Abstract
Exhaustive gene identification is a fundamental goal in all metagenomics projects. However, most metagenomic sequences are unassembled anonymous fragments, and conventional gene-finding methods cannot be applied. We have developed a prokaryotic gene-finding program, MetaGene, which utilizes di-codon frequencies estimated by the GC content of a given sequence with other various measures. MetaGene can predict a whole range of prokaryotic genes based on the anonymous genomic sequences of a few hundred bases, with a sensitivity of 95% and a specificity of 90% for artificial shotgun sequences (700 bp fragments from 12 species). MetaGene has two sets of codon frequency interpolations, one for bacteria and one for archaea, and automatically selects the proper set for a given sequence using the domain classification method we propose. The domain classification works properly, correctly assigning domain information to more than 90% of the artificial shotgun sequences. Applied to the Sargasso Sea dataset, MetaGene predicted almost all of the annotated genes and a notable number of novel genes. MetaGene can be applied to wide variety of metagenomic projects and expands the utility of metagenomics.
INTRODUCTION
Microorganisms form complex communities in natural environments and are responsible for most of the ecological cycles that shape those environments. Therefore, identifying all community members and genes in an environment is fundamental to obtaining a perspective on the ecological systems. However, only a fraction of the living microbes can be isolated in culture. The estimations based on 16S ribosomal RNA suggest that 99% of microbial species cannot be easily cultivated (1,2). Whole-genome shotgun sequencing of environmental-pooled DNA draws attention as a powerful method for revealing genomic sequences from various organisms in natural environments without isolation and cultivation of individual species. This metagenomic approach has been applied to various environmental samples including an acid mine biofilm (3), the Sargasso Sea (4), Minnesota farm soil (5), whale falls (5) and deep sea sediments (6). In exceptional cases, analyses of Pleistocene cave bears (7) and mammoth DNA (8) are interesting applications of metagenomics to paleogenomics. While the metagenomic approach allows us to capture representative DNA samples from many diverse organisms, many unidentified sequences are produced at the same time. Additionally, many sequence reads remain as unassembled one-pass sequences because of the variety of sizes of environmental genomes and their abundance. Indeed, half of the reads for the Sargasso Sea dataset and all of the reads for the Minnesota soil data were unassembled sequences of roughly 700 bp. Gene-finding is a fundamental goal in virtually all metagenomics projects, but the only realistic alternative is similarity searches of the sequence database or the metagenome itself (9). The sensitivity of similarity search depends strongly on the availability of the relevant sequences in the database, and many novel genes of interest, which are the key element in metagenomics, are missed. To overcome this problem and exhaustively extract genes in environmental genomes, we have developed the MetaGene gene-finding program, which is designed to predict genes from fragmented genomic sequences.
Computational gene-finding from genomic DNA sequences has a long history (10–12), and a number of prokaryotic gene-finding tools have been developed and widely applied to the annotation of the microbial genomes. Although a variety of algorithms, including the hidden Markov model (HMM), are employed in gene-finding tools (13–27), most of the algorithms require a sufficient number of experimentally identified gene sequences from which to learn the parameters for individual species. Non-supervised training procedures eliminate that problem and can be applied to an anonymous genome (18–22) that is either a complete genomic sequence or a large portion of the sequence that contains an adequate number of genes. However, most environmental genomic sequences are fragmented anonymous sequences that contain one or two partial genes, so almost all of these algorithms are inapplicable. Only a non-supervised procedure that employs a heuristically derived model (23) works well for finding genes in such fragmented anonymous sequences. In this method, codon frequencies are approximated by the GC content of a given sequence. Because strong correlation is observed between the codon frequencies and the GC content of the genomic sequence (23,28,29), the estimated models sufficiently represent the original codon usage of the genome. We extended the method to estimate di-codon frequencies and achieve a higher prediction accuracy than results from using mono-codon frequencies. In addition to the codon frequencies, other measures, such as the frequency distribution of open reading frame (ORF) lengths, the distance from leftmost start codons, and the distances between neighboring ORFs, are integrated in MetaGene. These additional measures remarkably improve the prediction accuracy, especially with respect to in specificity. Here, we report the results of a performance test of MetaGene applied to sequences of various lengths and GC% and its application to the contig sequences of the Sargasso Sea dataset as a case study.
