Abstract
Antisense oligodeoxynucleotides (oligos) are widely used for functional studies of both prokaryotic and eukaryotic genes. However, the identification of effective target sites is a major issue in antisense applications. Here, we study a number of thermodynamic and structural parameters that may affect the potency of antisense inhibition. We develop a cell-free assay for rapid oligo screening. This assay is used for measuring the expression of Escherichia coli lacZ, the antisense target for experimental testing and validation. Based on a training set of 18 oligos, we found that structural accessibility predicted by local folding of the target mRNA is the most important predictor for antisense activity. This finding was further confirmed by a direct validation study. In this study, a set of 10 oligos was designed to target accessible sites, and another set of 10 oligos was selected to target inaccessible sites. Seven of the 10 oligos for accessible sites were found to be effective (>50% inhibition), but none of the oligos for inaccessible sites was effective. The difference in the antisense activity between the two sets of oligos was statistically significant. We also found that the predictability of antisense activity by target accessibility was greatly improved for oligos targeted to the regions upstream of the end of the active domain for β-galactosidase, the protein encoded by lacZ. The combination of the structure-based antisense design and extension of the lacZ assay to include gene fusions will be applicable to high-throughput gene functional screening, and to the identification of new drug targets in pathogenic microbes. Design tools are available through the Sfold Web server at http://sfold.wadsworth.org.
INTRODUCTION
Antisense oligodeoxynucleotides (oligos), usually ∼20 nt in length, can modulate gene expression by hybridizing with cognate RNAs at complementary target sites (1). The potential of antisense oligos for fast and specific gene inhibition makes them useful tools for functional genomics studies and drug target validation (2–4). The promise of antisense oligos for therapeutic development has also been demonstrated (5–8).
It has been shown that usually only a small proportion of antisense oligos are functional and even fewer oligos are potent (9–11). The identification of effective target sites is a major issue in antisense applications. Experimental approaches to addressing this problem include the ‘gene walk’ approach, use of random or semi-random oligo libraries, and use of combinatorial oligonucleotide arrays (12–15). For example, the ‘gene walk’ approach empirically tests a large number of oligos targeted to various regions of the target mRNA, with a typical low success rate of 2–5% (16). These experimental approaches can identify effective target sites; however, they are time-consuming and costly, and are not easily adaptable for application to a large number of targets.
A number of sequence motifs have been reported to be correlated with antisense activity (17,18). However, such motif correlations are not supported by results from other studies (19–21). In addition, the GC content has been found to be a poor predictor of hybridization intensity (22). The essential step of the antisense process is the hybridization between the antisense oligo and its target mRNA; this can be simply viewed as a two-step process of nucleation at an accessible (single-stranded, unstructured) site and elongation by a ‘zippering’ process (15). There is compelling experimental evidence that the likelihood of successful hybridization is greatly influenced by secondary structural features of the target RNA (15,23–25).
Computational approaches to target-site selection are usually based on identification of accessible regions via predicted secondary structure of the target RNA. The mfold software (26) has been widely used for this purpose (21,22,27,28), but with limited success (14,16). For RNA folding prediction, the mfold software is based on free energy minimization. In a radical departure from free energy minimization, a statistical sampling approach to RNA folding prediction and antisense application has demonstrated promise and advantages (3,29–31). In comparisons with minimum free energy (MFE) predictions, this method has been shown to make better predictions of alternative mRNA structures (30) and antisense efficacy (3), and it enables an improved representation of the probable population of mRNA structures (32). This approach is the focus of two recent reviews on RNA secondary structure prediction algorithms (31,33). In this study, we further explore the value of the structure sampling algorithm for the rational design of antisense oligos using a cell-free assay system that we developed for rapid oligo screening. This assay system is used to measure expression of the Escherichia coli lacZ gene, the antisense target for experimental testing and validation. Based on testing results for a training set and two test sets of oligos, we found that the structural accessibility of the target is the most important predictor for antisense activity. This finding suggests a structure-based rational antisense design for prokaryotic applications.
MATERIALS AND METHODS
In vitro gene expression assay
The lacZ gene was expressed by in vitro transcription/translation from a fused nfo promoter in pRPC179 (provided by R. Cunningham, University at Albany, Albany, NY), using the E.coli S30 Extract System for Circular DNA (Promega). pRPC179 includes the promoter sequence of nfo and the beginning 144 nt of its open reading frame (ORF), which was fused to lacZ ORF at 19 nt, with a 18 nt linker sequence. Thus, β-galactosidase was expressed as a fusion protein that includes 48 amino acids from the N-terminal of EndoIV protein encoded by nfo and 6 amino acids encoded by the linker sequence. In vitro transcription/translation reactions were performed for 2 h at 37°C according to Promega's recommendations, using 6 μM 20 nt unmodified oligo [Integrated DNA Technologies (IDT)] and 70 ng/μl of pRPC179 DNA. Reactions were stopped by chilling on ice for 10 min. An equal volume of 80 μM 5-acetylamino-di-β-d-galactopyranoside (FDG) (Molecular Probes), which is cleaved by β-galactosidase to produce fluorescein, was added to the in vitro reactions to enable measurement of the activity of β-galactosidase (34). All reactions were done in 96-well microtiter plates, in triplicate. Fluorescence was measured using a CytoFluor multi-well plate reader. The inhibition percentage for an oligo is calculated as:
Western blot analysis
β-galactosidase expression levels were also measured by western blot analysis. A total of 40% of the volume of each in vitro reaction mixture was electrophoresed on a 10% SDS–PAGE gel at 200 V for 45 min in a Tris–SDS buffer (25 mM Tris, 0.19 M glysine, 1 g/l SDS). Proteins were transferred to PVDF membrane (Millipore) using a Semi-dry Transfer Cell (BioRad) according to manufacturer's recommendations. Western analysis was performed using rabbit anti-β-galactosidase antibody (Abcam) at 1:5000 dilution. Bands were detected using a horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody and were visualized using Kodak BioMax MR film (Sigma). Band intensities were quantified by ImageQuant software (Molecular Dynamics).
Secondary structure prediction
Sfold, the software implementing the structure sampling algorithm (35), was used to generate probable structures for the lacZ mRNA. The typical sample size of 1000 structures was used, which has been shown to be sufficient to guarantee statistical reproducibility (30,32). Due to the tight coupling of prokaryotic transcription and translation, the folding of the mRNA is considered to be a local event, and local folding has been previously used in a structural study of bacterial genes (36). We focused our analysis using a local folding constraint of window width 50 nt. Under this constraint, bases separated in the sequence by >50 nt cannot form a pair. Alternative window widths and global folding were also considered.
Thermodynamic parameters
The potency of an antisense oligo can be potentially affected by the secondary structure of the target (25), the self-folding potential of the oligo, and the stability of the oligo:target duplex (37). We define these terms in the current context and compute the total free energy change for modeling the hybridization process.
