Abstract
Here we describe the application of the in vitro virus mRNA display method, which involves covalent linkage of an in vitro-synthesized antibody (phenotype) to its encoding mRNA (genotype) through puromycin, for in vitro evolution of single-chain Fv (scFv) antibody fragments. To establish the validity of this approach to directed antibody evolution, we used random mutagenesis by error-prone DNA shuffling and off-rate selection to improve the affinity of an anti-fluorescein scFv as a model system. After four rounds of selection of the library of mRNA-displayed scFv mutants, we obtained six different sequences encoding affinity-matured mutants with five consensus mutations. Kinetic analysis of the mutant scFvs revealed that the off-rates have been decreased by more than one order of magnitude and the dissociation constants were improved ∼30-fold. The antigen-specificity was not improved by affinity maturation, but remained similar to that of the wild type. Although the five consensus mutations of the high-affinity mutants were scattered over the scFv sequence, analysis by site-directed mutagenesis demonstrated that the critical mutations for improving affinity were the two that lay within the complementarity determining regions (CDRs). Thus, mRNA display is expected to be useful for rapid artificial evolution of high-affinity diagnostic and therapeutic antibodies by optimizing their CDRs.
INTRODUCTION
Selection technologies, to obtain monoclonal antibodies with high affinity and specificity against defined antigens, are required for the development of diagnostic and therapeutic antibodies [reviewed in (1–4)], both to improve the detection limit for diagnostics and to decrease the required dose for therapeutics. In immunized animals, affinity maturation of antibodies occurs via repeated stimulation of antigen-specific proliferation of B cells and accumulation of point mutations introduced into the DNA (5–7). Therefore, it has been suggested that the affinity of antibodies can be improved by mimicking affinity maturation in the laboratory (8,9). For the in vitro evolution of recombinant antibodies such as single-chain Fv (scFv) and Fab antibodies, several display technologies such as phage display (10), yeast surface display (11), ribosome display (12–15) and DNA display (16) have been used to link an antibody (phenotype) and its encoding nucleic acid (genotype).
In this study, we have applied our in vitro virus (IVV) mRNA display system (17–19) for directed evolution of a single-chain antibody for the first time, although evolution of antibody mimics (fibronectin type III domains) using mRNA display has been reported previously (20,21). In mRNA display, an in vitro-synthesized polypeptide is covalently attached to its encoding mRNA through puromycin (17,19,22). Unlike phage display and yeast surface display, mRNA display (as well as ribosome display) is a totally in vitro system that does not require the transformation of living cells; thus, very large protein libraries (>1010 unique members) can easily be constructed and used for the selection of antibodies directed against antigens of interest. The covalent bond of the mRNA–protein complex in mRNA display should be more stable than the protein–ribosome–mRNA complex used in ribosome display with respect to thermal or physicochemical stress as a selection pressure. For the present study, we used an anti-fluorescein antibody as a model, because it has been well-characterized both structurally and kinetically (23,24). Further, laboratory evolution of the anti-fluorescein antibody was previously performed by yeast surface display (11) and ribosome display (14); hence, the antibody is a suitable model for evaluating our new method in comparison with the previous methods.
MATERIALS AND METHODS
DNA preparation
The oligonucleotide sequences used in this study are listed in Table 1. A DNA fragment that contains an SP6 promoter, the translational Ω enhancer from tobacco mosaic virus (25), a synthetic gene for anti-fluorescein scFv c12 (14) with a (Gly4Ser)4 linker, a FLAG-tag and a poly(A) sequence was constructed as follows. DNA fragments (FluscFv-1 through FluscFv-9) were assembled by overlap extension PCR with KOD-dash DNA polymerase (Toyobo) using FluscFv-F and FluscFv-R primers. The PCR product was cloned into pCR2.1-TOPO vector (Invitrogen) and the DNA sequence was confirmed with an ABI PRISM 3100 genetic analyzer (Applied Biosystems).
Table 1.
Oligonucleotide sequences
| Oligo | Sequence |
|---|---|
| FlaA-R | TTTTTTTTCTTGTCGTCATCGTCCTTGTAGTC |
| Flag-R | CTTGTCGTCATCGTCCTTGTAGTC |
| FluscFv-F | CGCTATGAGTGGCCTAAGTCAG |
| FluscFv-R | TTTTTTTTCTTGTCGTCGTCHTCCTTGTAGTCGCGTTTGAGTTCCAACTTAGTCCCTCCGC |
| FluscFv-1 | CGCTATGAGTGGCCTAAGTCAGATTTAGGTGACACTATAGAACAACAACAACAACAAACAACAACAAAATGGAGGTTCAGCTGCAGCAGTCTGGGCCAGAACTGGTGAAGCCTGGGGCCTCAGTGAAGATTTCCTGCAAAGCTACC |
| FluscFv-2 | GTCCAGGCCTCTGTTTGACCCACTCGATCCTGTAGGAGCTAAATGCGTAGCCGGTAGCTTTGCAGGAAATCTTCACTG |
| FluscFv-3 | TGGGTCAAACAGAGGCCTGGACAAGGCCTTGAATGGATTGGAGTCATCAACCCAGGATCCGGAGGCACTAACTACAACGAGAAGTTCAAGGGCAAGGCCGCCCTGACTGCAGACAAAAGC |
| FluscFv-4 | CCTTGCGCAGTAATAGACCGCAGAGTCCTCAGAGGTCAGGCTGCTGAGCTGGAGCTGCATGTAAGCGGTGCTGGAGCTTTTGTCTGCAGTCAGGGC |
| FluscFv-5 | GCGGTCTATTACTGCGCAAGGCGCGGAAACTACTACTTCGATTACTGGGGCCAAGGGACTACCCTTACTGTCTCTTCC |
| FluscFv-6 | CTCCGACTGAGGTGGACATGAATTTTTGAGACTGGGTCATCACAATGTCGCTGCCACCACCTCCGGATCCGCCACCGCCAGAGCCACCTCCGCCTGAACCGCCTCCACCGGAAGAGACAGTAAGGGTAGTCC |
| FluscFv-7 | CATGTCCACCTCAGTCGGAGACAGGGTCAGCGTCACCTGCAAGGCCAGCCAGAATGTGGACACTAATGTCGCCTGGTACCAACAGAAACCAGGGC |
| FluscFv-8 | GCAGAAGTACTCTGCCAAGTCTTCAGACTGCACATTGCTGATGGTGAGAGTGAAATCAGTCCCAGATCCGCTGCCGGTGAAGCGATCAGGGACTCCGGAGTACCTGTAGGATGCGGAGTGAATCAGGGCTTTAGGAGATTGCCCTGGTTTCTGTTGGTACC |
| FluscFv-9 | GAAGACTTGGCAGAGTACTTCTGCCGCCAATACAACAGCCACCCATGGACCTTCGGCGGAGGGACTAAGTTGGAAC |
| P1 | GTAAAACGACGGCCAGTG |
| P5 | CAGGAAACAGCTATGACC |
| pelB-F | CACCATGAAATACCTGCTGCCGACCG |
| pelBleader-F | CACCATGAAATACCTGCTGCCGACCGCTGCTGCTGGTCTGCTGCTCCTCGCTGCCCAGCCGGCGATGGCCATGGAGGTTCAGCTGCAGCAG |
| SP6-F | GTCAGATTTAGGTGACACTATAGAACAACAACAAC |
| T7-F | TAATACGACTCACTATAGGG |
| T7-R | TAGTTATTGCTCAGCGGTGG |
| W29ATG-F | GTGACACTATAGAACAACAACAACAACAAACAACAACAAAATG |
Oligonucleotide sequences are indicated in the 5′ to 3′ direction.