MATERIALS AND METHODS
Materials
We used the complete genomic sequences and the annotations of coding regions of 143 microorganisms that are available in GenBank. To avoid overfitting to well-studied genera and to form an accurate estimation of the prediction performance, one species per genus was selected for our study. A total of 116 bacterial and 15 archaeal genomes (Supplementary Table 1) were used to obtain the statistical values for model construction; the remaining 9 bacterial and 3 archaeal genomes (Supplementary Table 2) were used to evaluate the prediction performance. These genomic sequences were randomly split into fixed-length fragments (cardinally, 700 bp, the average length of the shotgun reads) that represent 1× genome for each species. The fragments were used as artificial shotgun sequences for the assessment. In these fragments, uninformative miniscule partial genes (less than 60 bp) were excluded from the annotations. The prediction performance was estimated by using the annotations as correct genes. The ratios of true-positives relative to all annotated genes (sensitivity) and to all predicted genes (specificity) were calculated. Both exactly matching predictions and partially matching predictions with correct reading frames were counted as true-positives. The contig sequences and annotations of the Sargasso Sea dataset were also obtained from the Venter Institute (http://www.venterinstitute.org/sargasso/) and used for the case study.
Architecture of MetaGene
MetaGene predicts genes in two stages. In the first stage, all possible ORFs are extracted from a given sequence and are scored by their base compositions and lengths. In this paper, an ORF is defined as a sequence of codons starting from a start codon and stopping at a stop codon. Partial ORFs are also extracted when they are located on the ends of the given sequence or are the entire sequence. In the second stage, an optimal (high-scored) combination of ORFs is calculated using the scores of orientations and distances of neighboring ORFs in addition to the scores for the ORFs themselves. This two-stage approach also allows us to predict overlapping genes with appropriate scores.
The MetaGene scoring scheme is based on a stochastic approach. Log-odds ratios are used for scoring throughout our algorithm. The frequency of an event observed in protein-coding ORFs is divided by the observed frequency in random ORFs, and a base-two logarithm of the ratio is used as the score for the event. The statistics used in MetaGene include di-codon frequencies, ORF length distributions, distance distributions from an annotated start codon to the leftmost start codon, and frequencies of orientations and orientation-dependent distances of neighboring ORFs. Details of the scoring scheme can be seen in the Supplementary Data section.
Estimation of the stochastic models for a given sequence
Codon frequency interpolation
Microbial genomes consist mainly of protein-coding regions, and the nucleotide frequencies in the three codon positions are strongly associated with those in the entire genome. In addition, Besemer and Borodovsky (23) reported that some amino acid residue frequencies are linearly related to the GC content of the genome. Since genes of certain species have almost the same codon usage or are classified into a few classes of codon usage, the usage is reflected in the local GC content of the genome. In this study, mono-codon and di-codon frequencies for the start, internal and stop codons are directly estimated by the GC% of a given sequence by using logistic regression analysis.
For the start codons, the proportions of ATG, GTG and TTG are estimated by the GC%. There is no strong correlation between the proportions of correct start codons and the GC% of the genomes, and ATG is used as the major start codon in most species. However, the proportions of incorrect start codons are strongly associated with the GC%, thus log-odds scores of the start codons vary according to the GC%. For the internal and stop codons, the conditional probability of a codon A, given a previous codon B, denoted P(A∣B), is estimated by using logistic regression analysis. We found that the di-codon frequencies are also related to the GC% like mono-codon frequencies and the relations can be successfully represented by a sigmoid function (see Supplementary Data). At the same time, we found that bacteria and archaea have slightly different trends in the relations between the di-codon frequencies (e.g. xxxATA and xxxGAG) and the GC%, which means the regression formulas derived from bacterial (or archaeal) codon frequencies are not suitable for predicting archaeal (or bacterial) genes. Actually, scores of protein-coding ORFs are degraded when codon frequencies of the wrong domain are used for the scoring (Supplementary Figure. S2). To overcome this limitation, we formulated a domain classification method for bacteria and archaea. For the domain classification, two sets of regression formulas for bacterial and archaeal codon frequencies are prepared, and both of them are applied to the scoring of ORFs in a given sequence. The definitive scores of the optimal paths of ORFs are independently calculated and compared, and the higher one is selected as the output for the given sequence. The domain classification method is just based on the differences of codon frequencies. However, some bacterial (archaeal) species have codon frequencies that are similar to archaeal (bacterial) genes. As a result, 5 of 116 bacterial (Anaplasma marginale, Aquifex aeolicus, Pelodictyon luteolum, Thermoanaerobacter tengcongensis and Thermotoga maritima) and 1 of 15 archaeal (Methanosphaera stadtmanae) genomes in the training data were misclassified by our domain classification method. The accuracies of gene-finding for the incorrectly classified genomes were still very high, because codon frequencies fitted to the species' genes were used. The aim of the domain classification is to improve gene-finding accuracy by using proper codon frequencies; our method achieves that purpose.
