Abstract
Sso7d is a chromosomal protein of hyperthermophilic Archaea. The crystal structure of Sso7d–d(GTAATIUAC)2 has been clarified at high resolution, showing that the protein binds in the minor groove of DNA, causing a sharp kink of ≈60°. Recently, we found that photoirradiation of Sso7d and 5-iodouracil-(IU)-containing 5′-d(GTAATIUAC)-3′ efficiently induced the abstraction of hydrogen from the methyl group of T5 at the kink. In the present study, we found that the photoreactivity of 5-bromouracil (BrU)-containing 5′-d(GTAATBrUAC)-3′ was enhanced in the presence of Sso7d. Using hypoxanthine (I)-containing 5′-d(ITAATBrUAC)-3′, we demonstrated that electron transfer occurs efficiently from Sso7d to DNA. Product analysis showed that Trp-24 of Sso7d, located at the surface of the DNA, is consumed to produce N′-formylkynurenine during photoirradiation, indicating that Trp-24 acts as an electron source. To explore the possibility of electron transfer between Sso7d and other DNA substrates, we examined the photochemical repair of the thymine dimer 5′-d(GTAAT<>TAC)-3′ by Sso7d. Sso7d effectively repaired 5′-d(GTAAT<>TAC)-3′ to 5′-d(GTAATTAC)-3′ under irradiation conditions. During this reaction, Trp-24 was not oxidized significantly, indicating that the anion radical of the repaired TT sequence is oxidized by the cation radical of Trp-24, and that a so-called “circular electron transfer” mechanism is operating in this system.
Keywords: Archaea, DNA repair, electron transfer, thymine dimer
Ultraviolet light from the sun causes various types of DNA damage that induce mutations, carcinogenesis, and cell death. Pyrimidine–pyrimidine photodimers are typical products of photoinduced damage (1). Photolyase repairs thymine dimers by using electron transfer from cofactor domains under light. Recently, Barton and colleagues (2–4) proposed an attractive hypothesis that some repair enzymes use electron transfer from redox cofactors for detection of oxidatively generated DNA damage. Evidence that photolyase can also repair thymine dimers without the involvement of a cofactor (1, 5) suggests that the amino acid itself has the potential to repair DNA by a photoinduced electron transfer mechanism. In the living cell, DNA is usually associated with various types of nucleoproteins, which implies the existence of DNA repair activities in nucleoproteins. In fact, Cullis et al. (6) demonstrated that the guanine radical formed in DNA is reduced by electron transfer from histone, even though the detailed mechanisms is not known. The Archaea, as representatives of one of the oldest lineages, offer special insight into the origin of cellular life and the ancestry of eukaryotes (7). Sso7d and Sac7d are nucleohistone-like proteins of archaea, and structures of their DNA complexes have been solved by x-ray crystallography and NMR (refs. 8–10 and Fig. 1). In this article, we demonstrate electron transfer from the archaeal nucleosomal protein Sso7d to DNA by using 5-bromouracil (BrU) residue. We also demonstrate that the electron transfer from Sso7d effectively repairs a T<>T dimer.
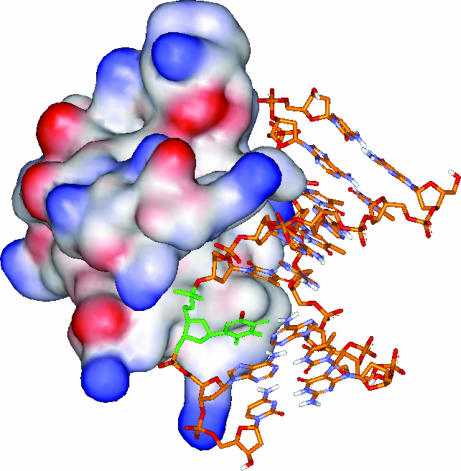
Fig. 1.
Structure of Sso7d-d(GTAATBrUAC)2 complex based on x-ray crystal structure (8). BrU residue and Br are colored in green and red, respectively.
Results and Discussion
Detection of Electron Transfer from Sso7d to DNA by Using BrU.
