RESULTS
Different parameter combinations of MultiGO were tested to see their effect on the overall P-values, i.e. the obtained P-value of the Fisher's combined probability test, and on the optimal cluster set selection (see Supplementary Data for more detailed description of the parameters). Similar to the original study first time applying HC to microarray data (4), different parameter combinations were compared using an expression tree created using average linkage and Pearson correlation coefficient. The analyses included the use of different maximum cluster sizes, statistical tests, correction methods, filtering of the low occurrence clusters and P-value cut-offs. The overall P-values of these analyses are shown in Supplementary Figures 2–6. Most parameters have no notable effect on the results. For example, analyses using different multiple hypothesis correction methods or analyses using different P-value cut-offs produce similar results with little variation. The only exceptions are analyses without multiple hypothesis correction and analyses using different maximum cluster sizes. The maximum cluster size parameter, which can be used to speed up calculations, filters clusters containing an improper number of genes. The use of small values seems to lead to unwanted filtering of significant clusters whereas the use of the maximum value did not affect the results.
Analyses of expression trees created using different HC parameters
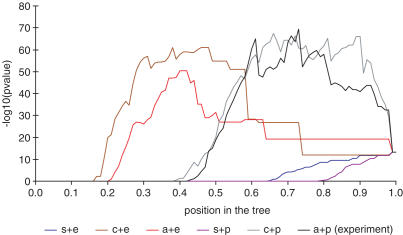
The effect of different linkage methods and distance metrics on the overall P-values, and on the expression tree cutting point selection, was investigated by clustering the expression data using different combinations of average, complete and single linkage, and of Euclidean and Pearson correlation coefficient distances. Parameters used to analyse these trees were selected based on the information of the parameter analyses and by using those statistical methods that are considered the most reliable. HD was used instead of BD, and FDR-correction was used with a rather conservative P-value cut-off (0.001), all genes were analysed and no filtering was performed. The performance of the different HC combinations is shown in Figure 2. For this expression data, each linkage method achieves its best result when Pearson coefficient correlation is used as the distance, agreeing well with the distance metric assumption made in the original HC study (4). From the different linkage methods, average linkage creates results with the most significant overall P-value. However, the difference between the complete and average linkage methods is almost indistinguishable, their most significant overall P-values were 4−68 and 3−70, indicating that these linkage methods perform equally well. This supports the previous study where it was shown that average and complete linkage methods outperform single linkage (6).
Figure 2.
The behaviour of the overall P-values in expression trees created using different linkage methods and distance metrics. In the figure a, c and s are average, complete and single linkage and e and p are Euclidean and Pearson coefficient correlation distances. The y-axis is the −log10 value of the overall P-value and the x-axis is the position at the tree.
Experimental data analysis
We analysed microarray data of mutants sensitive to cold stress and transgenic plants over-expressing a gene involved in ABA signalling. Additionally, microarray data from drought, cold and ABA experiments were included in the data set. The experimental analysis was done using the expression tree that was created using average linkage with Pearson coefficient correlation, the one containing the most significant overall P-value, and using parameters that were selected based on the information of the parameter estimation analyses (HD, FDR, 0.001 as the P-value cut-off, all genes as the maximum cluster size and no filtering). Results can be viewed at the group's web page (http://ekhidna.biocenter.helsinki.fi/poxo/multigo/arab_example).
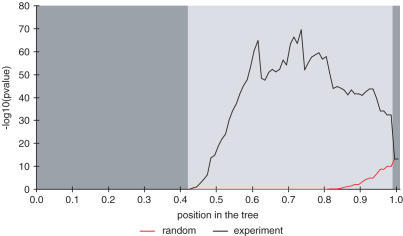
A set of interesting candidate clusters was obtained from the expression tree by cutting it at the location of the most significant overall P-value. The overall P-values in relation to their position in the tree are shown in Figure 3. The most significant overall P-value, obtained using Fisher's combined probability test, selects a set of clusters that would become merged together into biologically meaningless clusters, if one moves nearer to the root, whereas clusters with similar function would become split into several small clusters, if one moves farther from the root. The most significant overall P-value (3−70) is located at the height of 0.73 (Figure 3), where there are 42 clusters in total in the tree of which 14 have a significant GO-term associated with them (Table 1). Expression profiles of these 14 significant clusters are shown as heatmaps in Supplementary Figures 8–21. The 14 significant clusters contained genes that are involved in different biological functions that are likely to be the key functions affected by the experiment. For example, the experiments included ABA and environmental abiotic treatments that are listed in Table 1 as response to ABA (Node_11808) and as response to heat (Node_11805). Besides the affected abiotic stresses also clusters related to defence responses are listed, such as defense response (Node_11830) and response to wounding (Node_11791). Figure 3 indicates that there is a second potential cutting point in the expression tree located at the height of 0.61. At this position, the overall P-value is almost as significant (1e−65) as at the most significant position of the tree. Also the clusters that are located at this position are involved in functions that are mainly the same ones (9 out of the 14 functions reported at the 0.73 are reported here as well), including functions related to the probed experiments, such as response to ABA and response to heat. These findings indicate that a set of biologically meaningful clusters could indeed be caught by using overall P-values.
