Abstract
The Caenorhabditis elegans synthetic multivulva (synMuv) genes act redundantly to antagonize the specification of vulval cell fates, which are promoted by an RTK/Ras pathway. At least 26 synMuv genes have been genetically identified, several of which encode proteins with homologs that act in chromatin remodeling or transcriptional repression. Here we report the molecular characterization of two synMuv genes, lin-37 and lin-54. We show that lin-37 and lin-54 encode proteins in a complex with at least seven synMuv proteins, including LIN-35, the only C. elegans homolog of the mammalian tumor suppressor Rb. Biochemical analyses of mutants suggest that LIN-9, LIN-53, and LIN-54 are required for the stable formation of this complex. This complex is distinct from a second complex of synMuv proteins with a composition similar to that of the mammalian Nucleosome Remodeling and Deacetylase complex. The class B synMuv complex we identified is evolutionarily conserved and likely functions in transcriptional repression and developmental regulation.
Keywords: chromatin, transcription, vulval development, cell-fate specification, Ras
Cell differentiation requires coordinated changes in gene regulation. Such changes in gene expression are controlled by transcription factors and often are mediated through the modification of chromatin states. Biochemical techniques have identified many protein complexes that function in transcriptional repression and chromatin remodeling (1, 2). However, the in vivo functions of these complexes in organismal development have been unclear. Conversely, genetic studies of the nematode Caenorhabditis elegans have implicated genes likely to be involved in chromatin-mediated transcriptional repression as important in the development of the hermaphrodite vulva, but the biochemical properties of the products of these genes have been largely unknown.
The C. elegans vulva arises from three of six ectodermal blast cells that form the vulval equivalence group (3, 4). Each of these six cells, P3.p-P8.p, adopts one of three distinct cell fates, which are distinguished by their patterns of cell divisions and the descendant cell types they generate. The descendants of P5.p, P6.p, and P7.p form the vulva, through which sperm can enter and eggs can exit. Although P3.p, P4.p, and P8.p are competent to adopt vulval fates, they instead adopt a nonvulval fate, usually dividing once and fusing with the adjacent syncytial hypodermis. The specification of the vulval cell fates is antagonized by the synthetic multivulva (synMuv) genes (5, 6). The synMuv genes have been grouped into classes A, B, and C on the basis of genetic interactions (6, 7). Animals homozygous for loss-of-function mutations in any two synMuv gene classes have a multivulva (Muv) phenotype resulting from induction of more than three Pn.p cells, whereas animals homozygous for loss-of-function mutations in any single synMuv class are not Muv. Some class B synMuv genes encode proteins with homologs that function in other species in chromatin remodeling and transcriptional repression. These genes include lin-35, which encodes the only C. elegans homolog of the mammalian tumor-suppressor Rb, and efl-1 and dpl-1, which encode proteins homologous to the E2F and DP subunits of the heterodimeric transcription factor E2F (8, 9). The class B synMuv proteins LIN-53 RbAp48, HDA-1 HDAC1, and LET-418 Mi2 are homologous to components of the mammalian Nucleosome Remodeling and Deacetylase (NuRD) complex (8–16).
At least 12 additional class B synMuv genes have products that might act with LIN-35 Rb or components of a C. elegans NuRD-like complex in determining vulval cell fates (5, 6, 15, 17–19). However, genetic techniques are insufficient to determine how the synMuv proteins interact. In this study, we report the molecular identification of two class B synMuv genes, lin-37 and lin-54, and identify a previously undescribed class B synMuv protein complex that likely controls cell-fate specification through transcriptional repression.
Results
Class B synMuv Genes lin-37 and lin-54 Encode Proteins Conserved in Other Organisms.
Alleles of lin-37 and lin-54 were isolated in screens for mutations that cause a synMuv phenotype in combination with loss of function of a class A synMuv gene (6, 17). lin-37 and lin-54 are class B synMuv genes, as alleles of each gene cause a Muv phenotype with mutations in any of the four identified class A synMuv genes or with mutations in the class C synMuv gene trr-1 but not with mutations in other class B synMuv genes (refs. 6 and 17; Table 1, which is published as supporting information on the PNAS web site).