5-Halouracil, a photoreactive analogue of thymine, has been widely used for biological assays and studies of DNA chemistry (11–14). Recently, BrU has been used as an electron trap in DNA-mediated excess electron transfer processes (12, 13), because the formation of the anion radical of BrU rapidly eliminates bromo anions, to generate the uracil-5-yl radical (14, 15). Recently, we examined the photoreactions of 5-iodouracil (IU)-containing 5′-d(GTAATIUAC)-3′ and the Sso7d chromosomal protein from the hyperthermophilic archaeabacteria Sulfolobus solfataricus (8, 9) and demonstrated that the uracil-5-yl radical abstracts hydrogen from the methyl group of T5 at the sharp kink (16).
Because we recently found that photoinduced electron transfer occurs from guanine (G) to BrU through an A/T bridge in duplex DNA (17), evaluation of the contribution of the electron transfer process in this system might be important. Therefore, we examined the photoreactivity of 5′-d(GTAATBrUAC)-3′ (ODN1) in the presence and absence of Sso7d.
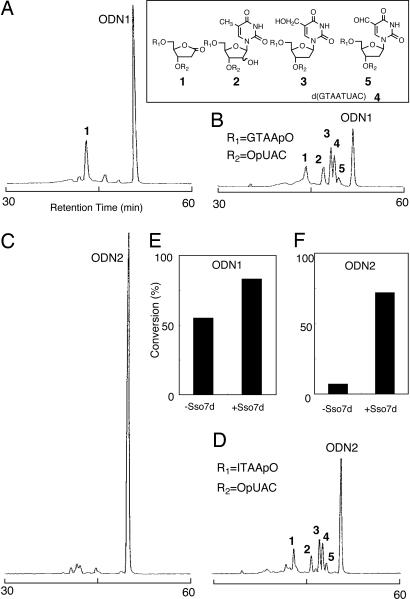
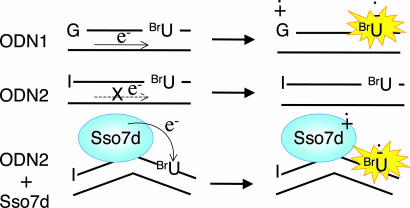
HPLC product analysis indicated that photoirradiation of ODN1 alone mainly gave 2-deoxyribonolactone-containing octamers, 1 (Fig. 2A). This result is consistent with previous observations (16, 17). Upon addition of Sso7d to the reaction mixture, the photoreactivity of ODN1 was enhanced, producing 1–5 (Fig. 2B), which are formed by intrastrand hydrogen abstraction at the T5 site by the uracil-5-yl radical, similar to that observed in the previous Sso7d-5′-d(GTAATIUAC)-3′ system (16). Enhancement of the photoreactivity of ODN1 upon addition of Sso7d suggests the possibility that some electrons come from Sso7d under these conditions. To eliminate the intrastrand electron transfer process from G to BrU in ODN1, we replaced the 5′ terminal G of ODN1 with the less electron-donating hypoxanthine (I); the oxidation potential of I is much higher than that of G (1.4 V vs. 1.2 V) (18). In fact, 5′-d(ITAATBrUAC)-3′ (ODN2) showed very low photoreactivity compared with ODN1 (Fig. 2C), confirming that the 5′ terminal I of ODN2 does not act as an electron donor for BrU. Interestingly, the addition of Sso7d greatly enhanced the photoreactivity of ODN2 by 10-fold, producing photoproducts 1–5 (Fig. 2D). The amounts of ODN1 and ODN2 consumed in the absence and the presence of Sso7d under irradiation are shown in Fig. 2 E and F. The results indicate that Sso7d activates the photoreactivity of BrU and suggest that Sso7d itself serves as an electron donor to activate BrU in ODN2 (Fig. 3).
Fig. 2.