ΔGdisruption is the energy cost for disrupting the mRNA structure so that the binding site becomes completely open (i.e. single-stranded). In light of the local folding nature of prokaryotic mRNAs, we consider a local disruption model, which assumes that the alteration of target structure due to oligo binding is local rather than global. Specifically, we assume that only the binding site is involved in structural alteration. Under this assumption, ΔGdisruption is the energy difference between the original mRNA structure and the new locally altered structure. For our statistical sample of structures, the energy of the original mRNA structure, ΔGbefore, can be represented by the average energy of the original sampled structures. The energy of the locally changed structure, ΔGafter, can be calculated by the average of all of the locally altered structures. We have ΔGdisruption = ΔGbefore − ΔGafter.
ΔGoligo is the self-folding energy of the antisense oligo. It is the energy of the MFE structure calculated by the mfold program. This term characterizes the potential of secondary structure formation for the antisense oligo. For short oligos of 20 nt in length, the MFE structure is a good representation of the rather small space of probable structures. Thus, we do not consider the sampling alternative for oligo folding to be appreciably advantageous.
ΔGhybrid is the energy gain due to the hybridization at the binding site. This parameter measures the affinity between the structure-free (i.e. completely single-stranded) antisense oligo and the structure-free target site. It is calculated as the sum of the stacking energies for the antisense oligo:target duplex (hybrid), with the penalty of an initiation energy:
where ΔGinitiation = 4.1 kcal/mol (28), and the sum is over DNA/RNA stacking energies (38) for the oligo:target duplex.
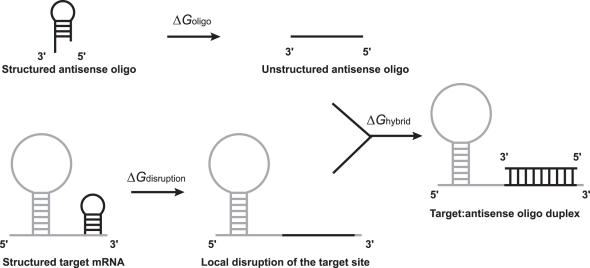
ΔGtotal is the total free energy change for the thermodynamic cycle of energy exchanges, as illustrated in Figure 1. It takes into account the intramolecular secondary structures of both the target and the antisense oligo, and the stability of the oligo:target hybrid. Thus, ΔGtotal is calculated by
Figure 1.
Thermodynamic cycle of free energy exchanges. ΔGdisruption is the target disruption energy, which measures the free energy cost to open the secondary structure at the target site; ΔGoligo is the self-folding minimal free energy (MFE) of the antisense oligo; ΔGhybrid is the binding energy (stability) for the hybrid formed between the antisense oligo and the target site.
Statistical analyses
Univariate linear regression and correlation analysis
The standard linear regression is employed for predicting antisense activity by each of the parameters listed above. The P-value measures the statistical significance of the parameter, and the R2 of the regression indicates the degree of variability in antisense activity that is attributed to the parameter. The correlation coefficient between a parameter and the antisense activity is computed. We note that the P- value of the correlation is the same as the P-value of the correlation is the same as the P-value of the parameter from the standard univariate regression analysis.
Multivariate regression by forward stepwise modeling
The R2 value may be substantially improved through inclusion of multiple parameters in a multivariate regression analysis. We adapt the forward stepwise method for model selection. At each step, a new and statistically significant parameter that most improves the R2 of the current model is included in the next regression model. This process is repeated either until all parameters are included in the final model, or until none of the excluded parameters can provide an appreciable improvement. The statistical package R (39) is used for the statistical analyses.
RESULTS
Development of in vitro assay
Evaluation of a substantial number of computationally designed antisense oligos requires an experimental system that can rapidly screen for oligo efficiency. We reasoned that an in vitro transcription/translation system would bypass problems associated with oligo entry into cells, and would provide an expression readout that directly corresponds to the accessibility and binding of the oligo to the mRNA target.
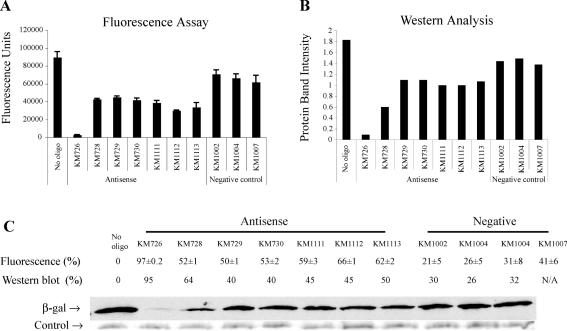
To establish such a highly sensitive in vitro assay, we started with a set of seven antisense oligos and three negative control oligos. In the fluorescence assay, the no-oligo control, which should represent the maximal expression of lacZ, gave the greatest number of fluorescence units (Figure 2A). The values in fluorescence units for the three negative control oligos were close to the value of the no-oligo control. All but one of the antisense oligos caused the number of fluorescence units to be reduced by about half, relative to the negative control oligos. The remaining antisense oligo, KM726, reduced the number of fluorescence units to 3% of the no-oligo control level, which indicated that the expression of lacZ, and consequently the activity of β-galactosidase, were almost completely inhibited by this oligo.
Figure 2.
Expression of β-galactosidase was quantified by both a FDG fluorescence assay and Western blot analysis. (A) Values in fluorescence units are shown for each reaction either with or without oligo, as specified. ‘No oligo’ was the control reaction without oligo. KM726, KM728, KM729, KM730, KM1111, KM1112 and KM1113 were designed antisense oligos. KM1002 and KM1004 were random negative controls. KM1007 was a scramble control of KM729. Results shown represent means of triplicate samples from one experiment. Every oligo was tested independently at least three times. Error bars show standard errors for multiple runs of the experiment. (B) Quantification of the Western blot. A total of 40% of the volume of each reaction was analyzed by Western blot. The vertical axis shows the relative intensity of each β-galactosidase protein band from the Western blot as quantified by ImageQuant 5.1 software. (C) Inhibition percentages for results in A (fluorescence) and in B (Western blot) are shown along with the corresponding protein bands from the Western blot. KM1008 was a mismatch control of KM726, which was tested in the fluorescence assay but not in the Western blot. The upper protein bands represent the β-galactosidase detected by chemiluminescence. The lower protein bands are from the Coomassie stained SDS–PAGE gel used for the Western blot assay, and serve as a uniform loading control.
The same reactions were next analyzed by Western blot using anti-β-galactosidase antibodies, a technique which more directly measures the amount of β-galactosidase protein produced in each reaction. The trend of the intensities of the β-galactosidase-specific bands produced in the presence of various oligos was very similar to what we observed for the results for the fluorescence units (Figure 2A and B). This agreement indicates that β-galactosidase activity assayed by FDG corresponds well with the actual amount of the protein in the reaction. Fluorescence units and the intensity of the western bands were both converted to inhibition percentages, with the no-oligo control used as the baseline (Figure 2C). The levels of inhibition calculated from the two assays were similar. Thus, we concluded that the fluorescence assay is reliable as a fast, alternative measure of relative β-galactosidase expression levels in this in vitro system.