Construction of a mutated scFv library
A randomized scFv library was constructed from the wild-type scFv c12 described above or from the recovered PCR products after each round of selection. Random point mutagenesis and recombination were performed by the combination of error-prone PCR and PCR-based DNA shuffling in a single PCR tube (26). The PCR was performed with Ex Taq DNA polymerase (Takara) in the presence of 0.5 mM MnCl2 and the DNA template (0.2 pmol) using primers W29ATG-F and FlaA-R (0.3 µM each). The PCR program was as follows: denaturation at 96°C for 5 min; 80 cycles at 96°C for 30 s and at 55°C for 5 s; then at 96°C for 30 s, at 58°C for 30 s and at 72°C for 15 min. The PCR product was separated on 1% low-melting temperature-agarose gel (Sigma) and gel-purified by using a Wizard PCR preps DNA purification kit (Promega). To add an SP6 promoter, the purified DNA (1 pmol) was re-amplified by PCR with KOD-plus DNA polymerase (Toyobo) using SP6-F and FlaA-R primers (10 cycles at 96°C for 30 s; at 58°C for 30 s; and at 72°C for 1 min). The PCR product was gel-purified again and used for the next round of selection.
In vitro transcription and translation
The IVV method was performed as described previously (27,28) with some modifications. Approximately 500 ng of the DNA library was in vitro-transcribed into mRNA using the RiboMAX large-scale RNA production systems-SP6 (Promega) for 2.5 h at 37°C, then DNase I (Promega) was added and incubation was continued for a further 0.5 h at 37°C. The mRNA was purified using an RNeasy mini kit (Qiagen) and ligated with a polyethyleneglycol (PEG) spacer [p(dCp)2-T(Cy5)p-PEGp-(dCp)2-puromycin] using T4 RNA ligase (Takara) at 16°C overnight. The ligation product was purified with the RNeasy mini kit and in vitro-translated in a wheat germ cell-free translation system (Promega) for 1 h at 25°C. The reaction mixture was centrifuged for 10 min at 15 000 g and the supernatant was used for off-rate selection.
Off-rate selection
The mRNA-displayed scFv library (2 pmol) was mixed with antigen (200 nM fluorescein-biotin, 5(6)-(biotinamidohexanoylamido)pentylthioureidylfluorescein; Sigma) in 1 ml of blocking solution (DIG wash and block buffer set; Roche) containing 20 mM EDTA and 100 µg/ml yeast total RNA (Sigma). The mixture was gently rotated for 1 h at 25°C, and then added to 50 µl of pre-blocked neutravidin–agarose beads (Pierce) and rotated for 15 min at 25°C. The beads were washed six times with 5 ml of PBST [PBS (pH 7.4) containing 0.2% Tween-20 (Bio-Rad)] at room temperature. For off-rate selection, 500 µl of PBST containing 1 mM free fluorescein (Nacarai tesque) and 100 µg/ml yeast total RNA were added and rotated for an appropriate time (round 1, 3 h; round 2, 20 h; round 3, 96 h; round 4, 336 h) at 4°C. Again, the beads were washed three times with 500 µl of PBST at room temperature to remove excess antigen. For elution of selected scFv genes, the peptide portion of the antigen-bound molecules was digested with proteinase K (Takara) in TE buffer (10 mM Tris–HCl, pH 7.6 and 1 mM EDTA) for 30 min at 37°C. The eluate was recovered with a 0.22 µm Ultrafree-MC filter (Millipore) and purified with the RNeasy mini kit. The purified mRNA was reverse-transcribed with ReverTra Ace reverse-transcriptase (Toyobo) using a FlaA-R primer for 1 h at 42°C. Subsequently, the RT product was directly amplified by PCR with KOD-plus DNA polymerase using W29ATG-F and FlaA-R primers (24 cycles at 96°C for 30 s; at 58°C for 30 s; and at 72°C for 1 min). The PCR product was purified using a QIAquick PCR purification kit (Qiagen).
Cloning and sequencing
The selected DNAs after four rounds were cloned using a TOPO TA cloning kit (Invitrogen) and sequenced with the ABI PRISM 3100 genetic analyzer. Subsequently, a signal sequence (pelB) for periplasmic secretion was added to each scFv gene by overlap extension PCR with KOD-plus DNA polymerase using 0.4 µM primers pelB-F and Flag-R and a 0.02 µM primer pelBleader-F (15 cycles at 96°C for 30 s; at 58°C for 30 s; and at 72°C for 1 min). The PCR products were purified using the QIAquick PCR purification kit and subcloned using a Champion pET directional TOPO expression kit (Invitrogen).