Photoreaction of BrU containing DNA in the presence or absence of Sso7d. (A–D) HPLC profiles of UV (280 nm)-irradiated ODN1 (A), ODN1 with Sso7d (B), ODN2 (C), and ODN2 with Sso7d (D). (E and F) Conversions of photoirradiated ODN1 and ODN2 in the absence or presence of Sso7d. The reaction mixture (total volume, 10 μl) contained BrU-containing octamer (84 μM strand concentration) and Sso7d (42 μM; B and D) in 50 mM sodium cacodylate buffer (pH 7.0) is shown. Irradiation was performed with a monochromatic light (280 nm) for 15 min at 0°C. The reaction mixtures were analyzed by HPLC on a Chemcobond 5-ODS-H column (4.6 × 150 mm) with detection at 254 nm; elution was performed with 0.05 M ammonium formate (pH 6.5) containing 0–10% acetonitrile over a linear gradient for 60 min at a flow rate of 1.0 ml/min at 40°C.
Fig. 3.
Proposed mechanism for the reaction of BrU in the absence or presence of Sso7d.
Photorecovery of Thymine Dimer by Sso7d.
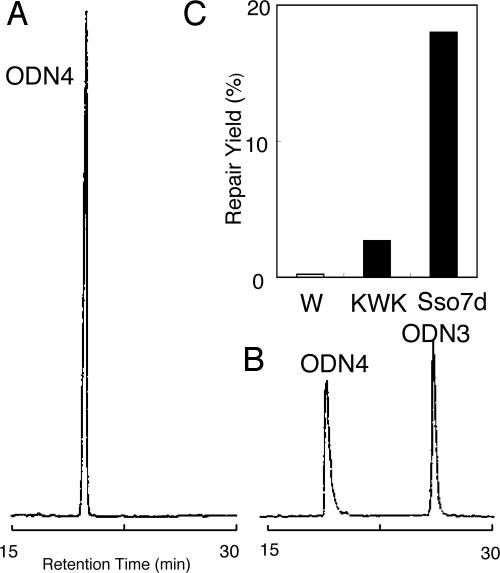
An intriguing question is whether this electron transfer occurs between Sso7d and other DNA substrates. To answer this question, we examined the photochemical repair of thymine dimer (T<>T) by Sso7d. It is well documented that a photoreactivating enzyme, photolyase, repairs T<>T by one electron reduction mechanism (1). Therefore, we prepared the T<>T-containing octamer 5′-d(GTAAT<>TAC)-3′ (ODN4) by photoirradiation of 5′-d(GTAATTAC)-3′ (ODN3) and examined the photoreactivation activity of Sso7d. Fluorescence quenching experiments (19) suggest that Sso7d binds to ODN4 similar to ODN2 and ODN3 (Fig. 9, which is published as supporting information on the PNAS web site). It was found that under UV irradiation at 295 nm for 1 h ODN4 was efficiently repaired by Sso7d to produce ODN3, indicating that electron transfer from Sso7d successfully repaired T<>T (Fig. 4B). For this experiment, a longer wavelength and a prolonged irradiation period were used to prevent background splitting of ODN2. Previously, Hélène and colleagues (20) reported that T<>T was repaired by Lys–Trp–Lys under irradiation. In fact, Lys–Trp–Lys repaired ODN4; however, the efficiency was significantly lower than Sso7d (Fig. 4C).
Fig. 4.
Photoreactivation of thymine dimer by Sso7d. (A and B) HPLC profile of ODN4 before (A) or after (B) photoirradiation in the presence of Sso7d. The reaction mixture (total volume, 10 μl) contained ODN4 (10 μM strand concentration) and Sso7d (20 μM) in 5 mM sodium cacodylate buffer (pH 7.0). Irradiation was performed with a monochrometor (295 nm) for 1 h at 0°C. The HPLC conditions were the same as described in Fig. 2 except for the acetonitrile gradient (3–10% over a linear gradient for 30 min). (C) Amount of ODN3 from photoirradiated ODN4 (10 μM) with Trp (W), Lys-Trp-Lys (KWK), or Sso7d (each concentrations are 5 μM).
Trp-24 of Sso7d Is a Source of Electron.
Next, we investigated which amino acid residue in Sso7d is responsible for electron transfer to DNA. Because the Härd group (19) has shown that fluorescence of the Trp residue (Trp-24) of Sso7d, which contains one Trp residue, is quenched when it binds to DNA, we anticipated that electron transfer from Trp-24 to DNA caused the efficient quenching of Trp-24.