Having established a reliable in vitro assay system, we then used the fluorescence-based in vitro β-galactosidase assay system to test a total of 38 oligos. In each experiment using standard reaction conditions, every oligo was tested with triplicate samples, and the average inhibition percentage was computed. Furthermore, the experiment for each oligo was repeated independently at least twice, on different dates.
Statistical analysis of a training oligo set
A training dataset of 18 antisense oligos for the E.coli lacZ mRNA (Table 1) was tested and analyzed. Six of the seven oligos in Figure 2, with the exception of KM1113, were included in the training dataset and were further tested. By univariate regression analysis, we found a significant correlation (at the 0.05 significance level) between ΔGdisruption and antisense activity (Table 2). The correlation for ΔGtotal is marginally significant (but insignificant at the 0.05 level). There is a lack of correlation for either ΔGoligo or ΔGhybrid. These findings indicate that the significance of ΔGtotal was due to the ΔGdisruption term in the calculation of ΔGtotal (see Materials and Methods). Thus, not surprisingly, multivariate stepwise regression did not result in a model with improved R2. For ΔGdisruption, a lack of significant correlation was observed when we used larger folding windows, e.g. with width of 75 nt (P-value of 0.31), 100 nt (P-value of 0.18), or 500 nt (P-value of 0.16). For the global folding option (i.e. with window width set equal to the length of the complete target sequence), the P-value was 0.32. These findings further support the choice of a window width of 50 nt for local folding of prokaryotic mRNAs. Among 10 sequence motifs that have been reported to be significantly correlated with antisense activity (18), the AAA motif is present in seven of the 18 training oligos, but was not significantly correlated with antisense activity (P-value of 0.39). None of the remaining nine motifs appeared in more than 3 oligos; we considered such low frequency of occurrence insufficient for analysis. Based on the results of the training oligos, our validation focuses on direct testing of an antisense design that utilizes ΔGdisruption.
Table 1.
Antisense oligos of the training set
| Oligo name | Target site position | Target sequence (5′→3′) | Inhibition (%) | Disruption energy (kcal/mol) | No. of test runs* |
|---|---|---|---|---|---|
| KM726 | 277–296 | AAACCCUGGCGUUACCCAAC | 89.7 ± 11.8 | −7.075 | 19 |
| KM1227 | 243–262 | CACUGGCCGUCGUUUUACAA | 59.1 ± 9.4 | −13.727 | 4 |
| KM1228 | 306–325 | UUGCAGCACAUCCCCCUUUC | 59.8 ± 10.5 | −7.009 | 4 |
| KM728 | 733–752 | UGAAUUUGACCUGAGCGCAU | 40.1 ± 15.6 | −10.3 | 12 |
| KM729 | 836–855 | CGGAUGAGCGGCAUUUUCCG | 48.9 ± 14.4 | −14.065 | 10 |
| KM730 | 1133–1152 | GCCGAAAUCCCGAAUCUCUA | 50.4 ± 12.4 | −8.065 | 8 |
| KM731 | 1357–1376 | GCUGAUGAAGCAGAACAACU | 27.0 ± 17.1 | −9.892 | 3 |
| KM734 | 2399–2418 | AACCUCAGUGUGACGCUCCC | 37.0 ± 9.2 | −7.495 | 3 |
| KM1110 | 514–533 | UGCGCCCAUCUACACCAACG | 39.4 ± 11.5 | −5.045 | 3 |
| KM1111 | 1468–1487 | UAUUGAAACCCACGGCAUGG | 53.4 ± 11.9 | −8.721 | 8 |
| KM1112 | 2262–2281 | UGAUUGAACUGCCUGAACUA | 58.8 ± 14.4 | −5.226 | 8 |
| KM1165 | 2928–2947 | GGCCGCAAGAAAACUAUCCC | 66.8 ± 7.8 | −11.258 | 2 |
| KM1159 | 322–341 | UUUCGCCAGCUGGCGUAAUA | 59.8 ± 4.2 | −6.953 | 2 |
| KM1160 | 674–693 | UUUCAUCUGUGGUGCAACGG | 60.2 ± 10.1 | −5.461 | 2 |
| KM1161 | 1299–1318 | AGCAUCAUCCUCUGCAUGGU | 39.8 ± 18.5 | −21.914 | 4 |
| KM1162 | 1818–1837 | GCCCGCUGAUCCUUUGCGAA | 53.4 ± 12.5 | −9.671 | 4 |
| KM1163 | 2793–2812 | ACCGGAUUGAUGGUAGUGGU | 23.1 ± 20.7 | −23.258 | 5 |
| KM1164 | 2818–2837 | GGCGAUUACCGUUGAUGUUG | 39.8 ± 15.0 | −15.113 | 4 |
*Number of independent tests performed. Each run of the test was done with triplicate samples. For KM726, KM728, KM729, KM730, KM111 and KM1112, inhibition data here include early runs presented in Figure 1 and additional runs after the Western blot analysis. For each run, an average inhibition percentage was computed for the triplicate samples. The standard error was calculated using data from multiple runs.
Table 2.
Correlation and univariate regression analyses for the training dataset*
| Parameter | Correlation coefficient | P-value | R2 |
|---|---|---|---|
| ΔGoligo | −0.08306 | 0.743156 | 0.0069 |
| ΔGdisruption | 0.474247 | 0.046766 | 0.224911 |
| ΔGhybrid | −0.03135 | 0.901733 | 0.000983 |
| ΔGtotal | −0.42346 | 0.079929 | 0.17932 |
*For standard univariate regression analysis, the P-value for a predictor is the same as that for the correlation coefficient.
A structure-based antisense design and validation
We first adapt two established empirical rules for antisense design. These include balanced base composition (40), and avoidance of four contiguous guanosine (G) residues which may cause potential non-specific effects by interacting with heparin-binding proteins (41). It is possible that the accessibility for some regions of the target mRNA is heavily influenced by tertiary folding of the target and by binding of proteins. Efficient modeling for RNA:protein interaction is beyond current computational means, even when the identity of an RNA-binding protein is known. To avoid potential regional bias due to these effects, we distributed oligos across the length of the mRNA sequence. Our structure-based design starts with implementation of the empirical rules as design filters:
For all possible target sites (here 20 nt in width) on lacZ mRNA, we remove sites with GC content below 40% or above 65%.
For the remaining sites from (1), we remove those that have four or more consecutive G's.
Sort the remaining sites from (2) by ΔGdisruption value, and select sites that have the most favorable ΔGdisruption values and are distributed evenly across the mRNA sequence.