Overexpression and purification
The BL21star(DE3)pLysS cells harboring the vector pET101/D-TOPO for the expression of scFv mutants with a C-terminal hexahistidine tag were grown in 1.5 liters of TB medium containing 100 µg/ml ampicillin at 30°C. When the culture attained an optical density of 0.8 at 600 nm, the cells were induced by the addition of isopropylthio-β-d-galactoside (final 1 mM). After an additional 3 h incubation, the cells were harvested by centrifugation and resuspended in 15 ml/g cell of osmotic buffer (30 mM Tris–HCl, pH 7.6, 1 mM EDTA and 0.5 mM sucrose). Again, the cells were harvested by centrifugation and resuspended in 5 ml/g cell of ice-cold Milli-Q water. The centrifuged supernatant was recovered and further centrifuged for 20 min at 15 000 g to separate an insoluble fraction. The supernatant was recovered as the periplasmic extract for competitive ELISA analysis. The protein concentration was evaluated by SDS–PAGE and Western blot analysis using HRP-conjugated anti-FLAG M2 antibody (Sigma).
For purification of proteins, the cells were grown as described above, harvested by centrifugation, and resuspended in 30 ml of TBS containing 100 U of DNase I (Promega), the EDTA-free protease inhibitor cocktail (Nacarai tesque) and 20% glycerol. The cells were lysed by sonication using a Bioruptor UCW-201 (Cosmo Bio) for 50 min at 30 s intervals. The resulting crude extracts were centrifuged for 20 min at 15 000 g and filtered through 0.22 µm PVDF Millex-GV filters (Millipore). The clear solution containing the histidine-tagged scFv protein was purified by immobilized metal affinity chromatography (IMAC) using TALON superflow metal affinity resin (Clontech) on an ÄKTA-FPLC system (Amersham Biosciences) with running buffer (10 mM HEPES–NaOH, pH 7.4, 150 mM NaCl and 20 mM imidazole) at a flow rate of 0.5 ml/min. The eluate obtained with a step gradient to 80 mM imidazole in running buffer was concentrated in a 30 000 MWCO PES Viva-spin 500 (Viva Science). Subsequently, the monomeric scFv was separated by size exclusion chromatography using Sephadex G-75 (Amersham Biosciences) with HBS buffer (10 mM HEPES–NaOH, pH 7.4 and 150 mM NaCl) at a flow rate of 0.2 ml/min, and then the buffer was changed to HBS-EP buffer (10 mM HEPES–NaOH, pH 7.4, 150 mM NaCl, 3 mM EDTA and 0.005% Tween-20) by using a MicroSpin G-25 column (Amersham Biosciences). The concentration and purity of scFv were confirmed by BCA assay using a Micro BCA protein assay kit (Pierce) and by CBB staining of SDS–PAGE gel, respectively.
Competitive ELISA
Streptavidin transparent C8 plates (Nunc) were incubated with 1 µg/ml fluorescein–biotin (Sigma) in 100 µl of blocking solution (Roche) per well for 1 h at 25°C, washed with TBST (TBS, pH 7.4 and 0.2% Tween-20), and blocked with 200 µl of blocking solution per well before use. Separately, the scFv-expressed periplasmic extract or wheat germ cell-free translated product was diluted 5-fold in blocking solution in the presence and absence of free fluorescein (10–1000 nM), and incubated for 1 h at 25°C. Then, the protein solution (100 µl) was transferred into wells on a plate with immobilized antigen and the plate was incubated for 15 min at 25°C. It was immediately washed five times with TBST and further incubated for 1 h with HRP-conjugated anti-FLAG M2 antibody (1:10 000 dilution in TBST; 100 µl per well). The plate was washed five times with TBST and then 100 µl of TMB substrate (Nacarai tesque) was added per well. The reaction was quenched with 1 N H2SO4 (100 µl per well) and the absorbance at 450 nm (reference wavelength at 655 nm) was measured. For each clone, the relative binding activity was calculated as the ratio of the ELISA signal in the presence of competitor (fluorescein) to that in the absence of competitor. All the experiments were performed in duplicate.
For the analysis of antigen-specificity, competitive ELISA was performed as described above, except for the use of various concentrations of fluorescein, Oregon green 488 carboxylic acid (Molecular Probes) or rhodamine 110 (Molecular Probes) as the competitor.
Surface plasmon resonance (SPR) spectroscopy
All the experiments were performed in duplicate at 25°C with a Biacore 3000 instrument (Biacore) using HBS-EP buffer. Fluorescein-conjugated BSA (BSA-FITC; Molecular Probes) or BSA (Pierce) as a background control was covalently immobilized onto a CM5 sensor chip (Biacore) by means of the amine coupling procedure according to the standard protocol. To prevent the rebinding of analytes to ligands immobilized on the CM5 sensor chip and to reduce themass transport limitation effect, the measurements were performed under conditions of low ligand amount (450 resonance units) and high flow rate (60 µl/min). To determine dissociation constants, five different concentrations (8–400 nM) of the purified scFvs were injected. The injection periods for association and dissociation were 1 and 10 min, respectively. The surface of the sensor chip was regenerated by pulse injection of 60 µl of 57 mM HCl (pH 1.2). The binding data were analyzed with the ‘1:1 binding with mass transfer’ model in the BIAevaluation software ver. 4.1 (Biacore).