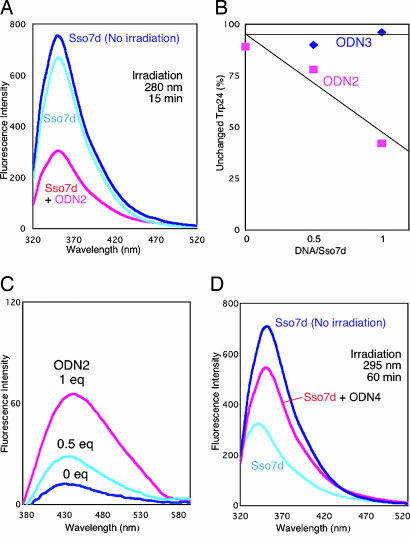
Therefore, we examined the fate of Trp-24 of Sso7d after electron injection. The photoirradiated Sso7d-ODN2 showed a significant reduction of emission from Trp-24 when Sso7d was irradiated at 280 nm for 15 min in the presence of ODN2, indicating the photodegradation of Trp-24 (Fig. 5A). In contrast, irradiation of Sso7d with ODN3 did not cause such a reduction (Fig. 5B), even though the binding of ODN3 causes a quenching of the fluorescence of Trp-24 (Fig. 9). This result suggests that electron transfer occurs to ODN3, but a rapid back electron transfer prevents oxidation of Trp-24, and indicates that BrU is essential for trapping an electron.
Fig. 5.
Fluorescence of damaged tryptophan residue of Sso7d after photoirradiation. (A) Fluorescence emission spectra of photoirradiated Sso7d with (pink) or without (light blue) ODN2 (excitation 295 nm, 5-nm slit width) are shown. (B) Fluorescence emission spectra (excitation 319 nm, 10-nm slit width) of photoirradiated Sso7d with different concentrations of ODN2 are shown. (C) Amount of undamaged Trp in photoirradiated Sso7d with different concentrations of ODN2 (pink rectangles) and ODN3 (blue diamonds) is shown. These values were determined from the fluorescence intensity at 360 nm of each photoirradiated sample (excitation 295 nm). (D) Fluorescence emission spectra of photoirradiated Sso7d with or without ODN4 is shown. Photoreaction procedure for A-C and D are same as in Figs. 2 and 4, respectively, except the reaction volume of Sso7d-ODN4 was 20 μl. For quantification of Trp-24, the reaction mixtures were diluted to 50 μl with a NaCl solution (final concentration of NaCl was 3 M).
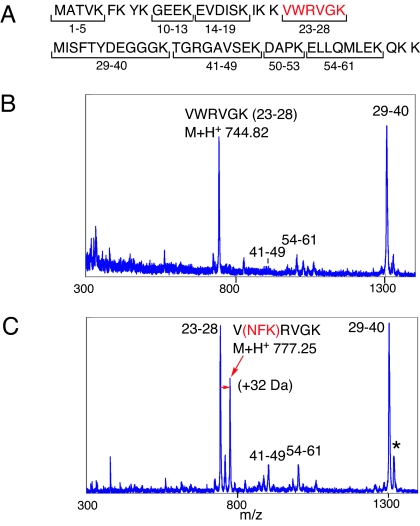
It is well documented that Trp residues in proteins are readily oxidized to N′-formylkynurenine (21, 22), which has a characteristic emission at 420 nm upon excitation at 319 nm. We confirmed that emission at 420 nm increased during irradiation of Sso7d with ODN2 (Fig. 5C). In addition, we investigated the lysyl endopeptidase (Lys-C) digests of Sso7d after photoirradiation with ODN2 by using MALDI-TOF mass spectrometry (Fig. 6). The mass of the Trp-24 containing peptide fragment VWRVGK was increased by 32 Da (Fig. 6C). This result indicates that Trp-24 in Sso7d is oxidized to N′-formylkynurenine during photoirradiation, which is consistent with the fluorescence data. We also observed the formation of the previously reported Met-29 oxidation fragment, which is formed by hydrogen abstraction by the uracil-5-yl radical (16).