For a test set of accessible sites, we selected 10 sites by the above design procedure. For the second test set of inaccessible sites, after steps (1) and (2) of the above design process, we selected 10 sites that have the most unfavorable ΔGdisruption values and are distributed evenly across the mRNA sequence. These two test sets of oligos (Tables 3–5) were tested under the same conditions and analyzed in the same way as for the antisense oligos of the training dataset. For the 20 antisense oligos combined from the two test sets, the ΔGdisruption parameter was again found to be significantly correlated with the antisense activity, with a P-value 0.00753 and a regression R2 of 0.335 (Table 6). The correlation of ΔGtotal was slightly less significant, and neither ΔGoligo nor ΔGhybrid showed a significant correlation. Thus, the pattern of significance parallels that for the training set, and also that for the combination of the training set and the two test sets (Table 7). Seven of the 10 oligos for accessible sites were found to be effective (>50% inhibition), but none of the oligos for inaccessible sites was effective. The average inhibition percentage was 56.0% for the accessible set, and 38.6% for the inaccessible set. The difference between the two sets, 17.4%, was statistically significant with a P-value 0.00502. These results further support the importance of effect of target secondary structure on antisense activity and validate the structure-based antisense design.
Table 3.
Test set 1 of antisense oligos with predicted accessible target sites
| Oligo name | Target site position | Target sequence (5′→3′) | Inhibition % | Disruption energy (kcal/mol) | No. of test runs |
|---|---|---|---|---|---|
| KM1425 | 369–388 | CCCAACAGUUGCGCAGCCUG | 63.1 ± 4.5 | −2.79 | 2 |
| KM1426 | 1007–1026 | UGGCAGGGUGAAACGCAGGU | 68.9 ± 5.1 | −7.61 | 2 |
| KM1427 | 1671–1690 | GCCCGGUGCAGUAUGAAGGC | 46.9 ± 5.7 | −7.62 | 2 |
| KM1428 | 1838–1857 | UACGCCCACGCGAUGGGUAA | 56.4 ± 4.8 | −6.31 | 2 |
| KM1429 | 1959–1978 | AUGAUGAAAACGGCAACCCG | 70.7 ± 10.3 | −3.08 | 2 |
| KM1430 | 2083–2102 | AGCAAAACACCAGCAGCAGU | 62.2 ± 18.2 | −3.79 | 3 |
| KM1431 | 2173–2192 | CGAGCUCCUGCACUGGAUGG | 24.2 ± 13.9 | −3.00 | 2 |
| KM1432 | 2332–2351 | ACCGAACGCGACCGCAUGGU | 68.8 ± 13.8 | −7.09 | 3 |
| KM1433 | 2548–2567 | ACAACUGCUGACGCCGCUGC | 43.2 ± 3.8 | −7.67 | 2 |
| KM1434 | 2839–2858 | AGUGGCGAGCGAUACACCGC | 55.6 ± 10.2 | −7.01 | 2 |
Table 5.
Non-overlapping of target sites for two test sets of oligos
| Oligo set | Separation (in nucleotide) between adjacent sites | |||
|---|---|---|---|---|
| Mean | Min | Max | Median | |
| Test set 1 | 254 | 70 | 644 | 147 |
| Test set 2 | 287 | 32 | 1090 | 104 |
| Test sets 1 and 2 combined | 127 | 14 | 433 | 89 |
Table 6.
Correlation and univariate regression analyses for 20 test sites
| Parameter | Correlation coefficient | P-value | R2 |
|---|---|---|---|
| ΔGoligo | −0.10046 | 0.673456 | 0.010092 |
| ΔGdisruption | 0.578547 | 0.007531 | 0.334716 |
| ΔGhybrid | −0.37592 | 0.102366 | 0.141318 |
| ΔGtotal | −0.57389 | 0.008148 | 0.329346 |
Table 7.
Correlation and univariate regression analyses for 20 test sites and 18 training sites
| Parameter | Correlation coefficient | P-value | R2 |
|---|---|---|---|
| ΔGoligo | −0.07624 | 0.649164 | 0.005812 |
| ΔGdisruption | 0.512524 | 0.001002 | 0.262681 |
| ΔGhybrid | −0.14917 | 0.371408 | 0.022251 |
| ΔGtotal | −0.49575 | 0.001551 | 0.245765 |
Greatly improved predictability of antisense activity for oligos with target sites upstream of the end of the active domain for β-galactosidase
We were interested in possible localization effects with respect to the active domain of β-galactosidase. This protein of 1023 amino acids folds into five sequential domains, with an α-complementation region at the amino terminus (42). The third domain contains the catalytic site, including the four residues Glu 461, Met 502, Tyr 503 and Glu 537 that are catalytically important (43–45).
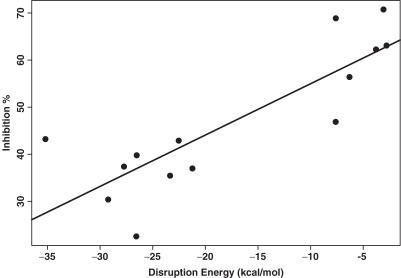
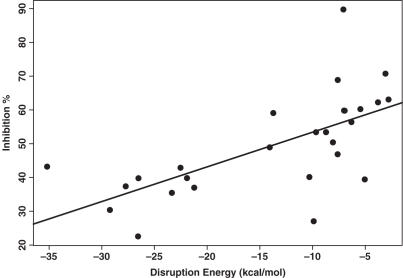
Six oligos of the accessible test set and eight oligos of the inaccessible test set have target sites located upstream of the end of the active domain. For these 14 oligos, the predictability of the antisense activity by ΔGdisruption improved greatly, with a P-value of 0.000168, and R2 of 0.706 (Table 8, Figures 3 and 4). For both the training set and the two test sets, there are 27 oligos that have target sites located within the first three domains. A substantial improvement was also observed for these 27 oligos (Table 9, Figure 5).
Table 8.
Correlation and univariate regression analyses for 14 oligos from test sets 1 and 2 having target sites within the first three domains of β-galactosidase
| Parameter | Correlation coefficient | P-value | R2 |
|---|---|---|---|
| ΔGoligo | −0.07688 | 0.793931 | 0.00591 |
| ΔGdisruption | 0.840307 | 0.000168 | 0.706116 |
| ΔGhybrid | −0.47544 | 0.085761 | 0.22604 |
| ΔGtotal | −0.83233 | 0.000221 | 0.692768 |
Figure 3.
Linear regression for prediction of the antisense activity (shown as inhibition % on the vertical axis) by ΔGdisruption for 14 oligos in the test sets that have target sites located upstream of the end of the active domain of β-galactosidase. The R2 for the regression is 0.706, and the P-value for ΔGdisruption is 0.000168.
Figure 4.
Five sequential domains I, II, III (active domain), IV, and V of β-galactosidase (PDB ID: 1DP0), with starting residue positions given (42). For each of the oligos in the two test sets, either a ‘+’ indicating agreement between prediction and observed inhibition, or a ‘−’ for disagreement is placed under the protein residue position corresponding to the target site on the lacZ mRNA. Here, prediction and observed inhibition are considered to be in agreement if either the oligo has the observed activity ≥50% and its ΔGdisruption ≥ −7 kcal/mol, or else the oligo has the observed activity < 50% and its ΔGdisruption < −20 kcal/mol. For the 20 oligos in the two test sets, agreement is observed for 12 of the 14 oligos (86%) targeted to domains I, II and III. In contrast, agreement is only observed for two of the six oligos (33%) targeted to domains IV and V.