Length distribution of ORFs
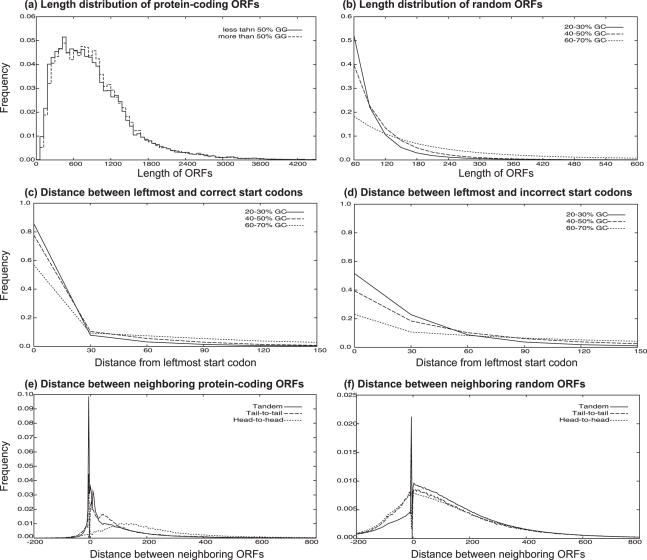
The ORF length is an important measure for distinguishing protein-coding ORFs from random ORFs. The average length of protein-coding (annotated) ORFs is about 950 bp, and the length distribution does not vary much among species (Figure 1a). Only slight decreases are observed in the frequencies of short ORFs from the high GC% genomes. On the other hand, the average lengths of random ORFs vary greatly according to the GC content of the genomes (Figure 1b). The average length of random ORFs (longer than 60 bp) in the low GC% genomes (<30%) is about 120 bp, while the average in the high GC% genomes (>60%) is about 300 bp. The difference in the lengths arises from the differences in the frequencies of incorrect start/stop codons in the genomes. We estimated the length distribution of random ORFs for a given sequence by linear interpolation using the distributions shown in Figure 1b. We used the distributions to calculate the log-odds scores of the ORF lengths for all extracted ORFs and added them to the codon-based scores. If an extracted ORF having a length l is a partial ORF, its true length will be greater than l. To appropriately evaluate the partial ORFs, which are the majority in metagenomic data, the upper probabilities derived from the distributions are used.
Figure 1.
Length distributions of (a) annotated ORFs and (b) random ORFs. ORFs are classified by the GC% of their genomes, and then the length distributions of each class are calculated. Distributions of distances between (c) leftmost start codons and annotated start codons and (d) leftmost start codons and incorrect start codons. Zero means the leftmost start codons are used as the start codons of the ORFs. (e) Orientation-dependent distributions of distances between neighboring ORFs. (f) The background distributions. Negative values mean overlapping of ORFs.
Distance distribution from the correct start codon to the leftmost start codon
It is known that the leftmost start codons are not always used as translation starts. The ratio of leftmost start codons to annotated start codons also varies greatly with the GC content of the genomes. Figure 1c and d show the distributions of the distance between the correct (or assumed) start codons and the leftmost ones. These GC%-dependent distributions arise from the incorrect start/stop codon frequencies like the random ORF lengths. However, a comparison between the distributions of the correct and the assumed (incorrect) start codons indicates that the protein-coding ORFs prefer the leftmost start codons to the others and that the appearance of upstream incorrect start codons is not common. Interestingly, the log-odds score of the leftmost start codon is larger in the high GC% genomes than in the low GC% genomes (data not shown), although the frequency of the leftmost start codon is lower in the high GC% genomes. Unfortunately, the annotated start codons are not always correct (22,30), and the obtained distributions may not be very accurate. The rates of the leftmost ‘real’ start codon may be especially lower in GC-rich genomes. On the other hand, these distributions show significant correlation between the translation start sites and the genome GC%, which means the annotations are not random, and the obtained distributions contain useful information inherent to the annotation data. The distributions for a given sequence are estimated by linear interpolation using these distributions, and the log-odds scores are added to the ORF scores as in the case of the ORF length distribution.
Orientation and distance of neighboring ORFs
Prokaryotic genes are frequently arranged in tandem in the genome because of the operon structures. In the genomes studied here, about 70% of neighboring genes are tandem, and the remaining genes are arranged head-to-head and tail-to-tail in equal proportion. The orientations of neighboring genes influence their distance distributions (Figure 1e). The tandem genes tend to be compactly arranged, and the mode of the distribution is −4 bp (overlaps of length four: ATGA, GTGA and TTGA). This packed arrangement is suitable for operon transcription for a single RNA. The distribution also indicates the existence of a RBS (ribosome binding site) at around 7 bp upstream of the start codons. The head-to-head genes prefer more distant arrangements because they have promoters in their upstream regions and avoid overlaps with coding regions. The tail-to-tail genes have a moderate but specific distribution compared to the others, probably indicating the existence of transcription stop signals. Such biased distributions can be used to evaluate an optimal sequence of ORFs in a given sequence. Because about half of the sequence reads (700 bp in length) have two partial genes per read, the information of neighboring ORFs is helpful in exacting gene prediction in environmental genomic sequences. We therefore integrated scores of the orientations and distances of neighboring ORFs into the calculation of a high-scoring path of ORFs. When the previous or next gene does not appear in a sequence, the upper probability derived from the distance distributions is used according to the gene orientation.