Fig. 6.
MALDI-TOF mass spectra of Lys-C digests of a mixture of Sso7d and ODN2 before and after photoirradiation are shown. (A) Amino acid sequence and putative Lys-C cleavage fragment of Sso7d. (B and C) Lys-C digests of Sso7d and ODN2 before photoirradiation (B) and after photoirradiation (C) with photoirradiation condition the same as in Fig. 2. ∗, Caused by the formation of Met-29 oxidation of the 29–40 fragment (M+H+ 1321.19).
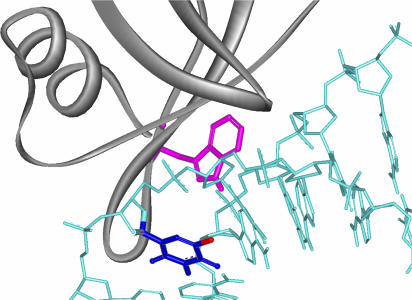
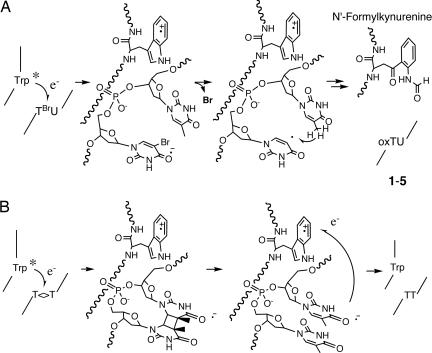
Inspection of the x-ray structure of the Sso7d-d(GTAATTAC)2 complex suggests that Trp-24 is located on the surface of the protein at the DNA contact site and 10 Å from the BrU residue (Fig. 7). These results clearly demonstrate that the Trp-24 of Sso7d is the electron source for BrU. The oxidation potential of Trp (1.0 V) (23), which is much lower than that of guanine (1.2 V), means that it is capable of donating electrons to BrU. The proposed mechanism for electron transfer from Trp-24 to BrU in ODN2 and the subsequent hydrogen abstraction of the uracil-5-yl radical and the oxidation of Trp-24 are shown in Fig. 8A. The initial electron transfer would occur from Trp-24 to the electron deficient BrU to provide the anion radical. Release of the halogen anion from BrU generates uracil-5-yl to abstract the hydrogen to give 1–5. The Trp-24 cation radical releases a proton and reacts with the molecular oxygen to provide N′-formylkynurenine.
Fig. 7.
Close-up view of the Sso7d-d(GTAATBrUAC)2 complex based on the x-ray crystal structure is shown. Trp-24, BrU6, and Br are drawn in violet, deep blue, and red, respectively.
Fig. 8.
Shown is the proposed scheme for electron transfer from the excited state of Trp-24 of Sso7d to BrU of ODN2 (A) and to T<>T in ODN4 (B).
In a photodimer-splitting experiment of ODN4, oxidation of the Trp-24 was significantly reduced compared with the irradiation in the absence of ODN4 (Fig. 5D), indicating that the anion radical of thymine is oxidized by the cation radical of Trp-24, and a circular electron transfer mechanism operates to some extent in this system (Fig. 8B). These results clearly indicate that Trp-24 in Sso7d injects electrons not only in the BrU-pyrimidine step but also in T<>T dimer. In this experiment, the condition of photoirradiation is the same as shown in Fig. 4.
Implications for Photoreactivation by Nucleoproteins.
In this study, we have demonstrated that electron transfer occurs from Sso7d to DNA by using two types substrates, BrU- and T<>T-containing DNA. In early earth, there is no ozone layer and strong UV light should reach the surface of Earth (24). DNA of the primitive life must be seriously suffered photodamage. Therefore, a defense system of the primitive organism against UV light was assumed to be crucially required. The present finding that this archaeal nucleoprotein processes photoreactivation activity suggests that the first life may have used coexisting RNAs or proteins to develop a repair system. Such system is less effective than the present-day DNA photolyases, which use cofactors, but photo-protection by nucleoproteins is a simple method for underdeveloped life. Our present findings lead to the hypothesis that such a primitive DNA repair system existed at the origin of life. Photolyase has Trp residues with potential to repair DNA without cofactor, and this Trp might be a trace of such function. Our results imply the possibility that the initial nucleoproteins might have evolved for the purpose of UV protection and DNA repair to allow survival in such conditions.