Using standard genetic techniques, we cloned lin-37 and demonstrated that it is equivalent to the gene ZK418.4 (Fig. 6 and Supporting Results in Supporting Text, which are published as supporting information on the PNAS web site). Mutations affecting the spliced mRNA were found in the two lin-37 alleles, n758 and n2234 (Supporting Results). A rescuing lin-37 cDNA is predicted to encode a hydrophilic protein of 275 aa with weak similarity to the Drosophila melanogaster protein Mip40 and related vertebrate proteins (20, 21).
We also cloned lin-54 and demonstrated that it is equivalent to JC8.6 (Supporting Results). To establish the null phenotype of lin-54, we isolated two deletions affecting JC8.6, n3423 and n3424 (Supporting Results). In addition to causing a class B synMuv phenotype (Table 1), both deletions caused a fully penetrant sterile phenotype, as do null mutations in some other class B synMuv genes (6, 8, 13, 17, 22–24).
Both lin-54 splice forms (Supporting Results) encode proteins rich in cysteines, which are clustered in two domains that share a nearly identical pattern and spacing. Domains with this signature CXCX4CX4CXCX6CX2–3CXCX2C sequence are found in proteins from plants, insects, and mammals (ref. 25; Fig. 7C, which is published as supporting information on the PNAS web site). Some of these proteins have similarity to LIN-54 outside of the cysteine-rich domains, including the Drosophila protein Mip120 and the soybean protein CPP1, which have sequence-specific DNA-binding activities (21, 25). The DNA-binding activity of CPP-1 was mapped to the cysteine-rich domains. Recently, yeast-one hybrid and chomatin immunoprecipitation studies have shown that LIN-54 can bind to the promoters of multiple genes expressed in the C. elegans digestive tract (26). In addition, Drosophila Mip120 interacts with the homologs of other class B synMuv proteins (20, 21, 27, 28).
LIN-37 and LIN-54 Are Broadly Expressed in Nuclei Throughout Development.
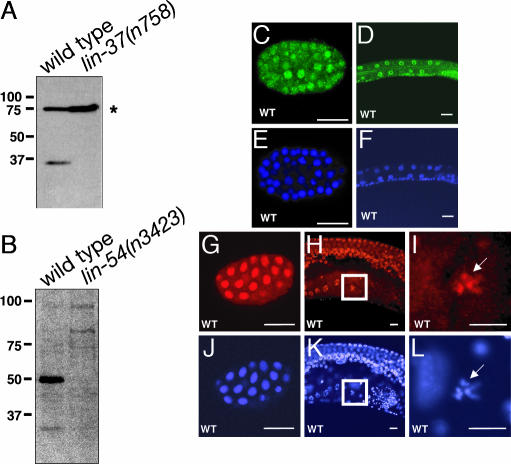
We generated antibodies that specifically recognize LIN-37 and LIN-54 in Western blots and immunostaining. Antibodies raised against LIN-37 recognized a protein of ≈35 kDa on Western blots of WT protein extracts but not of protein extracts from lin-37(n758) mutant animals (Fig. 1A). Antibodies raised against LIN-54 recognized a protein of ≈50 kDa on Western blots of WT protein extracts, but not of lin-54(n3423) mutant extracts (Fig. 1B). In immunostained whole mounts, LIN-37 was detected in most if not all nuclei of WT animals from the one-cell embryo through the adult (Fig. 1 C and D and data not shown) and was absent in lin-37(n758) mutant animals (data not shown). Immunostained whole mounts of WT animals demonstrated that, like LIN-37, LIN-54 was present in the nuclei of all or almost all cells from the embryo through the adult (Fig. 1 G–I and data not shown). In the hermaphrodite germ line, LIN-54 was localized to condensed chromosomes during the diakinesis phase of meiosis (Fig. 1 H and I). LIN-54 was not detected in lin-54(n3423) mutant animals (data not shown).
Fig. 1.
LIN-37 and LIN-54 are expressed broadly in nuclei throughout development. (A) Anti-LIN-37 antibodies were used to blot extracts from WT and lin-37(n758) mutant animals. The asterisk denotes nonspecific immunoreactivity. (B) Anti-LIN-54 antibodies were used to blot extracts from both WT and lin-54(n3423) mutant animals, which were derived from heterozygous mothers. (C and D) Whole-mount staining with anti-LIN-37 antisera of an embryo and the midbody of an L1 larva, respectively. (E and F) DAPI staining of the animals shown in C and D. (G and H) Whole-mount staining with anti-LIN-54 antisera of an embryo and the midbody of an adult hermaphrodite, showing the germ line, respectively. (J and K) DAPI staining of the animals shown in G and H. (I and L) Enlargements of the boxed portions of H and K, respectively. (Scale bars: 10 μm.)