The definition of the background distribution is difficult in the intergenic regions. If the distances from one to all possible ORFs are used, a uniform distribution of extremely low frequency (depending on the genome size) is obtained. This problem can be avoided by limiting the range of ORF distances, but the background frequency depends on the range setting. In this study, the number of random ORFs between neighboring protein-coding ORFs was counted, and the distribution of the number was used to determine the neighbors. Although the number of random ORFs depends on the GC% of the genome, the distance distribution derived from the number is independent of the GC%. This is obvious because the occurrence rate of random ORFs depends on the GC%. We decided to use the upper quartile of the distribution, which means using the distances from one to the first 10 candidate ORFs to derive the background distribution. The obtained distributions are shown in Figure 1f.
RESULTS AND DISCUSSION
Testing on complete genomes
Although our main target is raw sequence reads and relatively short contigs, we tested the prediction performance of MetaGene on anonymized complete genomic sequences to assess the validity of our method. MetaGene just estimates codon frequencies and length (distance) distributions by the GC% of a given sequence, and does not use any other non-supervised procedure adapted to complete (long) genome using statistics from a sufficient number of candidate ORFs in the tested genome. In addition, all genomic sequences and annotations of the tested organisms were completely excluded from the training data for the codon frequency interpolation by the GC%. Despite this implementation, prediction accuracies of MetaGene for the complete genomes were extremely high (Table 1) and comparable to those of GeneMark.hmm, which learned its parameters using individual genomes and annotations (supervised learning). The prediction performance of MetaGene was independent of the GC content of the genomes, and the whole range of genes is precisely predicted. Only the sensitivity of Chlorobium tepidum and the specificity of Wolbachia were remarkably low both in MetaGene and GeneMark.hmm. Some archaea-like and eukaryote-like genes exist in the C.tepidum genome and degraded the sensitivity of the prediction because their codon biases differ from those of bacteria. This problem was partly improved in the MetaGene prediction for the fragmented sequences by using the domain classification method (see next section). In the Wolbachia genome, the annotated gene content is significantly lower than in the other microorganisms, and the length distribution of annotated genes is much different from that in the others. The length distribution of predicted genes was also different from that of the others, and a large number of small genes (about 200 bp) were predicted by MetaGene (and Gene Mark.hmm). This probably means that there are many pseudogenes in the genome or that sequencing (or assembling) errors occurred.
Table 1.
Gene prediction accuracies for nine bacterial and three archaeal complete genomes
| Organisms | GC% | Known genes | MetaGene | GeneMark.hmm | ||
|---|---|---|---|---|---|---|
| Sn (%) (exact) | Sp (%) | Sn (%) (exact) | Sp (%) | |||
| Archaea | ||||||
| M.jannaschii DSM 2661 | 31.4 | 1724 | 98.4 (69.9) | 96.1 | 98.9 (63.1) | 95.4 |
| A.fulgidus DSM 4304 | 48.6 | 2407 | 96.2 (73.5) | 94.8 | 96.9 (72.0) | 94.0 |
| N.pharaonis DSM 2160 | 63.4 | 2661 | 96.9 (80.7) | 97.7 | 95.8 (84.8) | 98.8 |
| Bacteria | ||||||
| Buchnera aphidicola str. APS | 26.3 | 563 | 99.6 (88.5) | 94.6 | 99.8 (88.6) | 95.6 |
| Prochlorococcus marinus str. MIT 9312 | 31.2 | 1809 | 95.9 (86.6) | 94.9 | 97.1 (87.7) | 95.7 |
| Wolbachia endosymbiont strain TRS | 34.2 | 805 | 95.2 (76.5) | 75.9 | 98.9 (85.7) | 75.0 |
| Helicobacter pylori J99 | 39.2 | 1477 | 96.3 (74.2) | 96.3 | 98.3 (88.2) | 95.1 |
| Bacillus subtilis subsp. subtilis str. 168 | 43.5 | 4102 | 94.0 (61.8) | 96.9 | 97.9 (86.2) | 95.3 |
| E.coli K12 | 50.8 | 4236 | 94.7 (72.3) | 97.3 | 97.2 (74.3) | 96.8 |
| C.tepidum TLS | 56.5 | 2252 | 82.4 (59.9) | 95.1 | 84.1 (58.1) | 93.4 |
| Corynebacterium jeikeium K411 | 61.4 | 2137 | 94.6 (70.0) | 97.0 | 95.5 (72.3) | 97.7 |
| Burkholderia pseudomallei K96243 chr.1 | 67.7 | 3398 | 97.4 (71.9) | 93.9 | 96.6 (61.0) | 95.0 |
| B.pseudomallei K96243 chr.2 | 68.5 | 2328 | 97.7 (70.3) | 91.5 | 95.9 (62.1) | 89.4 |
| Average | 95.3 (73.5) | 94.0 | 96.4 (75.7) | 93.6 | ||
Sn: sensitivity, Sp: specificity, exact: only an exact match is treated as correct. Results of GeneMark.hmm were obtained using the web interface of the GeneMark program (version 2.4) with same length threshold as MetaGene. The only result of GeneMark.hmm for P.marinus was obtained from the RefSeq database of NCBI.