Figure 3.
Reliability of the overall P-values estimated using random permutations. The most significant overall P-values were chosen from the randomised data for each position at the tree. The light grey area indicates those positions of the tree where random analyses yielded less significant overall P-values and the dark grey is an area where all random analyse yielded equal or more significant overall P-values. The left y-axis is the −log10 value of the overall P-value and the x-axis is the position at the tree.
Table 1.
Significant clusters at the most significant overall P-value position of the expression tree
| GO-term | Annotation | P-value | Cluster | Genes |
|---|---|---|---|---|
| GO:0006412 | Protein biosynthesis | 1E−40 | Node_11771 | 182 |
| GO:0042254 | Ribosome biogenesis and assembly | 1E−10 | Node_11802 | 1664 |
| GO:0006952 | Defense response | 3E−10 | Node_11830 | 446 |
| GO:0015979 | Photosynthesis | 2E−09 | Node_11822 | 1393 |
| GO:0009657 | Plastid organization and biogenesis | 1E−07 | Node_11832 | 707 |
| GO:0009408 | Response to heat | 9E−07 | Node_11805 | 243 |
| GO:0009737 | Response to abscisic acid stimulus | 1E−06 | Node_11808 | 598 |
| GO:0044249 | Cellular biosynthesis | 1E−06 | Node_11820 | 379 |
| GO:0015979 | Photosynthesis | 1E−06 | Node_11801 | 80 |
| GO:0016070 | RNA metabolism | 2E−06 | Node_11813 | 1167 |
| GO:0009611 | Response to wounding | 6E−06 | Node_11791 | 129 |
| GO:0042221 | Response to chemical stimulus | 2E−04 | Node_11810 | 804 |
| GO:0030163 | Protein catabolism | 4E−04 | Node_11800 | 180 |
| GO:0043412 | Biopolymer modification | 9E−04 | Node_11829 | 862 |
The position contained 14 significant and totally 42 clusters.
Another viewpoint to the data can be obtained by sorting the GO-terms according to the number of times they occur as the best GO-term of a cluster in the whole expression tree. This number corresponds roughly to the number of genes involved in the function and embodies the level of co-expression of genes, i.e. functions involving a large number of tightly co-expressed genes will occur on top. Table 2 lists the top 20 most common best GO-terms. This listing illustrates that several of the most common GO-terms are related to abiotic stresses. For example, Table 2 contains functions such as response to ABA, response to heat, response to temperature stimulus, response to cold and response to water deprivation. Intriguingly, also in here response to wounding is reported.
Table 2.