RESULTS AND DISCUSSION
Strategy for in vitro selection of scFv by mRNA display
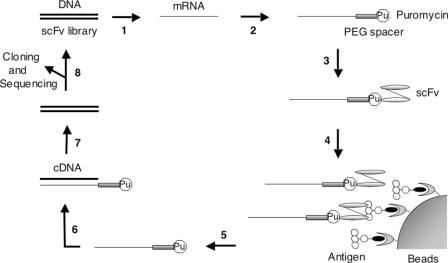
The scheme for in vitro selection of anti-fluorescein scFv using the IVV method is shown in Figure 1. First we constructed the template DNA for IVV, containing an SP6 promoter and Ω enhancer upstream of the anti-fluorescein scFv gene (VH-linker-VL), and the FLAG-tag and poly(A) sequence in the downstream region. The scFv region was subjected to random mutagenesis by error-prone PCR (the error rate was 0.8%; 6–7 nt per scFv gene). The transcribed mRNA (1.2 × 1012) molecules were attached to a PEG spacer containing puromycin at the 3′ end and in vitro-translated in 10 µl of wheat germ cell-free translation system, resulting in the covalent bonding of the C-terminus of the full-length scFv protein (phenotype) with its coding mRNA (genotype) through the puromycin-PEG spacer. We estimated that the number of resultant mRNA-displayed scFv molecules in the initial library was 6 × 1010 (ligation efficiency of mRNA with the puromycin-PEG spacer and efficiency of IVV formation were 50 and 10%, respectively). The library was captured on fluorescein-immobilized beads and the mRNA portion of selected molecules was amplified by RT–PCR. In each round of selection, the recovered DNA was further mutagenized by error-prone DNA shuffling (26,29).
Figure 1.
Schematic representation of the in vitro selection of scFv antibodies by the IVV method. Step 1: a DNA library of scFv mutants is in vitro-transcribed into mRNA. Step 2: the mRNA is enzymatically ligated with puromycin (Pu) through a PEG spacer. Step 3: the ligated product is in vitro-translated by a cell-free translation system, resulting in covalent bonding of puromycin with the full-length protein at the C-terminus. Step 4: the mRNA-displayed scFv library is captured on antigen-immobilized beads and unbound molecules are washed away. Step 5: the bound molecules are eluted by protease digestion. Step 6: the mRNA portion of eluted molecules is reverse-transcribed into cDNA. Step 7: the cDNA is amplified by PCR. Step 8: the DNA can be used for the next round of selection after mutagenesis or can be analyzed by cloning and sequencing after several rounds of selection.
Off-rate selection of the randomized scFv library
We applied off-rate selection (10,14) as a selection pressure for affinity maturation of anti-fluorescein scFv. In off-rate selection, mRNA-displayed scFvs that bound to fluorescein-immobilized beads were washed with a large excess of free fluorescein to prevent the rebinding of antibodies to the beads. Since the off-rate (koff) depends on the half-life (τ1/2) of the antigen–antibody complex, higher-affinity binders with low koff can be retained on the beads for a longer washing time. We set the time periods of off-rate selection so as to decrease koff about several-fold per round (3, 20, 96 and 336 h). Although mRNA is considered to be a labile molecule, it is stable for at least 2 weeks at low temperature (14).
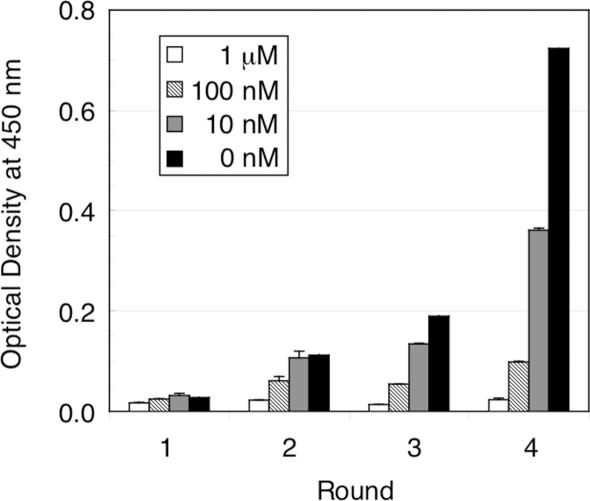
After four rounds of off-rate selection, the total binding activity of in vitro-translated products of the library DNA at each round was analyzed by competitive ELISA. As shown in Figure 2, the binding activity gradually increased in successive rounds of selection (Figure 2; 0 nM), whereas little or no activity was observed in the presence of competitor (Figure 2; 10 nM, 100 nM and 1 µM), indicating the enrichment of specific binders with high affinity in the library.
Figure 2.
Competitive ELISA for monitoring the fraction of the mutant scFv library that bound to fluorescein at each round of selection. The wheat germ cell-free translated products containing mutant scFv libraries were pre-incubated with a competitor (0–1000 nM free fluorescein) and allowed to bind to fluorescein-immobilized plates. After washing of the plates, remaining scFvs were detected by HRP-conjugated anti-FLAG M2 antibody and TMB substrate with absorbance measurements at 450 nm (reference at 655 nm). All measurements were performed in duplicate.
Competitive ELISA for the second screening
The library DNA from the fourth round of selection was cloned and sequenced. The rate of amino acid substitutions was 3.5%. Seventy-seven randomly chosen genes were subcloned into an Escherichia coli expression vector, their expression was induced, and the binding activity of scFv mutants in the periplasmic extracts was analyzed by ELISA. Clones, whose ELISA signal (OD at 450 nm) was <0.2, were defined as ELISA-negative and those clones, whose signal was in the range of 0.2–1.0, were defined as ELISA-positive. Out of the 77 clones, 45 clones (58%) were ELISA-positive. In SDS–PAGE analysis, all ELISA-positive clones exhibited protein expression in the periplasm, whereas 27 out of 32 ELISA-negative clones showed little or no expresssion in the periplasm (data not shown). Although the other five ELISA-negative clones were expressed in periplasm, three of them showed low ELISA signals (<0.2) and two had no binding activity (data not shown).
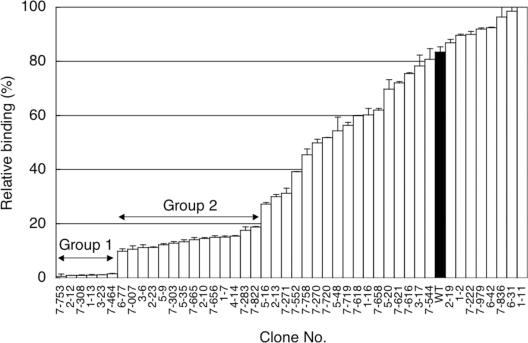
We estimated the affinity of the 45 ELISA-positive clones by competitive ELISA (Figure 3). The relative binding was defined as the ratio of the ELISA signal in the presence of free fluorescein as a competitor to that in the absence of the competitor. The lower relative binding of 37 clonesthan that of wild type indicates that these clones have higher affinity than the wild type. The six highest-affinity clones with relative binding of <5% and 14 clones with <20% were classed as Group 1 and Group 2, respectively. The amino acid changes are listed in Table 2. All six Group 1 mutants with the highest affinity contain five consensus mutations, W47C in FR H2, D101N in CDR H3, S15F in the linker, V21I in FR L1 and H94Y in CDR L3 (Table 2, boldface), whereas a majority of the 14 Group 2 mutants with moderate affinity contain four consensus mutations, A68V in FR H3, G96D in CDR H3, R18M in FR L1 and H94Y in CDR L3. Notably, the mutation H94Y in CDR L3 was contained in both Group 1 and Group 2 mutants. It is likely that the advantageous mutations were accumulated among the affinity-matured scFv genes during the repetition of DNA shuffling and off-rate selection in the selection cycles.