For the complete genomes, our domain classification method worked perfectly, and the predictions for all species were performed with regression formulas of the appropriate domain. Examples of score distributions of ORFs derived from the archaeal and the bacterial set of regression formulas are shown in Figure 2. The separation performances varied significantly with the species and the selected set of regression formulas. The scores for correct genes showed especially remarkable improvement from the use of the appropriate set of the regression formulas. This means the background frequencies for bacteria and archaea are almost the same, while the codon frequencies are significantly different.
Figure 2.
Distributions of log-odds scores of ORFs calculated with (a) archaeal models and (b) bacterial models. Score distributions of the annotated genes and false positives for Archaeoglobus fulgidus and Escherichia coli are indicated.
Testing on artificial shotgun sequences
To assess the prediction performance of MetaGene on small fragments of anonymous genomic sequences, the complete genomic sequences of 12 organisms were randomly split into 700 bp fragments of 1× genome coverage. About 1.4 genes per fragment were annotated in the artificial shotgun sequences, and 92% of the annotated genes were partial genes. Here, miniscule partial genes, which are less than 60 bp in length, had been excluded from the annotations of the fragments. Table 2 shows the accuracies of MetaGene on the artificial shotgun sequences. The sensitivities for the fragments were high enough and equivalent to those for the complete genomes, while the specificities were significantly degraded. The result shows the difficulty of distinguishing fragmented short genes from background noise. Despite the degradation of the specificity, the absolute values were still high enough, and the performance of MetaGene on the shotgun sequences is sufficient for practical use in metagenomic gene-finding. Interestingly, the sensitivities based on the exact match criterion were higher than those on the complete genomes because many ORFs for the fragments lack their 5′ ends, including start codons.
Table 2.
Prediction accuracies of MetaGene on artificial shotgun sequences (700 bp long)
| Organisms | Sensitivity (%) (exact) | Specificity (%) | Correct domain (%) |
|---|---|---|---|
| Archaea | |||
| M.jannaschii DSM 2661 | 97.8 (82.4) | 94.1 | 70.2 |
| A.fulgidus DSM 4304 | 95.8 (81.5) | 93.7 | 99.3 |
| N.pharaonis DSM 2160 | 97.1 (86.2) | 93.0 | 80.8 |
| Bacteria | |||
| B.aphidicola str. APS | 98.2 (90.9) | 92.7 | 98.6 |
| P.marinus str. MIT 9312 | 95.5 (87.6) | 92.7 | 90.9 |
| W.endosymbiont strain TRS | 93.1 (80.8) | 76.0 | 72.8 |
| H.pylori J99 | 92.6 (77.7) | 92.7 | 95.1 |
| B.subtilis subsp. subtilis str. 168 | 92.3 (73.5) | 92.5 | 92.9 |
| E.coli K12 | 95.3 (81.2) | 93.2 | 97.9 |
| C.tepidum TLS | 88.1 (73.2) | 89.6 | 78.4 |
| C.jeikeium K411 | 94.0 (78.5) | 91.4 | 85.8 |
| B.pseudomallei K96243 chr.1 | 96.8 (81.2) | 87.9 | 93.3 |
| B.pseudomallei K96243 chr.2 | 96.6 (80.5) | 85.7 | 93.0 |
| Average | 94.9 (81.2) | 90.4 | 88.4 |
In about 90% of the fragmented sequences, the correct domain (i.e. the same domain as the original genome) was selected to predict genes (Table 2). Interestingly, more annotated genes were successfully detected for the fragmented genomic sequences of C.tepidum TLS than for the complete genomic sequences by using the archaeal codon frequencies as well as the bacterial codon frequencies. In the C.tepidum genome, the existence of many archaea-like proteins (about 12% of the annotated proteins) was reported (31). Many of these archaea-like genes, which may result from lateral gene transfer, were missed in the prediction for the complete genome because the bacterial codon frequencies were selected. For the fragmented sequences, all genes, including these archaea-like genes, were correctly predicted by using the proper codon frequencies for each fragment. As a result, the sensitivity increased significantly despite the short lengths of the input sequences. In the prediction for the two archaeal genomes, Methanocaldococcus jannaschii and Natronomonas pharaonis, 25–30% of the fragments were misclassified, which means the bacterial codon frequencies were applied to them. However, the prediction accuracies on these genomes were adequately high. This result suggests that these species have a considerable number of genes that are similar to bacterial genes. In fact, it is known that M.jannaschii shares the majority of genes related to energy production, cell division and metabolism with bacteria (32).