Top 20 most common GO-terms of being the most significant GO-term of the clusters
| GO-term | Annotation | min P-value | max P-value | Clusters |
|---|---|---|---|---|
| GO:0006412 | Protein biosynthesis | 5E−84 | 6E−04 | 42 |
| GO:0015979 | Photosynthesis | 4E−12 | 4E−04 | 33 |
| GO:0042254 | Ribosome biogenesis and assembly | 4E−31 | 3E−04 | 29 |
| GO:0009725 | Response to hormone stimulus | 4E−11 | 2E−04 | 12 |
| GO:0009737 | Response to abscisic acid stimulus | 3E−10 | 7E−04 | 11 |
| GO:0016070 | RNA metabolism | 2E−06 | 8E−04 | 8 |
| GO:0009408 | Response to heat | 4E−12 | 4E−06 | 8 |
| GO:0044249 | Cellular biosynthesis | 5E−11 | 2E−04 | 8 |
| GO:0009266 | Response to temperature stimulus | 8E−10 | 1E−08 | 7 |
| GO:0006396 | RNA processing | 1E−05 | 7E−04 | 6 |
| GO:0009409 | Response to cold | 3E−12 | 3E−08 | 6 |
| GO:0030163 | Protein catabolism | 3E−07 | 6E−04 | 6 |
| GO:0009657 | Plastid organization and biogenesis | 3E−08 | 1E−07 | 5 |
| GO:0009611 | Response to wounding | 6E−07 | 3E−04 | 5 |
| GO:0009414 | Response to water deprivation | 2E−05 | 4E−04 | 4 |
| GO:0009684 | Indoleacetic acid biosynthesis | 2E−06 | 8E−05 | 4 |
| GO:0009658 | Chloroplast organization and biogenesis | 1E−06 | 5E−04 | 4 |
| GO:0006520 | Amino acid metabolism | 1E−06 | 6E−05 | 3 |
| GO:0006468 | Protein amino acid phosphorylation | 2E−06 | 7E−05 | 3 |
| GO:0043412 | Biopolymer modification | 9E−05 | 9E−04 | 3 |
Random data analyses
Randomisation analyses were performed to assess the reliability of the experimental analysis, of the overall P-values and of the GO-terms of the single clusters. The most significant overall P-values of these analyses are shown in Figure 3 (Supplementary Table 2 and Figure 7). The random analyses were performed using the random-mode of MultiGO. The random-mode of the tool retains the original number of clusters and their cluster sizes, but assigns the submitted genes randomly into these. The analysis was repeated 1000 times using previous parameters of the experimental data analysis (HD, FDR, 0.001 as the P-value cut-off, all genes as the maximum cluster size and no filtering).
As illustrated in Figure 3, hardly any cluster sets had a more significant overall P-value in the randomised analyses in comparison with the cluster sets of the experimental analysis. The only exceptions are cluster sets near the root and near the leaves. In these parts of the trees, randomly generated data yielded cluster sets that have as significant overall P-values as the corresponding cluster sets have in the experimental analysis (Supplementary Table 2). Near the leaves, both random and experimental analyses yielded insignificant overall P-values for the cluster sets, i.e. P-values are 1.00, whereas near the root overall P-values are the same in both cases. Another verification of the correctness is that the most significant overall P-value (7−14) of the 1000 random repeats is much less significant than the most significant overall P-value (3−70) of the experimental data.
The most significant GO-terms of a single cluster are also much less significant in the random analyses than in the experimental analysis and typically are located near the root (Supplementary Table 3). For example, the P-value of the most significant GO-term obtained in the random analyses is 7−14 and it can be found in a cluster near the root (relative position 0.99). On the other hand, the most significant P-value obtained in the experimental analysis is 5−84 and the cluster with this GO-term is located in the first fifth of the tree (relative position 0.18).
BIOLOGICAL RESULTS
Two interesting, and fairly unexpected, biological phenomena that are reported in Table 1 are defense response (Node_11830) and response to wounding (Node_11791). These functions indicate that biotic defence responses are also affected in these abiotic stress experiments. It can be noted that these biotic defence functions are segregated and are distinctly expressed (Supplementary Figures 8 and 13). Node_11830 enclosing defense response as the most significant GO-term is clearly separated from the response to wounding cluster. This cluster comprises genes induced early by drought (30 min to 1 h) and cold (3 h) as well as by over-expression of ERD15. The gene set contains a series of transcription factors related to defence responses such as WRKY18, WRKY22, WRKY33, WRKY40, WRKY46, WRKY53, WRKY54 and WRKY70, as well as disease resistance proteins with toll-interleukin-resistance domains. Genes in the cluster are repressed late by ABA (3 h) and cold (24 h), and are repressed in sfr2, sfr3 and sfr6 mutant experiments in cold conditions. In contrast, Node_11791 contains genes induced by drought (30 min to 12 h) and cold (3 h), repressed by sfr2, sfr3 and sfr6 mutants in cold and comprises jasmonic acid and ethylene responsive genes, such as JR1, AOC4, ATMYC2 and VSP1. It is interesting that the defense response cluster contains a series of transcription factors but no enrichment of GO-terms of the known hormone signals salicylic acid, ethylene and jasmonate. This might point to a set of early biotic stress-responsive genes, manifested even as a response to abiotic stresses. Recent evidence has shown that ABA has a role both in positive and negative regulation of defence gene signalling (33), and the present analysis clearly points to a tight co-regulation of different types of stress-regulated genes independent of the stress stimulus.