Materials and Methods
Preparation of Oligonucleotides.
Oligonucleotide strands were synthesized on an ABI DNA synthesizer (Applied Biosystem, Foster City, CA). After purification by RP-HPLC, DNA concentrations were determined by using enzymatic digestion. The thymine dimer-containing ODN4 was prepared by photoirradiation of ODN3 at 302 nm using a transilluminator (Fig. 10, which is published as supporting information on the PNAS web site). It was confirmed that acetophenone photosensitization of ODN3 also gave ODN4 (Fig. 10). The product was characterized by direct photo splitting of the dimer-containing strand with light at 254 nm to regenerate the undamaged strand (24). The thymine dimer-containing strand (ODN4) was purified by HPLC and tested for the presence of cyclobutane [rather than the (6–4)] thymine dimer by direct photoreversal experiments at 254 nm. In addition, ODN4 was identified by electrospray ionization mass spectra on a PE Sciex (Thornhill, Canada) API 165 mass spectrometer (negative mode). Electrospray ionization-mass for ODN4 was: calculated 2407.5, found 2407.0.
HPLC Analyses of Photoirradiated BrU and Thymine Dimer-Containing Deoxyoctanucleotide.
The reaction mixture (total volume 10 μl in a microcentrifuge tube) contained deoxynucleotide in 5 mM sodium cacodylate buffer (pH 7.0) in the presence or absence of Sso7d protein. All of the photoreactions were performed under aerobic conditions. After irradiation with monochromatic UV light (HM-5 hypermonochromator, Jasco, Tokyo, Japan) at 0°C, reaction mixture was analyzed by HPLC. HPLC analysis was carried out with the PU-980 HPLC system (Jasco) equipped with a Chemcobond 5-ODS-H column (4.6 × 150 mm). Detection was carried out at 254 nm. Yields of reaction products were determined by comparison of their HPLC peak areas.
Fluorescence Measurements.
Steady-state fluorescence measurements on photoirradiated Sso7d were conducted with a Jasco FP-6300 spectrofluorometer. Measurements were performed by using a fluorescence cell with a 0.5-cm path length. As the fluorescence intensity of Sso7d is quenched when it forms a complex with DNA (20), it was difficult to estimate the conversion of the Trp in Sso7d. To solve this problem, we added a high concentration of NaCl (3 M final concentration) to the reaction mixture after the photoirradiation. As Sso7d cannot bind to DNA in such high-salt conditions, the fluorescence intensity of the Trp is recovered (>90%). The fluorescence intensity of the Trp of Sso7d was not affected in these conditions.
MALDI-TOF Mass Measurements of Enzymatic Digest of Sso7d.
The samples were digested by Lys-C (Wako Chemicals, Osaka, Japan) in 50 mM Tris·HCl, pH 9.2 for 4 h. The digested samples were desalted with a Millipore (Billerica, MA) microC18 Zip Tip. The mass spectra were measured by using an Applied Biosystems Voyager DE-PRO MALDI-TOF in the positive-ion, linear mode using an α-cyano-4-hydroxycinnamic acid matrix.
Supplementary Material
Acknowledgments
We thank Dr. T. Oyoshi (Shizuoka University, Shizuoka, Japan) for helpful discussions and Prof. K. Miki and Dr. T. Nonaka (Kyoto University) for MALDI-TOF MS measurements. This study was partly supported by a Grant-in-Aid for Priority Research from the Ministry of Education, Science, Sports, and Culture and a grant from Solution Oriented Research for Science and Technology of Japan Science and Technology. R.T. is a Research Fellow of the Japan Society for the Promotion of Science.