To examine the effectiveness of the measures employed by MetaGene to precisely predict genes on the fragmented sequences, three additional prediction models were constructed and applied to the artificial shotgun sequences (Table 3). One model used all of the ORF scores (the ORF model), such as the codon frequencies, the ORF length distributions and the distance distributions of start codons, but did not use the orientation and distance distributions of neighboring ORFs. Another model used only the codon frequencies (di-codon model), and no other measures were integrated. The other (mono-codon model) was an alternative the di-codon model that substituted mono-codon frequencies for di-codon frequencies. Surprisingly, the di-codon model was sufficiently accurate, suggesting that the GC%-dependent di-codon models were the predominant gene-finding measures in MetaGene. The mono-codon model also showed high sensitivities, but the specificities were low. In the predictions of the ORF model, both sensitivities and specificities were improved. Most of the annotated genes were partial in the artificial shotgun sequences, and statistically-accurate scoring for such partial genes worked well. In addition, the length/distance scores facilitated an exact prediction of ORFs. In the original MetaGene, the orientation and orientation-dependent distance distributions helped to predict an optimal gene set in a genomic sequence, and significantly improved the specificities of the predictions. All of the measures certainly improved the prediction performance, especially the specificity. By integrating all of the measures, MetaGene achieves reliable gene prediction for small fragments of genomic sequences.
Table 3.
Effectiveness of the measures integrated in MetaGene
| Prediction models | Sensitivity (exact) | Specificity |
|---|---|---|
| MetaGene (original) | 94.9 (81.2) | 90.4 |
| ORF model | 94.8 (79.7) | 88.3 |
| Di-codon model | 94.0 (70.0) | 86.7 |
| Mono-codon model | 93.8 (67.7) | 84.3 |
Raw shotgun sequences vary in length, although the average is about 700 bp. We applied MetaGene to various fixed-length fragments ranging from 100 to 1000 bases (1× genome) and inspected the change of the prediction performance with the length of the input sequence (Figure 3). The prediction accuracies naturally decreased along with the shortening of the input sequences. However, MetaGene retained relatively high accuracies on smaller fragments, and extreme degradation of accuracy was observed only for the 100 bp fragments. Generally, most raw sequence reads are larger than 500–600 bp, which is to say that MetaGene can predict genes on the metagenomic data with high reliability.
Figure 3.
Sensitivity and specificity of MetaGene for the sets of fixed-length artificial shotgun sequences. The average values for 12 species are indicated.
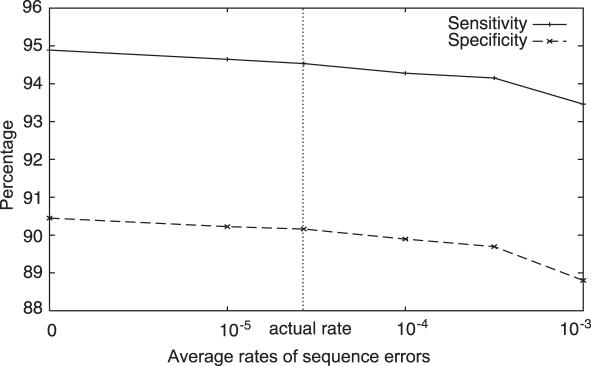
Another problem of raw data are the occurrence of sequence errors. Shotgun sequences include more errors than the complete genomes because of the one-pass status of the shotgun sequences. Most low-quality nucleotides concentrate near the ends of the sequences, and such nucleotides are ordinarily cut out from the sequences (3,5). As a result, the shotgun sequences have a relatively high average of quality values, which are assigned to nucleotides by a base-call program (the average is usually more than 40, or less than one error per 10 000 bp). We tested the effect of sequence errors on gene-finding using artificial sequences with variable error rates (Figure 4). Position-specific error rates were obtained from actual trace data (sequence + quality files), and the average was modulated. Degradation of the prediction performance was sufficiently low, suggesting reliable predictions can be performed by MetaGene on real metagenomic sequences.
Figure 4.
Effect of sequence errors on gene-finding. Nucleotides of the artificial sequences (700 bp) were changed according to position-specific error rates derived from actual data. The percentages are plotted against the averages of the position-specific error rates.