The more expected biological phenomenon reported in both tables is response to ABA. Finding this GO-term is expected, since it is considered to be an important signal compound in the abiotic stress responses that are probed in the experiments here (21,22). The largest cluster in the data having response to ABA stimulus as its most significant GO-term (Node_11861) comprises a series of protein phosphatases 2C (e.g. AT4G26080 or AT1G07430) and transcription factors like the ABA-responsive elements-binding factor ABF3 (AT4G34000). These genes are induced early (0.5–1 h) by ABA and drought in the chosen set of experiments. Cold treatment also leads to up-regulation of a series of genes in this cluster in wild type and sfr2, sfr3 and sfr6 mutants (Supplementary Figure 22). This cluster also contains effector-like ABA-related genes such as the dehydrin Rab18 (AT5G66400) as well as lipid transfer proteins, which here are only induced late (3 h) by ABA. It is tempting to speculate that uncharacterised protein phosphates C and unknown proteins in these clusters have a specific role in early or later transcriptional responses to abiotic stresses.
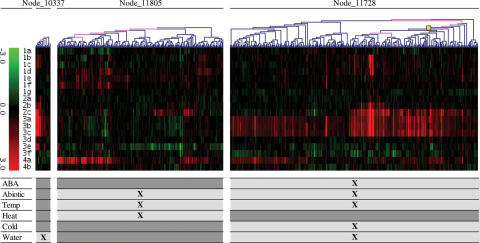
Another GO-term involved in abiotic stress responses and listed in both tables 1 and 2 is response to heat. This listing can then be expanded by collecting other related processes from Table 2: response to temperature stimulus, response to cold and response to water deprivation. Expression of genes involved in these abiotic stresses can be viewed in Figure 4. Figure 4 shows those clusters (Node_11728, Node_11805 and Node_10337) that are the largest clusters having the corresponding term (response to temperature stimulus, response to heat and response to water deprivation) as the best GO-term, and contain most of the other clusters sharing the same best GO-term. Note that the largest cluster having response to cold function as its most significant term (Node_10952) is a child of Node_11728 and is represented as a yellow square in the figure.
Figure 4.
Expression of genes involved in the reported abiotic stress responses. Figure shows those clusters that are the largest clusters having the given GO-term as their best GO-term (response to temperature stimulus is the best GO-term of Node_11728, response to heat of Node_11805 and response to water deprivation of Node_10337) (response to cold of Node_10952 is represented by a yellow square). Clusters sharing the same best GO-term as the largest cluster are coloured in purple in the expression tree. The table shows GO-terms that are related to abiotic functions (ABA is response to abscisic acid stimulus, Abiotic is response to abiotic stimulus, Temp is response to temperature stimulus, Heat is response to heat, Cold is response to cold and Water is response to water) and that are detected as significant within the set of clusters. Notations of the monitored experimental data sets are explained in Supplementary Table 1.
By further investigating these abiotic clusters it can be found that Node_11728 comprises genes that are up-regulated late by ABA (3 h), by cold in sfr2, sfr3 and sfr6 mutants and by over-expression of ERD15. On the other hand, cold seems to down-regulate these genes in wild-type plants. This cluster contains ABA-responsive genes that are interestingly also induced by low temperature and/or dehydration. Genes belonging to the response to heat (Node_11805) cluster are expressed in a different fashion. This gene set contains various heat shock proteins that are induced late by drought (3–12 h) and by over-expression of ERD15. In turn, these genes are repressed by wild-type and by cold in sfr2, sfr3 and sfr6 mutants. It is interesting to note that whereas functions, such as response to temperature stimulus and response to abiotic stimulus, are observed as significant in both Node_11728, Node_11805 and in their child clusters, the response to cold and response to heat are specifically listed only in within their own set of clusters (Figure 4). These findings point to a specific co-expression of genes involved in either cold or heat stress responses and suggest, because of the shared higher order functions, that the co-expression could be orchestrated in a more advanced level that is common for these abiotic stresses.
CONCLUSION
We have developed a novel tool called MultiGO that can be used to discover functionally enriched gene clusters from hierarchically organized expression trees. We have also demonstrated, with an example, how the tool can be used to discover the biologically meaningful key gene sets from the vast amount of data. In the example, MultiGO was able to discover relevant biological functions that were expected to be found, according to the experimental setting. We conclude that the performed analyses are thus reliable and can form an overview of the various simulated functions and of their connections. Importantly, the tool was also able to highlight novel gene sets that have not been previously linked to abiotic stresses or ABA-activated functions. Therefore the tool has the capability to discover functions that could otherwise have been missed, leading to novel biological insights concerning the experiments of interest.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Christopher Wilton for fixing the language. This work was supported by grant no. 40672/01 from the Tekes and a grant from the Finnish Ministry of Education to MK. Funding to pay the Open Access publication charges for this article was provided by a grant from the Academy of Finland.