Abbreviations
- IU
5-iodouracil
- BrU
5-bromouracil.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Sancar A. Chem Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 2.DeRosa MC, Sancar A, Barton JK. Proc Natl Acad Sci USA. 2005;102:10788–10792. doi: 10.1073/pnas.0503527102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boon EM, Livingston AL, Chimiel NH, David SS, Barton JK. Proc Natl Acad Sci USA. 2003;100:12543–12547. doi: 10.1073/pnas.2035257100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yavin E, Boal AK, Stemp EDA, Boon EM, Livingston AL, O'Shea VL, David SS, Barton JK. Proc Natl Acad Sci USA. 2005;102:3546–3551. doi: 10.1073/pnas.0409410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim S, Li Y, Sancar A. Proc Natl Acad Sci USA. 1992;89:900–904. doi: 10.1073/pnas.89.3.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullis PM, Jones GDD, Symons MCR, Lea JS. Nature. 1987;330:773–774. doi: 10.1038/330773a0. [DOI] [PubMed] [Google Scholar]
- 7.Whiteman WB, Pfeifer F, Baum P, Klein A. Genetics. 1999;152:1245–1248. doi: 10.1093/genetics/152.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao YG, Su SY, Robinson H, Padmanabhan S, Lim L, McCrary BS, Edmondson SP, Shriver JW, Wang AH. Nat Struct Biol. 1998;5:782–786. doi: 10.1038/1822. [DOI] [PubMed] [Google Scholar]
- 9.Agback P, Baumann H, Knapp S, Ladenstein R, Härd T. Nat Struct Biol. 1998;5:579–584. doi: 10.1038/836. [DOI] [PubMed] [Google Scholar]
- 10.Robinson H, Gao Y-G, McCrary BS, Edmondson SP, Shriver JW, Wang AH-J. Nature. 1998;392:202–205. doi: 10.1038/32455. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y, Sugiyama H. Angew Chem Int Ed. 2006;45:1354–1362. doi: 10.1002/anie.200501962. [DOI] [PubMed] [Google Scholar]
- 12.Ito T, Rokita DS. J Am Chem Soc. 2004;126:15552–15559. doi: 10.1021/ja045637n. [DOI] [PubMed] [Google Scholar]
- 13.Kaden P, Mayer-Enthart E, Trifonov A, Fiebig T, Wagenknecht H-A. Angew Chem Int Ed. 2005;44:1636–1639. doi: 10.1002/anie.200462592. [DOI] [PubMed] [Google Scholar]
- 14.Tashiro R, Sugiyama H. J Am Chem Soc. 2003;125:15282–15283. doi: 10.1021/ja0380291. [DOI] [PubMed] [Google Scholar]
- 15.Sugiyama H, Tsutsumi Y, Saito I. J Am Chem Soc. 1990;112:6720–6721. [Google Scholar]
- 16.Oyoshi T, Wang A H-J, Sugiyama H. J Am Chem Soc. 2002;124:2086–2087. doi: 10.1021/ja016968s. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe T, Bando T, Xu Y, Tashiro R, Sugiyama H. J Am Chem Soc. 2005;127:44–45. doi: 10.1021/ja0454743. [DOI] [PubMed] [Google Scholar]
- 18.Lewis FD, Wu Y. J Photochem Photobiol C Photochem Rev. 2001;2:1–16. [Google Scholar]
- 19.Lundbäck T, Hansson H, Knapp S, Ladenstein R, Härd T. J Mol Biol. 1998;276:775–786. doi: 10.1006/jmbi.1997.1558. [DOI] [PubMed] [Google Scholar]
- 20.Behmoaras T, Toulme JJ, Hélène C. Nature. 1981;292:858–859. doi: 10.1038/292858a0. [DOI] [PubMed] [Google Scholar]
- 21.Ma Y-S, Chao C-C, Stadtman ER. Arch Biochem Biophys. 1999;363:129–134. doi: 10.1006/abbi.1998.1076. [DOI] [PubMed] [Google Scholar]
- 22.Buxton T, Guilbault GG. ClinChem. 1974;20:765–768. [PubMed] [Google Scholar]
- 23.Harriman A. J Phys Chem. 1987;91:6102–6104. [Google Scholar]
- 24.Chinnapen DJ-F, Sen D. Proc Natl Acad Sci USA. 2004;101:65–69. doi: 10.1073/pnas.0305943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.