Application to metagenomic sequences
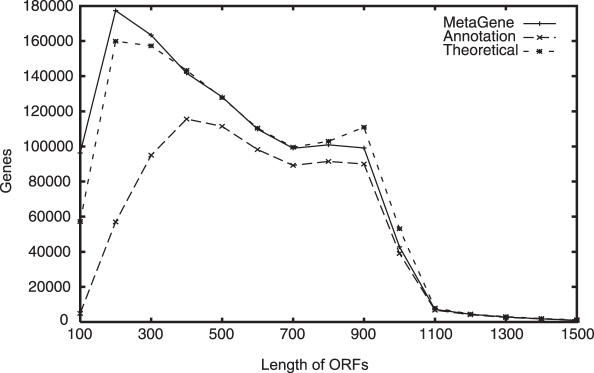
We applied MetaGene to the contig sequences in the Sargasso Sea dataset. The dataset consists of about 0.8 million contigs having an average length of about 1 kb (about 820 Mb in total) and having about one million genes annotated. The annotated genes were determined mainly by their similarity to the known bacterial genes. Some hypothetical genes were also determined based on the presence of conserved ORFs (4). For the contig sequences, about 1.4 million genes were predicted by MetaGene. Almost all annotated genes (96% of known genes and 92% of hypothetical genes) appeared in our predictions, and about 0.4 million genes were additionally predicted. These genes are strong candidates for novel genes from uncultured microorganisms, and they were five times more numerous than the hypothetical genes in the annotation. This means homology-based methods miss on notable number (>30%) of genes, and thus reduce the advantage of the metagenomic approach. The numbers of estimated genes disaggregated by length are shown in Figure 5. The MetaGene predictions were almost identical to the theoretical numbers, while those of the annotated genes were much lower in a wide range of lengths. Not only short ORFs but also relatively long ones were captured by MetaGene as novel genes. Many short ORFs exist in the Sargasso data, and MetaGene also predicted many short ORFs. However, these distributions suggest that most of the false positives may be accumulated in these short ORFs. The number of ORFs having 900 bases was lower than the theoretical number. The Sargasso sequences have a peak at around 900 bp, and many partial ORFs whose lengths are identical to the input sequences (900 bp) were predicted in theory. Because of the sequencing errors, some of such long ORFs may be broken by false stop codons and/or frame shifts in the real sequence data.
Figure 5.
Length distributions of ORFs predicted by various methods. The theoretical distribution was calculated by using gene densities and the length distributions of complete ORFs.
In the Sargasso Sea dataset, about 90% of the genes were predicted by the bacterial model, and only a fraction was classified into archaeal genes. Although this value does not directly reflect the abundances of bacterial and archaeal species, the result is consistent with the estimate based on 16S rRNA sequences. Our domain classification method was designed to precisely predict all genes and not to correctly estimate the domain of the given sequence. However, we believe that the additional domain information is useful in obtaining an overview of the microbial community in the environmental sample.
Availability of MetaGene
MetaGene has a very simple architecture and does not require a long calculation time. It took only about 15 min to test all of the contigs of the Sargasso Sea dataset on a single core of a Pentium D (3.4 GHz)/Linux system. The software implemented in C++ can be downloaded from http://metagene.cb.k.u-tokyo.ac.jp/. A Web interface to the software is also available at the web site.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors are grateful to Takehiko Itoh at Mitsubishi Research Institute, Inc. for preparing the test dataset and to Natsu Ishii for help with manuscript preparation. Funding to pay the Open Access publication charges for this article was provided by Grant-in-Aid for Scientific Research on Priority Areas “Systems Genomics” from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-2-reviews0003. reviews0003.1--0003.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rappe M., Giovannoni S. The uncultured microbial majority. Annu. Rev. Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 3.Tyson G.W., Chapman J., Hugenholtz P., Allen E.E., Ram R.J., Richardson P.M., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F. Community structure and metabolism through reconsruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 4.Venter J.C., Remington K., Heidelberg J.F., Halpern A.L., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 5.Tringle S.G., Mering C.V., Kobayashi A., Salamov A.A., Chen K., Chang H.W., Podar M., Short J.M., Mathur E.J., Detter J.C., et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 6.Hallam S.J., Putnam N., Preston C.M., Detter J.C., Rokhsar D., Richardson P.M., DeLong E.F. Reverse Methanogenesis: testing the hypothesis with environmental genomics. Science. 2004;305:1457–1462. doi: 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- 7.Noonan J.P., Hofreiter M., Smith D., Priest J.R., Rohland N., Rabeder G., Krause J., Detter C.D., Paabo S., Rubin E.M. Genomic sequencing of Pleistocene Cave Bears. Science. 2005;309:597–599. doi: 10.1126/science.1113485. [DOI] [PubMed] [Google Scholar]