Conflict of interest statement. None declared.
REFERENCES
- 1.Quackenbush J. Microarray data normalization and transformation. Nature Genet. 2002;32:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- 2.Allison D.B., Cui X., Page G.P., Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nature Rev. Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 3.Quackenbush J. Computational analysis of microarray data. Nature Rev. Genet. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- 4.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tavazoie S., Hughes J.D., Campbell M.J., Cho R.J., Church G.M. Systematic determination of genetic network architecture. Nature Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 6.Gibbons F.D., Roth F.P. Judging the quality of gene expression-based clustering methods using gene annotation. Genome Res. 2002;10:1574–1581. doi: 10.1101/gr.397002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Törönen P. Selection of informative clusters from hierarchical cluster tree with gene classes. BMC Bioinformatics. 2004;5:32. doi: 10.1186/1471-2105-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raychaudhuri S., Chang J.T., Imam F., Altman R.B. The computational analysis of scientific literature to define and recognize gene expression clusters. Nucleic Acids Res. 2003;31:4553–4560. doi: 10.1093/nar/gkg636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raychaudhuri S., Schutze H., Altman R.B. Using text analysis to identify functionally coherent gene groups. Genome Res. 2002;12:1582–1590. doi: 10.1101/gr.116402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masys D.R., Welsh J.B., Lynn F.J., Gribskov M., Klacansky I., Corbeil J. Use of keyword hierarchies to interpret gene expression patterns. Bioinformatics. 2001;17:319–326. doi: 10.1093/bioinformatics/17.4.319. [DOI] [PubMed] [Google Scholar]
- 11.Jenssen T.K., Laegreid A., Komorowski J., Hovig E. A literature network of human genes for high-throughput analysis of gene expression. Nature Genet. 2001;28:21–28. doi: 10.1038/ng0501-21. [DOI] [PubMed] [Google Scholar]
- 12.Berriz G.F., King O.D., Bryant B., Sander C., Roth F.P. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–2504. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
- 13.Hosack D.A., Dennis G., Jr, Sherman B.T., Lane H.C., Lempicki R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Shahrour F., Diaz-Uriarte R., Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 15.Boyle E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., Sherlock G. GO:TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher R.A. Edinburgh: Oliver and Boyd; 1932. Statistical Methods for Research Workers. [Google Scholar]
- 18.Zhu J. Salt and drought stress signal transduction in plants. Ann. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong L., Zhu J.K. Regulation of abscisic acid biosynthesis. Plant Physiol. 2003;133:29–36. doi: 10.1104/pp.103.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shinozaki K., Yamaguchi S.-K., Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- 21.Himmelbach A., Yang Y., Grill E. Relay and control of abscisic acid signaling. Curr. Opin. Plant Biol. 2003;6:470–479. doi: 10.1016/s1369-5266(03)00090-6. [DOI] [PubMed] [Google Scholar]
- 22.Yamaguchi–Shinozaki K., Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 23.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 24.Kiyosue T., Yamaguchi S.K., Shinozaki K. ERD15, a cDNA for a Dehydration-Induced Gene from Arabidopsis thaliana. Plant Physiol. 1994;106:1707. doi: 10.1104/pp.106.4.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craigon D.J., James N., Okyere J., Higgins J., Jotham J., May S. NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res. 2004;32:D575–D577. doi: 10.1093/nar/gkh133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhee S.Y., Beavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M., et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 28.Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu W.M., Mei R., Di X., Ryder T.B., Hubbell E., Dee S., Webster T.A., Harrington C.A., Ho M.H., Baid J., et al. Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics. 2002;18:1593–1599. doi: 10.1093/bioinformatics/18.12.1593. [DOI] [PubMed] [Google Scholar]
- 30.Baldi P., Long A.D. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 31.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 33.Mauch-Mani B., Mauch F. The role of abscisic acid in plant-pathogen interactions. Curr. Opin. Plant Biol. 2005;8:409–414. doi: 10.1016/j.pbi.2005.05.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.