Table 9.
Correlation and univariate regression analyses for 27 oligos from both the training and the test sets having target sites within the first three domains of β-galactosidase
| Parameter | Correlation coefficient | P-value | R2 |
|---|---|---|---|
| ΔGoligo | −0.11058 | 0.582935 | 0.012229 |
| ΔGdisruption | 0.651955 | 0.000229 | 0.425045 |
| ΔGhybrid | −0.21194 | 0.288565 | 0.044918 |
| ΔGtotal | −0.64089 | 0.000316 | 0.410745 |
Figure 5.
Linear regression for prediction of the antisense activity (shown as inhibition % on the vertical axis) by ΔGdisruption for 27 oligos in the training set and the two test sets that have target sites located upstream of the end of the active domain of β-galactosidase. The R2 for the regression is 0.425, and the P-value for ΔGdisruption is 0.000229.
It should be emphasized that antisense activity in this study was monitored through β-galactosidase activity. It has been shown that a fragment of β-galactosidase has the potential to fold into the correct conformation (46). An antisense oligo targeted to the fourth or the fifth domain could still give rise to a functional β-galactosidase that may lack one or both of the ending domains due to possible translation stoppage or target cleavage, as long as the third domain is intact and correctly folded. Since the third domain contains the active site, a partially synthesized β-galactosidase with this domain corrected folded can still retain its functionality and exhibit a negative antisense effect. Due to the limited understanding of protein folding, it is extremely difficult, if not impossible, to determine whether or not a partial β-galactosidase protein sequence can form the active conformation and perform its physiological functions. This uncertainty negatively affects the predictability of antisense activity at the protein level when the active domain is not known, and might explain why the predictive power of our method increases dramatically when the target sites downstream of the active domain were excluded in the analysis.
Computational costs and software availability
The CPU times and memory requirements for our implementation of partition function calculation and for the sampling of 1000 structures have been previously reported (47). The calculation of ΔGdisruption for each individual site is a linear operation. However, the calculation for all possible sites on a target mRNA becomes time-consuming. Thus, simple design filters including GC content are used to make computation by a web server manageable. The tools for implementing the structure-based design are available through the application module Soligo of the Sfold software for the folding and design of nucleic acids. Sfold is available through our Web server at http://sfold.wadsworth.org. Sample output for a folded sequence is available at http://sfold.wadsworth.org/demo.
DISCUSSION
Cell-free assay for antisense screening
The novel oligo assay system developed for this study has several advantages over common oligo screening strategies that are based on modified oligos, such as phosphorothioate oligos, morpholino oligos, or locked nucleic acid oligos. Vehicles for delivering such modified oligos into living cells include cationic lipids, liposomes, and peptide conjugation. Effects of the antisense oligos are usually monitored by Western blot analysis for protein expression, Northern blot analysis or RT PCR for mRNA expression, or the measurement of a specific phenotype associated with the expression of a target gene. The interpretation of data for these screening strategies can be complicated by the uncertainties associated with the experimental processes. The use of cell-free extracts is advantageous, because this allows addition of oligos directly to the reaction mixture and avoids problems associated with penetration of oligos through the cell membrane into the cytoplasm. Thus, the potency of an oligo is not affected by the permeability of the specific type of the oligo. Further, since oligos are present prior to mRNA synthesis and have ready access to the mRNA during transcription, background expression of the target gene prior to oligo addition is avoided. This represents an improvement over other assay systems in which the expression of the target before entry of oligos complicates the analysis. In addition, available mRNAs are limited to the target and other plasmid genes in our cell-free system, thereby reducing non-specific binding of oligos to other messages normally present in whole cells.
We observed consistent results with unmodified oligos in our cell-free assay, but we cannot rule out the possibility of oligo degradation in the reaction. Improved stability of oligos may increase the potency of antisense inhibition. The use of unmodified antisense oligos appears to contribute to the moderate level of activity for some of the tested oligos. It is well understood that unmodified oligos are susceptible to DNase degradation, which lowers their effective concentration and compromises activity. Indeed, phosphorothioate modification of several of the most potent antisense oligos from the test dataset led to significant improvement (preliminary data not shown). Thus, modified oligos will be considered for future in vivo experiments. We also tested non-specific effects often associated with high oligo concentrations. We found that the use of a concentration of 10 μM of negative control oligos increased the non-specific effects to unacceptable levels (data not shown).
The fluorescence-based assay of β-galactosidase activity was shown to be as reliable as Western blot analysis in these studies, and it is faster and much more amenable to high-throughput applications. The FDG assay takes 2 h and requires few manipulations, beyond set-up of the initial expression reactions. In contrast, Western analysis takes a full day, an entails many manipulations. Our fluorescence-based assay uses a 96-well-plate, which allowed us to routinely assay 31 oligos with a no-oligo control in triplicate, in 5 h. Thus, our fluorescence-based cell-free assay allows rapid and efficient screening of many antisense candidates to identify the best inhibitors. lacZ is an attractive target for initial testing, because it is characterized. The ∼3 kb length of its mRNA provides ample opportunities for identification of potent target sites. This lacZ reporter system can also be easily adapted for screening oligos against other targets.
Structure-based antisense design
In this work, we have computed energetic parameters for modeling oligo:target hybridization. In particular, the calculation of the target disruption energy ΔGdisruption takes advantage of our structure sampling algorithm that has been shown to effectively represent the likely population of mRNA structures, thus overcoming limited representation by the optimal folding or a heuristic set of suboptimal foldings for antisense target prediction. As a measure of target structural accessibility, ΔGdisruption was found to be the most important predictor for antisense activity. This finding is in agreement with previous reports based on experimental methods (15,25) or alternative computational approaches (21,22,48).
In our study, we selected non-overlapping test sites that are distributed evenly across the sequence of the target mRNA. This strategy not only avoids potential auto-correlation among overlapped sites which could introduce a bias for data analysis, but also ensures the independence of the sites, which is an implicit assumption for valid regression analysis. In a previous study, the oligo binding energy ΔGhybrid was found to be correlated with antisense activity (37).
Such a correlation was not found in our study. It is possible that the contradictory results are partly due to overlapping of some target sites in the previous study. However, the raw data in that study were not made available to enable us to assess the extent of overlap.
It has been shown that the secondary structure of antisense RNAs of ∼100 nt is important for the kinetics of the antisense activity (49). In our study, the stability of 20 nt antisense oligo was not correlated with antisense activity. This suggests that, for short oligos, the effect of oligo folding is too small to have a significant impact on antisense binding.
The window width of 50 nt was used by Sfold for local folding of lacZ mRNA. The significance of ΔGdisruption diminished for larger folding windows. This finding suggests that in prokaryotes, where transcription and translation are tightly coupled so that the mRNA is bound by multiple translational machineries, the folding of mRNA is likely constrained within local regions. Our observation is consistent with the local folding consideration in a structure study on bacterial mRNAs (36).