Figure 3.
Competitive ELISA for estimating the affinities of 45 selected clones after the fourth round of selection. The periplasmic extracts for wild type and mutant scFv clones were pre-incubated in the presence or absence of 10 nM fluorescein, and then allowed to bind to fluorescein-immobilized plates. After the plates were washed, remaining scFvs were detected by HRP-conjugated anti-FLAG M2 antibody and TMB substrate with absorbance measurements at 450 nm (reference at 655 nm). The relative binding indicates the ratio of the ELISA signal in the presence of the competitor to that in the absence of the competitor; thus, lower relative binding of clones compared with the wild-type scFv (WT; filled bar) indicates that these clones have higher affinity than the wild type. All the measurements were performed in duplicate.
Table 2.
Mutations in scFv fragments selected by the IVV method
| Clone | VH | VL | Linker |
|---|---|---|---|
| Group 1 | |||
| 7-753 | W47C, D101N | V21I, S76T, H94Y | S15F |
| 2-12 | W47C, A68T, K73Q, D101N | V21I, H94Y | S15F |
| 7-308 | W47C, D101N | V21I, A51V, H94Y | S15F |
| 1-13 | K19R, S21T, W47C, S74N, S76G, S82bG, E85D, D101N, Y102N, S112F | V21I, S80T, H94Y, E105K | G12S, S15F |
| 3-23 | K19R, W47C, S74N, Q81H, S82bG, D101N, Y102N | V21I, S80T, H94Y | G12S, S15F |
| 7-464 | W47C, K62N, K73I, D101N | V21I, Q38H, H94Y | S15F |
| Group 2 | |||
| 6-77 | G96D | M11L, P59S, H94Y | G19D |
| 7-007 | A68V, G96D, S112P | R18M, H94Y | |
| 3-6 | A68V, G96D | R18M, H94Y | S5L |
| 2-23 | A68V, G96D | R18M, Y53H, H94Y | |
| 5-9 | A68V, Q81R, D101N | V21I, D30H | S15F |
| 7-303 | A68V, G96D | T13I, R18M, D70S, H94Y, L104M | |
| 5-35 | A68V, G96D | R18M, P59H, S67Y, H94Y | |
| 7-665 | A68V, G96D | R18M, K24R, L83S, H94Y | S20I |
| 2-10 | A68V, G96D | R18M, H94Y | |
| 7-656 | S30T, M80L, Y99H | T22I, A84T, H94Y, K103M | S15P |
| 1-7 | N58I, G96D | I2V, V21I, N32K, Y53C, D60G | |
| 4-14 | Q3R, W47C, S76R, M80L, Y99N | H94Y | |
| 7-283 | W47C, S76R, M80L, Y99N | G68R, H94Y | |
| 7-822 | K64N, A68V, G96D | R18M, H94Y | G11C |
Amino acid mutations of the VH, VL and linker regions of selected mutants. Residue numbering is according to Kabat et al. (32). The original sequence of the wild type is shown in (14). Group 1 and Group 2 indicate 6 clones with high activity and 14 clones with moderate activity, respectively (see also Figure 3). Boldface residues indicate five consensus mutations.
Characterization of the affinity-matured mutants
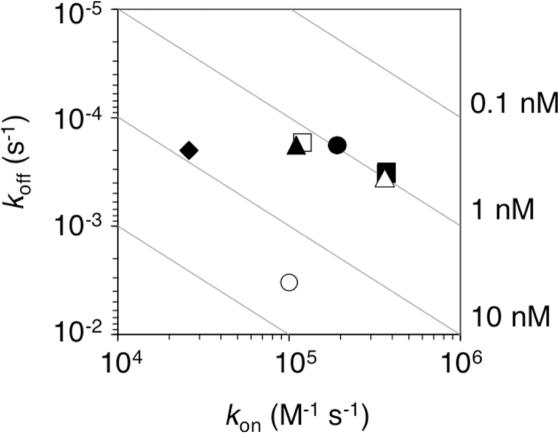
The binding affinities of purified scFvs (five Group 1 mutants and a Group 2 mutant, 2-10) were measured by SPR spectroscopy (Table 3 and Figure 4). Although the on-rates of the mutants were similar to that of the wild type, the off-rates were decreased by more than one order of magnitude, suggesting that the application of selection pressure by off-rate selection had been successful. The dissociation constants of Group 1 mutants were improved by ∼30-fold, which is comparable with that obtained by ribosome display (14), probably because the selection conditions were similar to each other.
Table 3.
Kinetic parameters of selected clones
| Clone | kon (M−1 s−1 × 105) | koff (s−1 × 10−4) | KD (nM) | τ1/2 (min) |
|---|---|---|---|---|
| Wild type | 1.0 ± 0.05* | 33 ± 0.62 | 32 | 3.5 |
| 2-12 | 1.9 ± 0.02 | 1.8 ± 0.007 | 0.95 | 64 |
| 3-23 | 3.7 ± 0.06 | 3.3 ± 0.17 | 0.88 | 35 |
| 7-308 | 3.6 ± 0.02 | 3.6 ± 0.01 | 0.99 | 33 |
| 7-464 | 1.2 ± 0.002 | 1.7 ± 0.009 | 1.4 | 70 |
| 7-753 | 1.1 ± 0.02 | 1.8 ± 0.008 | 1.7 | 63 |
| 2-10 | 0.26 ± 0.002 | 2.0 ± 0.18 | 7.5 | 59 |
*Mean ± SEM.