LIN-37, LIN-54, and Other Class B synMuv Proteins form a High Molecular Mass Protein Complex.
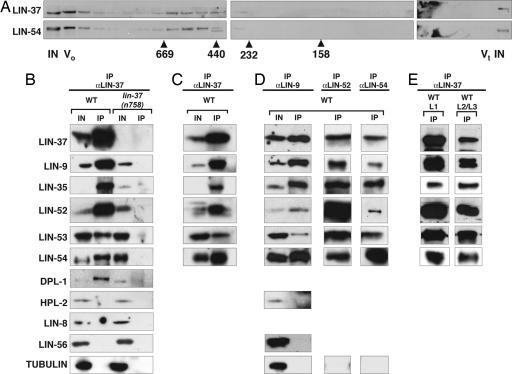
Previous studies demonstrated that some class B synMuv proteins can interact in vitro and in vivo (8, 9, 15, 29). We performed gel-filtration experiments with embryonic protein extracts and detected LIN-37 and LIN-54 in fractions corresponding to an apparent molecular mass between 440 and 669 kDa. As both LIN-37 and LIN-54 coelute above their monomeric molecular mass, these proteins might be members of multisubunit complexes (Fig. 2A).
Fig. 2.
A subset of class B synMuv proteins form a complex in vivo. (A) Embryo extracts were subjected to Superose 6 gel filtration. Antibodies specific to LIN-37 and LIN-54 were used for immunoblotting the fractions. Elution peaks for protein standards (in kilodaltons) are indicated below the fractions. (B) Embryo extracts from WT and lin-37(n758) mutant animals were precipitated with anti-LIN-37 antibodies. Proteins were separated by SDS/PAGE and antibodies specific to the antigens indicated on the left were used for immunoblotting. (C) Immunoprecipitations were performed by using WT embryonic extracts in the presence of 50 μg/ml ethidium bromide. (D) Immunoprecipitations with antibodies that recognized LIN-9, LIN-52, and LIN-54 were performed by using WT embryonic extracts. Immunoblots of coprecipitating LIN-52 from L1 larval extracts with anti-LIN-54 antibodies is shown. (E) Extracts from WT L1 or late L2/early L3 larvae were precipitated as in B. IN, 2% of input for immunoprecipitations; 10% of input for gel filtration chromatography. IP, 100% of the immunoprecipitate. V0, void volume. Vt, total volume.
To identify components of these complexes we performed coimmunoprecipitation experiments. We used anti-LIN-37 antibodies to immunoprecipitate proteins from WT and lin-37(n758) mutant embryo extracts and determined the presence of coimmunoprecipitating synMuv proteins by using Western blots. The class B synMuv proteins LIN-9, LIN-35 Rb, LIN-52, LIN-53 RbAp48, LIN-54, and DPL-1 DP were specifically coimmunoprecipitated by anti-LIN-37 antiserum from WT but not lin-37(n758) extracts (Fig. 2B). Neither the class B synMuv protein HPL-2, the class A synMuv protein LIN-56, nor tubulin coimmunoprecipitated with LIN-37 from either WT or mutant protein extracts, suggesting that the identified interactions are specific (Fig. 2B). The same set of proteins also coimmunoprecipitated with LIN-37 in the presence of 50 μg/ml ethidium bromide. Because ethidium bromide can disrupt complexes that depend on DNA structure and DNA binding (30), the observed interactions likely are not DNA-dependent (Fig. 2C). The same class B synMuv proteins also were precipitated by antibodies that recognize LIN-9, LIN-52, or LIN-54 (Fig. 2D) but not by antibodies that recognize the synMuv proteins LIN-61 or LIN-56 (data not shown). We failed to detect DPL-1 in some of the coimmunoprecipitates, but because we also failed to detect DPL-1 in the input, we believe this result reflects poor recognition by the antibody and not the absence of DPL-1. Our analyses of immunoprecipitates from extracts of synchronized cultures demonstrated that these proteins remain associated throughout larval development, including the late L2 and early L3 stages when the fates of the Pn.p cells are specified (Fig. 2E).