The proposed design method assumes that oligo:target hybridization causes only local disruption of the target structure. In view of the local nature of the folding of prokaryotic mRNAs, we consider the local disruption model to be a reasonable approximation for prokaryotic applications. For eukaryotes, mature mRNAs are transported from the nucleus into the cytoplasm where translation occurs and where regulatory mechanisms are present. Thus, the folding of a eukaryotic mRNA may be global. For eukaryotic applications, the alternative to the local disruption model is the global disruption model, which assumes that the binding of an oligo induces global alteration of target structure. This global model has been considered previously for modeling of antisense binding based on MFE predictions (22,48). The application of our structure sampling in this context will require re-folding of the target, with the binding site constrained to be unpaired, so as to generate a new sample of probable mRNA structures. The modified computing method will need to be tested in a eukaryotic system.
The combination of the computational design and the rapid cell-free assay renders our method applicable to high-throughput antisense applications. The lacZ system can be extended by gene fusion approaches (50,51). Thus, the approach we have outlined can be adapted for use with any gene of interest, and should be especially useful for functional screening of genes, and for identification of new drug targets in pathogenic microbes. We are currently developing functional genomics applications for Mycobacterium tuberculosis and Yersinia pestis.
The goal of an antisense experiment is to down-regulate the expression of the target protein. For the multiple-domain enzyme β-galactosidase, a partly synthesized peptide (in the case of successful antisense binding and target cleavage) has the potential to fold correctly (46) and behaves functionally as an intact protein. Thus, to maximize the degree of correspondence between antisense binding and the protein expression in bacterial functional genomics applications of the lacZ reporter, it is advisable to select target sites upstream of the end of the known active domain of the target protein. As indicated by the results for β-galactosidase, the antisense activity is less predictable for target sites downstream of the active domain. For these sites, data interpretation is further complicated by our currently limited understanding of protein folding. We emphasize that the finding of the regional effects with respect to the active domain of the target protein is based on one system tested for one target. Generalization of this finding will require large scale testing of other targets with known active domains.
The concentration of the target is relevant for bi-molecular interaction, but is unknown and thus is difficult to take into account in equilibrium equations of reactions. Also, it is unknown whether the target concentration in the cell-free system is comparable to that in an in vivo system. Clearly, the ratio of the oligo concentration and the target concentration is important. For the same oligo concentration, it is difficult to assess whether the above ratio for the cell-free system is comparable to that for an in vivo system. In the cell-free system, the oligos have ready access to the target, whereas inefficient uptake across the cell wall is expected to be an issue for an in vivo bacterial system (52). Thus, the effective oligo concentration for the cell-free system is expected to be higher than that for an in vivo system. In this case, the equilibrium between unbound oligo and unbound target, and oligo:target hybrid, may be shifted toward oligo:target hybridization, effectively reducing the effect of the target structure. In other words, for the same oligo concentration, the effect of target structure may be greater for an in vivo system in comparison to our cell-free system. Clearly, the effects of target structure and other parameters studied here need to be re-examined in an in vivo system. This will require large scale in vivo testing.
The target sites of all tested oligos are down stream from the translation initiation site. The initiation codon AUG starts at nucleotide position 74 of the lacZ mRNA, whereas the target site closest to AUG (oligo KM1227), starts at nucleotide position 243. Thus, blockage of the translation initiation complex (53) is unlikely to be the antisense mechanism for the oligos described in this study. Either translation arrest through blockage of the translational machinery, or RNAse associated RNA degradation, or both, are more likely mechanisms. RNase H, the ribonuclease that specifically degrades the RNA strand of DNA:RNA hybrid, is expected to be present in the cell extracts (Promega). More generally, members of RNase H family can be found in nearly all organisms including prokaryotes.
In a study of hybridization experiments with large numbers of oligo probes, oligo self-folding and the stability of the oligo:target hybrid, i.e. the ΔGhybrid here, were found to be significantly correlated with the hybridization intensity (54). Similar findings were reported for a large number of antisense experiments in eukaryotic systems (37). We could not confirm these findings in our test system. Our study measured prokaryotic gene expression rather than intensity of hybridization between interacting nucleic acids, or eukaryotic gene expression. In addition to these fundamental differences, for the oligos tested in our study, the GC content ranges from 40 to 65%, with an average of 55%. It is possible that the balanced GC content in our study does not provide sufficient variation in the stability of the oligo or the oligo:target duplex to allow for detection of correlation. The target structure was not considered in the analysis of probing data. For the other study, a heuristic set of suboptimal structures was used, but no correlation between target structure and antisense activity was found. The disruption energy based on a statistical sample of structures generated by global folding presents a novel tool for an alternative structural analysis of these data.
Table 4.
Test set 2 of antisense oligos with predicted inaccessible target sites
| Oligo name | Target site position | Target sequence (5′→3′) | Inhibition % | Disruption energy (kcal/mol) | No. of test runs |
|---|---|---|---|---|---|
| KM1435 | 403–422 | CUUUGCCUGGUUUCCGGCAC | 43.2 ± 14.1 | −35.19 | 2 |
| KM1436 | 856–875 | UGACGUCUCGUUGCUGCAUA | 37.0 ± 11.6 | −21.23 | 2 |
| KM1437 | 959–978 | CAGAUGUGCGGCGAGUUGCG | 37.4 ± 5.5 | −27.73 | 2 |
| KM1438 | 1232–1251 | CGGAUUGAAAAUGGUCUGCU | 42.9 ± 7.0 | −22.53 | 2 |