Figure 4.
Kinetic distribution plot. The kinetic parameters of purified scFvs were determined by SPR spectroscopy (see Materials and Methods and Table 3). The wild type (open circle); Group 1 mutants 2-12 (filled circle), 7-753 (open square), 3-23 (filled square), 7-308 (open triangle) and 7-464 (filled triangle); and Group 2 mutant 2-10 (filled diamond). The dissociation constants (Kd = koff/kon) are represented as gray lines (0.1–10 nM). Group 1 mutants with highest affinity were clustered in the upper right-hand corner of the plot.
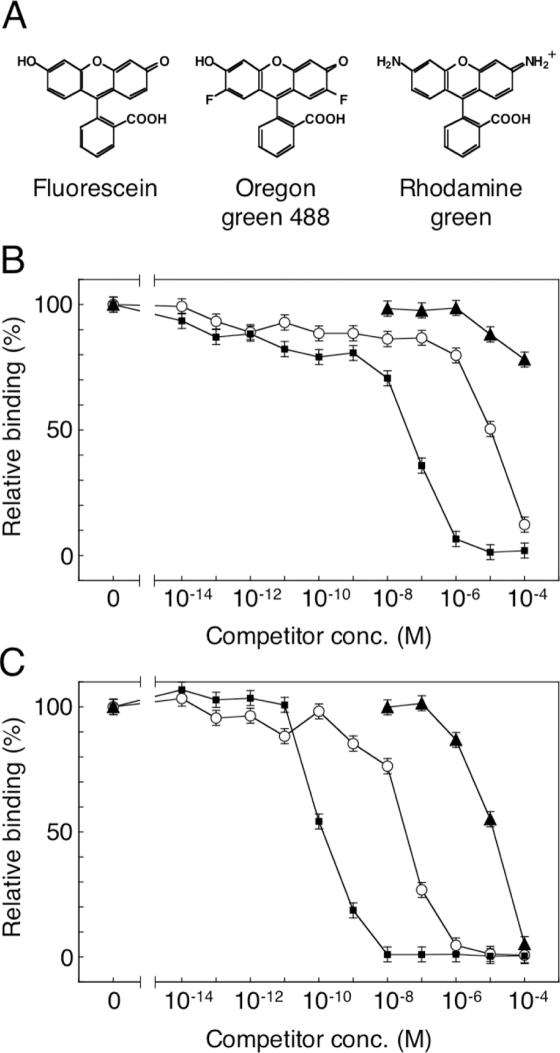
Next, we examined the antigen-specificity of the affinity-matured mutants by competitive ELISA. Two fluorescein analogs, Oregon green 488 and rhodamine green, as well as fluorescein were used as competitors (Figure 5A). The affinity of Group 1 mutant 2-12 (Figure 5C) was higher than that of wild type (Figure 5B), not only for fluorescein but also for the two fluorescein analogs, with an almost identical ratios of increase. Similar results were obtained for the other five Group 1 mutants (data not shown). The results suggest that the specificity of the affinity-matured mutants was not altered, but remained similar to that of the wild type.
Figure 5.
Competitive ELISA for estimating the antigen-specificity of the affinity-matured mutants. (A) The structures of fluorescein, Oregon green 488 and rhodamine green. For immobilization on the ELISA plates, biotin is attached to the benzene moiety of fluorescein; thus, differences of the xanthene moiety would be critical for scFv binding. The periplasmic extracts of the wild type (B) and a typical mutant 2-12 (C) were pre-incubated in the presence of various concentrations of fluorescein (filled squares), Oregon green 488 (open circles) or rhodamine green (filled triangles). The other five mutants in Group 1 gave similar curves. For experimental details, see the legend of Figure 3 and Materials and Methods.
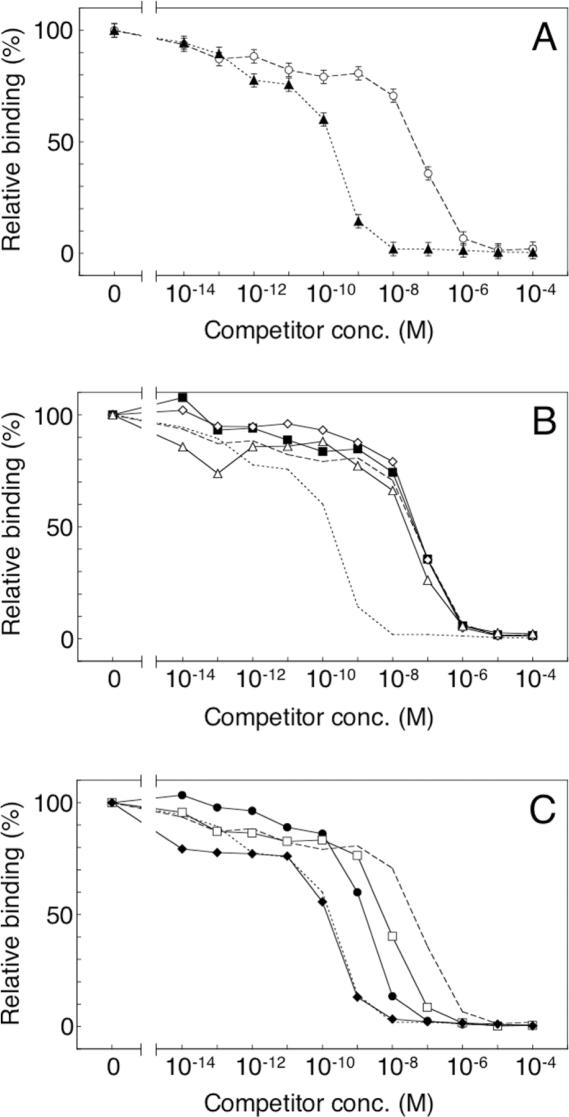
To determine the most important mutations for affinity improvement, we first constructed an scFv mutant that contains only the five consensus mutations in Group 1 mutants (named CM5). As shown in Figure 6A (filled triangles), the activity of the CM5 mutant was similar to that of Group 1 mutants (such as 2-12 shown in Figure 5C, filled squares), indicating that only the five mutations in Group 1 contribute significantly to the increased affinity. Subsequently, we constructed and examined five scFv mutants, each with a single amino acid mutation, W47C, D101N, S15F, V21I or H94Y. Three mutations, W47C and V21I in the framework and S15F in the linker, did not influence binding activity (Figure 6B), whereas two, D101N or H94Y in the CDR, resulted in intermediate binding activity between CM5 and wild type (Figure 6C). Moreover, the double mutant containing both D101N and H94Y showed binding activity similar to that of CM5 (Figure 6C, filled diamonds). These results clearly demonstrated that the critical mutations for improving affinity are D101N and H94Y in CDR3 of VH and VL, respectively. This result is consistent with the previous findings in laboratory evolution of high-affinity scFv mutants with CDR mutations (11,14). The locations of the CDR mutations on the three-dimensional structure were very close to the fluorescein site (14). It has been argued that the mutation H94Y in VL directly affects the interaction with the antigen in the antigen-binding site, and that mutations D101G, D101A and D101S in VH break the salt bridge between D101 and R94 in VH and increase the flexibility of CDR H3 (14). Our results suggest that the mutation D101N in VH also affects the affinity via a mechanism similar to that of the mutations D101G, D101A and D101S. The framework mutations W47C in VH and V21I in VL, as well as the linker mutation S15F, may be neutral mutations fixed in an early round of selection by random genetic drift.