- 8.Poinar H.N., Schwarz C., Qi J., Shapiro B., MacPhee R.D.E., Buigues B., Tikhonov A., Huson D.H., Tomsho L.P., Auch A., et al. Metagenomics to Paleogenomics: large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- 9.Chen K., Pachter L. Bioinformatics for whole-genome shotgun sequencing of microbial communities. PLoS Comput. Biol. 2005;1:106–112. doi: 10.1371/journal.pcbi.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fickett J.W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1981;10:5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gribskov M., Devereux J., Burgess R.R. The codon preference plot: graphic analysis of protein coding sequences and prediction of gene expression. Nucleic Acids Res. 1984;12:539–549. doi: 10.1093/nar/12.1part2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staden R. Measurements of the effects of that coding for a protein has on a DNA sequence and their use for finding genes. Nucleic Acids Res. 1984;12:551–567. doi: 10.1093/nar/12.1part2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borodovsky M.Y., Sprizhitskii Y.A., Golovanov E.I., Aleksandrov A.A. Statistical patterns in primary structures of functional regions in the E.coli genome: III. Computer recognition of coding regions. Mol. Biol. 1986;20:1145–1150. [PubMed] [Google Scholar]
- 14.Borodovsky M.Y., McIninch J.D. GeneMark: parallel gene recognition for both DNA strands. Comput. Chem. 1993;17:123–153. [Google Scholar]
- 15.Salzberg S.L., Delcher A.L., Kasif S., White O. Microbial gene identification using interpolated Markov mode. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krogh A., Mian I.S., Haussler D. A hidden Markov model that finds genes in E.coli DNA. Nucleic Acids Res. 1994;22:4768–4778. doi: 10.1093/nar/22.22.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukashin A.V., Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delcher A.L., Harmon D., Kasif S., White O., Salzberg S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes W.S., Borodovsky M. How to interpret an anonymous bacterial genome: machine learning approach to gene identification. Genome Res. 1998;8:1154–1171. doi: 10.1101/gr.8.11.1154. [DOI] [PubMed] [Google Scholar]
- 20.Audic S., Claverie J.M. Self-identification of protein-coding regions in microbial genomes. Proc. Natl Acad. Sci. USA. 1998;95:10026–10031. doi: 10.1073/pnas.95.17.10026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baldi P. On the convergence of a clustering algorithm for protein-coding regions in microbial genomes. Bioinformatics. 2000;16:367–371. doi: 10.1093/bioinformatics/16.4.367. [DOI] [PubMed] [Google Scholar]
- 22.Besemer J., Lomsadze A., Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Besemer J., Borodovsky M. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 1999;27:3911–3920. doi: 10.1093/nar/27.19.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frishman D., Mironov A., Mewes H.-W., Gelfand M. Combining diverse evidence for gene recognition in completely sequenced bacterial genomes. Nucleic Acids Res. 1998;26:2941–2947. doi: 10.1093/nar/26.12.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shmatkov A.M., Melikyan A.A., Chernousko F.L., Borodovsky M. Finding prokaryotic genes by the ‘frame-by-frame’ algorithm: targeting gene starts and overlapping genes. Bioinformatics. 1999;15:874–886. doi: 10.1093/bioinformatics/15.11.874. [DOI] [PubMed] [Google Scholar]
- 26.Yada T., Nakao M., Totoki Y., Nakai K. Modeling and predicting transcriptional units of Escherichia coli genes using hidden Markov model. Bioinformatics. 1999;15:987–993. doi: 10.1093/bioinformatics/15.12.987. [DOI] [PubMed] [Google Scholar]
- 27.Yada T., Totoki Y., Takagi T., Nakai K. A novel bacterial gene-finding system with improved accuracy in locating start codons. DNA Res. 2001;8:97–106. doi: 10.1093/dnares/8.3.97. [DOI] [PubMed] [Google Scholar]
- 28.Karlin S. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 2001;9:335–343. doi: 10.1016/s0966-842x(01)02079-0. [DOI] [PubMed] [Google Scholar]
- 29.Tu Q., Ding D. Detcting pathogenicity islands and anomalous gene clusters by iterative discriminant analysis. FEMS Microbiol. Lett. 2003;221:269–275. doi: 10.1016/S0378-1097(03)00204-0. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen P., Krogh A. Large-scale prokaryotic gene prediction and comparison to genome annotation. Bioinformatics. 2005;21:4322–4329. doi: 10.1093/bioinformatics/bti701. [DOI] [PubMed] [Google Scholar]
- 31.Eisen J.A., Nelson K.E., Paulsen I.T., Heidelberg J.F., Wu M., Dodson R.J., Deboy R., Gwinn M.L., Nelson W.C., Haft D.H., et al. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl Acad. Sci. USA. 2002;99:9509–9514. doi: 10.1073/pnas.132181499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bult C.J., White O., Olsen G.J., Zhou L., Fleischmann R.D., Sutton G.G., Blake J.A., FitzGerald L.M., Clayton R.A., Gocayne J.D., et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.