| KM1739 | 1329–1348 | AUGAGCAGACGAUGGUGCAG | 39.8 ±17.7 | −26.52 | 2 |
| KM1440 | 1438–1457 | CUACGGCCUGUAUGUGGUGG | 30.4 ± 26.3 | −29.23 | 3 |
| KM1441 | 1562–1581 | CGCGAUCGUAAUCACCCGAG | 22.6 ± 18.0 | −26.56 | 3 |
| KM1442 | 2012–2031 | CCGAACGAUCGCCAGUUCUG | 35.5 ± 14.4 | −23.35 | 2 |
| KM1443 | 3122–3141 | CAACUGAUGGAAACCAGCCA | 49.5 ± 3.0 | −26.88 | 2 |
| KM1444 | 3174–3193 | GGCUGAAUAUCGACGGUUUC | 47.6 ± 36.7 | −27.59 | 3 |
Acknowledgments
The Computational Molecular Biology and Statistics Core at the Wadsworth Center is acknowledged for providing computing resources for this work. The pRPC179 construct was provided by R. Cunningham of University at Albany. This work was supported in part by National Science Foundation grant DMS-0200970 and National Institutes of Health (NIH) grant R01 GM068726 to Y.D. Funding to pay the Open Access publication charges for this article was provided by NIH grant R01 GM068726.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zamecnik P.C., Stephenson M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl Acad. Sci. USA. 1978;75:280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor M.F., Wiederholt K., Sverdrup F. Antisense oligonucleotides: a systematic high-throughput approach to target validation and gene function determination. Drug Discov. Today. 1999;4:562–567. doi: 10.1016/s1359-6446(99)01392-6. [DOI] [PubMed] [Google Scholar]
- 3.Ding Y., Lawrence C.E. Statistical prediction of single-stranded regions in RNA secondary structure and application to predicting effective antisense target sites and beyond. Nucleic Acids Res. 2001;29:1034–1046. doi: 10.1093/nar/29.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Backer M.D., Nelissen B., Logghe M., Viaene J., Loonen I., Vandoninck S., de Hoogt R., Dewaele S., Simons F.A., Verhasselt P., et al. An antisense-based functional genomics approach for identification of genes critical for growth of Candida albicans. Nat. Biotechnol. 2001;19:235–241. doi: 10.1038/85677. [DOI] [PubMed] [Google Scholar]
- 5.Ball H.A., Sandrasagra A., Tang L., Van Scott M., Wild J., Nyce J.W. Clinical potential of respirable antisense oligonucleotides (RASONs) in asthma. Am J Pharmacogenomics. 2003;3:97–106. doi: 10.2165/00129785-200303020-00003. [DOI] [PubMed] [Google Scholar]
- 6.Waters J.S., Webb A., Cunningham D., Clarke P.A., Raynaud F., di Stefano F., Cotter F.E. Phase I clinical and pharmacokinetic study of bcl-2 antisense oligonucleotide therapy in patients with non-Hodgkin's lymphoma. J. Clin. Oncol. 2000;18:1812–1823. doi: 10.1200/JCO.2000.18.9.1812. [DOI] [PubMed] [Google Scholar]
- 7.Bowen-Yacyshyn M.B., Bennett C.F., Nation N., Rayner D., Yacyshyn B.R. Amelioration of chronic and spontaneous intestinal inflammation with an antisense oligonucleotide (ISIS 9125) to intracellular adhesion molecule-1 in the HLA-B27/beta2 microglobulin transgenic rat model. J. Pharmacol. Exp. Ther. 2002;302:908–917. doi: 10.1124/jpet.102.036053. [DOI] [PubMed] [Google Scholar]
- 8.Marwick C. First ‘antisense’ drug will treat CMV retinitis. JAMA. 1998;280:871. [PubMed] [Google Scholar]
- 9.Chiang M.Y., Chan H., Zounes M.A., Freier S.M., Lima W.F., Bennett C.F. Antisense oligonucleotides inhibit intercellular adhesion molecule 1 expression by two distinct mechanisms. J. Biol. Chem. 1991;266:18162–18171. [PubMed] [Google Scholar]
- 10.Dean N.M., McKay R., Condon T.P., Bennett C.F. Inhibition of protein kinase C-alpha expression in human A549 cells by antisense oligonucleotides inhibits induction of intercellular adhesion molecule 1 (ICAM-1) mRNA by phorbol esters. J. Biol. Chem. 1994;269:16416–16424. [PubMed] [Google Scholar]
- 11.Monia B.P., Johnston J.F., Geiger T., Muller M., Fabbro D. Antitumor activity of a phosphorothioate antisense oligodeoxynucleotide targeted against C-raf kinase. Nature Med. 1996;2:668–675. doi: 10.1038/nm0696-668. [DOI] [PubMed] [Google Scholar]
- 12.Allawi H.T., Dong F., Ip H.S., Neri B.P., Lyamichev V.I. Mapping of RNA accessible sites by extension of random oligonucleotide libraries with reverse transcriptase. RNA. 2001;7:314–327. doi: 10.1017/s1355838201001698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho S.P., Britton D.H., Stone B.A., Behrens D.L., Leffet L.M., Hobbs F.W., Miller J.A., Trainor G.L. Potent antisense oligonucleotides to the human multidrug resistance-1 mRNA are rationally selected by mapping RNA-accessible sites with oligonucleotide libraries. Nucleic Acids Res. 1996;24:1901–1907. doi: 10.1093/nar/24.10.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho S.P., Bao Y., Lesher T., Malhotra R., Ma L.Y., Fluharty S.J., Sakai R.R. Mapping of RNA accessible sites for antisense experiments with oligonucleotide libraries. Nat. Biotechnol. 1998;16:59–63. doi: 10.1038/nbt0198-59. [DOI] [PubMed] [Google Scholar]
- 15.Milner N., Mir K.U., Southern E.M. Selecting effective antisense reagents on combinatorial oligonucleotide arrays. Nat. Biotechnol. 1997;15:537–541. doi: 10.1038/nbt0697-537. [DOI] [PubMed] [Google Scholar]
- 16.Sohail M., Southern E.M. Selecting optimal antisense reagents. Adv. Drug Deliv. Rev. 2000;44:23–34. doi: 10.1016/s0169-409x(00)00081-8. [DOI] [PubMed] [Google Scholar]
- 17.Tu G.C., Cao Q.N., Zhou F., Israel Y. Tetranucleotide GGGA motif in primary RNA transcripts. Novel target site for antisense design. J. Biol. Chem. 1998;273:25125–25131. doi: 10.1074/jbc.273.39.25125. [DOI] [PubMed] [Google Scholar]
- 18.Matveeva O.V., Tsodikov A.D., Giddings M., Freier S.M., Wyatt J.R., Spiridonov A.N., Shabalina S.A., Gesteland R.F., Atkins J.F. Identification of sequence motifs in oligonucleotides whose presence is correlated with antisense activity. Nucleic Acids Res. 2000;28:2862–2865. doi: 10.1093/nar/28.15.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sohail M., Hochegger H., Klotzbucher A., Guellec R.L., Hunt T., Southern E.M. Antisense oligonucleotides selected by hybridisation to scanning arrays are effective reagents in vivo. Nucleic Acids Res. 2001;29:2041–2051. doi: 10.1093/nar/29.10.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lloyd B.H., Giles R.V., Spiller D.G., Grzybowski J., Tidd D.M., Sibson D.R. Determination of optimal sites of antisense oligonucleotide cleavage within TNFalpha mRNA. Nucleic Acids Res. 2001;29:3664–3673. doi: 10.1093/nar/29.17.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patzel V., Steidl U., Kronenwett R., Haas R., Sczakiel G. A theoretical approach to select effective antisense oligodeoxyribonucleotides at high statistical probability. Nucleic Acids Res. 1999;27:4328–4334. doi: 10.1093/nar/27.22.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walton S.P., Stephanopoulos G.N., Yarmush M.L., Roth C.M. Prediction of antisense oligonucleotide binding affinity to a structured RNA target. Biotechnol. Bioeng. 1999;65:1–9. [PubMed] [Google Scholar]