Figure 6.
Competitive ELISA for analysis of the contributions of the five consensus mutations in Group 1 mutants. The periplasmic extracts of the wild-type and mutant scFvs were pre-incubated in the presence of various concentrations of fluorescein. For experimental details, see the legend of Figure 3 and Materials and Methods. (A) The wild type (open circles, dashed line) and the mutant CM5 (filled triangles, dotted line). (B) The single mutants W47C (filled squares), V21I (open diamonds) and S15F (open triangles); the wild type (a dashed line) and CM5 (a dotted line). (C) The single mutants D101N (filled circles) and H94Y (open squares); the double mutant D101N/H94Y (filled diamonds); the wild type (dashed line) and CM5 (dotted line).
In summary, completely in vitro evolution of a single-chain antibody by using the IVV mRNA display method was accomplished for the first time. By means of the combination of off-rate selection and DNA shuffling, the affinity of scFv mutants was improved ∼30-fold without affecting the specificity. The critical mutations D101N in VH and H94Y in VL located within the CDRs are similar to those previously found by ribosome display (14). Although both mRNA display and ribosome display are totally in vitro selection systems that allow the selection of antibodies against novel antigens, including toxic molecules, from a huge library, only fewer applications of mRNA display than that of ribosome display have been reported so far (30). Our results demonstrate that mRNA display can be expected to be useful for rapid and efficient evolution of high-affinity antibodies for diagnostic and therapeutic applications by optimizing the CDRs. Furthermore, in comparison with ribosome display, the use of a covalent bond in mRNA display should allow in vitro selection of antibodies with high stability at a higher temperature or in the presence of denaturants of ribosomes, as well as the selection of metal-dependent antibodies (31) in the presence and absence of EDTA. We believe that this method should also be applicable to in vitro selection of scFv antibodies directed against various antigens fromnaive or synthetic scFv libraries.
Acknowledgments
The authors thank Nobutaka Matsumura, Michiko Onimaru, Toru Tsuji, Masamichi Ishizaka, Seiji Tateyama and Kenichi Horisawa for experimental advice and useful discussions. This research was supported in part by the Industrial Technology Research Grant Program in 2005 from the NEDO of Japan, and by a Grant-in-Aid for Scientific Research and a Special Coordination Fund grant from the MEXT of Japan. Funding to pay the Open Access publication charges for this article was provided by Keio University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hoogenboom H.R. Selecting and screening recombinant antibody libraries. Nat. Biotechnol. 2005;23:1105–1116. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- 2.Holliger P., Hudson P.J. Engineered antibody fragments and the rise of single domains. Nat. Biotechnol. 2005;23:1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 3.Wu A.M., Senter P.D. Arming antibodies: prospects and challenges for immunoconjugates. Nat. Biotechnol. 2005;23:1137–1146. doi: 10.1038/nbt1141. [DOI] [PubMed] [Google Scholar]
- 4.Adams G.P., Weiner L.M. Monoclonal antibody therapy of cancer. Nat. Biotechnol. 2005;23:1147–1157. doi: 10.1038/nbt1137. [DOI] [PubMed] [Google Scholar]
- 5.French D.L., Laskov R., Scharff M.D. The role of somatic hypermutation in the generation of antibody diversity. Science. 1989;244:1152–1157. doi: 10.1126/science.2658060. [DOI] [PubMed] [Google Scholar]
- 6.Nossal G.J.V. The molecular and cellular basis of affinity maturation in the antibody response. Cell. 1992;68:1–2. doi: 10.1016/0092-8674(92)90198-l. [DOI] [PubMed] [Google Scholar]
- 7.Berek C. Somatic mutation and memory. Curr. Opin. Immunol. 1993;5:218–222. doi: 10.1016/0952-7915(93)90007-f. [DOI] [PubMed] [Google Scholar]
- 8.Marks J.D., Hoogenboom H.R., Griffiths A.D., Winter G. Molecular evolution of proteins on filamentous phage: mimicking the strategy of the immune system. J. Biol. Chem. 1992;267:16007–16010. [PubMed] [Google Scholar]
- 9.Irving R.A., Coia G., Roberts A., Nuttall S.D., Hudson P.J. Ribosome display and affinity maturation: from antibodies to single V-domains and steps towards cancer therapeutics. J. Immunol. Methods. 2001;248:31–45. doi: 10.1016/s0022-1759(00)00341-0. [DOI] [PubMed] [Google Scholar]
- 10.Hawkins R.E., Russell S.J., Winter G. Selection of phage antibodies by binding affinity: mimicking affinity maturation. J. Mol. Biol. 1992;226:889–896. doi: 10.1016/0022-2836(92)90639-2. [DOI] [PubMed] [Google Scholar]
- 11.Boder E.T., Midelfort K.S., Wittrup K.D. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc. Natl Acad. Sci. USA. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanes J., Plückthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl Acad. Sci. USA. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He M., Taussig M.J. Antibody–ribosome–mRNA (ARM) complexes as efficient selection particles for in vitro display and evolution of antibody combining sites. Nucleic Acids Res. 1997;25:5132–5134. doi: 10.1093/nar/25.24.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jermutus L., Honegger A., Schwesinger F., Hanes J., Plückthun A. Tailoring in vitro evolution for protein affinity and stability. Proc. Natl Acad. Sci. USA. 2001;98:75–80. doi: 10.1073/pnas.011311398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zahnd C., Spinelli S., Luginbuhl B., Amstutz P., Cambillau C., Plückthun A. Directed in vitro evolution and crystallographic analysis of a peptide-binding single chain antibody fragment (scFv) with low picomolar affinity. J. Biol. Chem. 2004;279:18870–18877. doi: 10.1074/jbc.M309169200. [DOI] [PubMed] [Google Scholar]
- 16.Reiersen H., Løbersli I., Løset G.Å., Hvattum E., Simonsen B., Stacy J.E., McGregor D., Fitzgerald K., Welschof M., Brekke O.H., et al. Covalent antibody display: an in vitro antibody-DNA library selection system. Nucleic Acids Res. 2005;33:e10. doi: 10.1093/nar/gni010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nemoto N., Miyamoto-Sato E., Husimi Y., Yanagawa H. In vitro virus: bonding of mRNA bearing puromycin at the 3′-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Lett. 1997;414:405–408. doi: 10.1016/s0014-5793(97)01026-0. [DOI] [PubMed] [Google Scholar]
- 18.Miyamoto-Sato E., Nemoto N., Kobayashi K., Yanagawa H. Specific bonding of puromycin to full-length protein at the carboxyl terminus. Nucleic Acids Res. 2000;28:1176–1182. doi: 10.1093/nar/28.5.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyamoto-Sato E., Takashima H., Fuse S., Sue K., Ishizaka M., Tateyama S., Horisawa K., Sawasaki T., Endo Y., Yanagawa H. Highly stable and efficient mRNA templates for mRNA–protein fusions and C-terminally labeled proteins. Nucleic Acids Res. 2003;31:e78. doi: 10.1093/nar/gng078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu L., Aha P., Gu K., Kuimelis R.G., Kurz M., Lam T., Lim A.C., Liu H., Lohse P.A., Sun L., et al. Directed evolution of high-affinity antibody mimics using mRNA display. Chem. Biol. 2002;8:933–942. doi: 10.1016/s1074-5521(02)00187-4. [DOI] [PubMed] [Google Scholar]
- 21.Parker M.H., Chen Y., Danehy F., Dufu K., Ekstrom J., Getmanova E., Gokemeijer J., Xu L., Lipovsek D. Antibody mimics based on human fibronectin type three domain engineered for thermostability and high-affinity binding to vascular endothelial growth factor receptor two. Protein Eng. Des. Sel. 2005;9:435–444. doi: 10.1093/protein/gzi050. [DOI] [PubMed] [Google Scholar]
- 22.Roberts R.W., Szostak J.W. RNA–peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl Acad. Sci. USA. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitlow M., Howard A.J., Wood J.F., Voss E.W., Jr, Hardman K.D. 1.85 Å structure of anti-fluorescein 4-4-20 Fab. Protein Eng. 1995;8:749–761. doi: 10.1093/protein/8.8.749. [DOI] [PubMed] [Google Scholar]
- 24.Schwesinger F., Ros R., Strunz T., Anselmetti D., Güntherodt H.-J., Honegger A., Jermutus L., Tiefenauer L., Plückthun A. Unbinding forces of single antibody–antigen complexes correlate with their thermal dissociation rates. Proc. Natl Acad. Sci. USA. 2000;97:9972–9977. doi: 10.1073/pnas.97.18.9972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doi N., Takashima H., Kinjo M., Sakata K., Kawahashi Y., Oishi Y., Oyama R., Miyamoto-Sato E., Sawasaki T., Endo Y., et al. Novel fluorescence labeling and high-throughput assay technologies for in vitro analysis of protein interactions. Genome Res. 2002;12:487–492. doi: 10.1101/gr.218802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao H., Giver L., Shao Z., Affholter J.A., Arnold F.H. Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat. Biotechnol. 1998;16:258–261. doi: 10.1038/nbt0398-258. [DOI] [PubMed] [Google Scholar]
- 27.Horisawa K., Tateyama S., Ishizaka M., Matsumura N., Takashima H., Miyamoto-Sato E., Doi N., Yanagawa H. In vitro selection of Jun-associated proteins using mRNA display. Nucleic Acids Res. 2004;32:e169. doi: 10.1093/nar/gnh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyamoto-Sato E., Ishizaka M., Horisawa K., Tateyama S., Takashima H., Fuse S., Sue K., Hirai N., Masuoka K., Yanagawa H. Cell-free cotranslation and selection using in vitro virus for high-throughput analysis of protein–protein interactions and complexes. Genome Res. 2005;15:710–717. doi: 10.1101/gr.3510505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stemmer W.P. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 30.Lipovsek D., Plückthun A. In vitro protein evolution by ribosome display and mRNA display. J. Immunol. Methods. 2004;290:51–67. doi: 10.1016/j.jim.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 31.Blake D.A., Chakrabarti P., Khosraviani M., Hatcher F.M., Westhoff C.M., Goebel P., Wylie D.E., Blake R.C., II Metal binding properties of a monoclonal antibody directed toward metal–chelate complexes. J. Biol. Chem. 1996;271:27677–27685. doi: 10.1074/jbc.271.44.27677. [DOI] [PubMed] [Google Scholar]
- 32.Kabat E.A., Wu T.T., Perry H.M., Gottesmann K.S., Foeller C. Sequences of Proteins of Immunological Interest. 5th edn. Bethesda, MD: National Institutes of Health Publication No. 91-3242; 1991. [Google Scholar]