- 23.Lima W.F., Monia B.P., Ecker D.J., Freier S.M. Implication of RNA structure on antisense oligonucleotide hybridization kinetics. Biochemistry. 1992;31:12055–12061. doi: 10.1021/bi00163a013. [DOI] [PubMed] [Google Scholar]
- 24.Mir K.U., Southern E.M. Determining the influence of structure on hybridization using oligonucleotide arrays. Nat. Biotechnol. 1999;17:788–792. doi: 10.1038/11732. [DOI] [PubMed] [Google Scholar]
- 25.Vickers T.A., Wyatt J.R., Freier S.M. Effects of RNA secondary structure on cellular antisense activity. Nucleic Acids Res. 2000;28:1340–1347. doi: 10.1093/nar/28.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 28.Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 29.Ding Y., Lawrence C.E. A bayesian statistical algorithm for RNA secondary structure prediction. Comput. Chem. 1999;23:387–400. doi: 10.1016/s0097-8485(99)00010-8. [DOI] [PubMed] [Google Scholar]
- 30.Ding Y., Lawrence C.E. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 2003;31:7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Y. Statistical and Bayesian approaches to RNA secondary structure prediction. RNA. 2006;12:323–331. doi: 10.1261/rna.2274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding Y., Chan C.Y., Lawrence C.E. Clustering of RNA Secondary Structures with Application to Messenger RNAs. J. Mol. Biol. 2006;359:554–571. doi: 10.1016/j.jmb.2006.01.056. [DOI] [PubMed] [Google Scholar]
- 33.Mathews D.H. Revolutions in RNA secondary structure prediction. J. Mol. Biol. 2006;359:526–532. doi: 10.1016/j.jmb.2006.01.067. [DOI] [PubMed] [Google Scholar]
- 34.Rowland B., Purkayastha A., Monserrat C., Casart Y., Takiff H., McDonough K.A. Fluorescence-based detection of lacZ reporter gene expression in intact and viable bacteria including Mycobacterium species. FEMS Microbiol. Lett. 1999;179:317–325. doi: 10.1111/j.1574-6968.1999.tb08744.x. [DOI] [PubMed] [Google Scholar]
- 35.Ding Y., Chan C.Y., Lawrence C.E. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32:W135–141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katz L., Burge C.B. Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res. 2003;13:2042–2051. doi: 10.1101/gr.1257503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matveeva O.V., Mathews D.H., Tsodikov A.D., Shabalina S.A., Gesteland R.F., Atkins J.F., Freier S.M. Thermodynamic criteria for high hit rate antisense oligonucleotide design. Nucleic Acids Res. 2003;31:4989–4994. doi: 10.1093/nar/gkg710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugimoto N., Nakano S., Katoh M., Matsumura A., Nakamuta H., Ohmichi T., Yoneyama M., Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 39.Team R.D.C. 2004. R: A language and environment for statistical computing (ISBN 3-900051-07-0) [Google Scholar]
- 40.Phillips M.I., Zhang Y.C. Basic principles of using antisense oligonucleotides in vivo. Meth. Enzymol. 2000;313:46–56. doi: 10.1016/s0076-6879(00)13004-6. [DOI] [PubMed] [Google Scholar]
- 41.Stein C.A. Two problems in antisense biotechnology: in vitro delivery and the design of antisense experiments. Biochim. Biophys. Acta. 1999;1489:45–52. doi: 10.1016/s0167-4781(99)00143-8. [DOI] [PubMed] [Google Scholar]
- 42.Jacobson R.H., Zhang X.J., DuBose R.F., Matthews B.W. Three-dimensional structure of beta-galactosidase from E.coli. Nature. 1994;369:761–766. doi: 10.1038/369761a0. [DOI] [PubMed] [Google Scholar]
- 43.Ring M., Huber R.E. Multiple replacements establish the importance of tyrosine-503 in beta-galactosidase (Escherichia coli) Arch Biochem. Biophys. 1990;283:342–350. doi: 10.1016/0003-9861(90)90652-f. [DOI] [PubMed] [Google Scholar]
- 44.Gebler J.C., Aebersold R., Withers S.G. Glu-537, not Glu-461, is the nucleophile in the active site of (lac Z) beta-galactosidase from Escherichia coli. J. Biol. Chem. 1992;267:11126–11130. [PubMed] [Google Scholar]
- 45.Cupples C.G., Miller J.H., Huber R.E. Determination of the roles of Glu-461 in beta-galactosidase (Escherichia coli) using site-specific mutagenesis. J. Biol. Chem. 1990;265:5512–5518. [PubMed] [Google Scholar]
- 46.Celada F., Ullmann A., Monod J. An immunological study of complementary fragments of beta-galactosidase. Biochemistry. 1974;13:5543–5547. doi: 10.1021/bi00724a014. [DOI] [PubMed] [Google Scholar]
- 47.Ding Y., Chan C.Y., Lawrence C.E. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA. 2005;11:1157–1166. doi: 10.1261/rna.2500605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathews D.H., Burkard M.E., Freier S.M., Wyatt J.R., Turner D.H. Predicting oligonucleotide affinity to nucleic acid targets. RNA. 1999;5:1458–1469. doi: 10.1017/s1355838299991148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patzel V., zu Putlitz J., Wieland S., Blum H.E., Sczakiel G. Theoretical and experimental selection parameters for HBV-directed antisense RNA are related to increased RNA-RNA annealing. Biol. Chem. 1997;378:539–543. doi: 10.1515/bchm.1997.378.6.539. [DOI] [PubMed] [Google Scholar]
- 50.Barletta R.G., Kim D.D., Snapper S.B., Bloom B.R., Jacobs W.R., Jr Identification of expression signals of the mycobacteriophages Bxb1, L1 and TM4 using the Escherichia-Mycobacterium shuttle plasmids pYUB75 and pYUB76 designed to create translational fusions to the lacZ gene. J. Gen. Microbiol. 1992;138:23–30. doi: 10.1099/00221287-138-1-23. [DOI] [PubMed] [Google Scholar]
- 51.Timm J., Lim E.M., Gicquel B. Escherichia coli-mycobacteria shuttle vectors for operon and gene fusions to lacZ: the pJEM series. J. Bacteriol. 1994;176:6749–6753. doi: 10.1128/jb.176.21.6749-6753.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harth G., Zamecnik P.C., Tang J.Y., Tabatadze D., Horwitz M.A. Treatment of Mycobacterium tuberculosis with antisense oligonucleotides to glutamine synthetase mRNA inhibits glutamine synthetase activity, formation of the poly-L-glutamate/glutamine cell wall structure, and bacterial replication. Proc. Natl Acad. Sci. USA. 2000;97:418–423. doi: 10.1073/pnas.97.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harth G., Horwitz M.A., Tabatadze D., Zamecnik P.C. Targeting the Mycobacterium tuberculosis 30/32-kDa mycolyl transferase complex as a therapeutic strategy against tuberculosis: Proof of principle by using antisense technology. Proc. Natl Acad. Sci. USA. 2002;99:15614–15619. doi: 10.1073/pnas.242612299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matveeva O.V., Shabalina S.A., Nemtsov V.A., Tsodikov A.D., Gesteland R.F., Atkins J.F. Thermodynamic calculations and statistical correlations for oligo-probes design. Nucleic Acids Res. 2003;31:4211–4217. doi: 10.1093/nar/gkg476. [DOI] [PMC free article] [PubMed] [Google Scholar]