Antibodies raised against the class B synMuv protein EFL-1 can detect EFL-1 in immunostained whole mounts (31) but not in Western blots, so we could not assess immunoprecipitates for the presence of EFL-1. All known DP-like proteins exist as heterodimers with E2F proteins (32). For this reason, we believe it likely that at least one of the two C. elegans members of the E2F family, EFL-1 or EFL-2 (8, 31), associates with the precipitated proteins in vivo. EFL-1 is a probable candidate, because efl-1 is a class B synMuv gene (8). Because this complex contains homologs of the known transcriptional regulators Rb and DP and mutations in members of the complex cause a Muv phenotype we refer to this complex, containing LIN-9, LIN-35, LIN-37, LIN-52, LIN-53, LIN-54, DPL-1, and EFL-1, as the DP, Rb, and MuvB (DRM) complex. This complex includes only a subset of class B synMuv proteins, because LIN-37 fails to coimmunoprecipitate with LIN-36::GFP (Fig. 8, which is published as supporting information on the PNAS web site) and HPL-2 does not coimmunoprecipitate with LIN-37 or LIN-9 (Fig. 2 B and D). It is possible that LIN-37 and/or LIN-54 were present in the extracts we studied not only in the DRM complex, but also in one or more protein complexes.
The DRM Complex Is Distinct from a NuRD-Like Complex.
The mammalian homologs of the synMuv proteins HDA-1 HDAC1 (14), LET-418 Mi2 (13), and LIN-53 RbAp48 (9) are components of the mammalian NuRD complex (10, 12, 33). By analogy to mammalian systems and because HDA-1 and LET-418 associate in vivo (15), it has been proposed that HDA-1, LET-418, and LIN-53 form a NuRD-like complex in C. elegans (16). Because mammalian HDAC1 interacts with Rb, it has been suggested that this C. elegans NuRD-like complex is recruited to promoters of vulval genes by the class B synMuv proteins LIN-35 Rb, EFL-1 E2F, and DPL-1 DP (8, 13–16). Because we identified LIN-35 and DPL-1 as components of the DRM complex, we tested whether this complex also includes the chromatin-modifying enzymes HDA-1 HDAC1 and LET-418 Mi2.
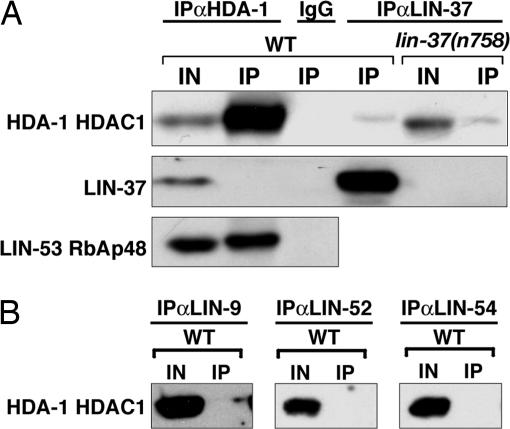
HDA-1 failed to specifically coprecipitate with LIN-37 (Fig. 3A). A small amount of HDA-1 nonspecifically coprecipitated with LIN-37 from both WT and lin-37(n758) embryonic extracts, similar to the nonspecific HDA-1 precipitation noted by Unhavaithaya et al. (15). Similarly, HDA-1 did not coimmunoprecipitate with LIN-37 in extracts from L3 larvae (data not shown). In addition, HDA-1 did not precipitate with LIN-9, LIN-52, or LIN-54 (Fig. 3B). Reciprocally, LIN-37 did not coimmunoprecipitate with HDA-1 (Fig. 3A). By contrast, we found that LIN-53 coimmunoprecipitated with HDA-1 (Fig. 3A). Previous data suggested that LET-418 associates with HDA-1 (15), and in agreement with these data, we observed LET-418 in HDA-1 immunoprecipitates (data not shown). Together, these results suggest that HDA-1, LET-418, and LIN-53 are found in a complex distinct from the DRM complex. We propose that LIN-53 is present in both the DRM complex and a NuRD-like complex. RbAp48, the mammalian homolog of LIN-53, has been found similarly in multiple chromatin-modifying complexes, including the chromatin assembly complex Caf1, the NuRD complex, and the Sin3 complex (10, 11, 34, 35).
Fig. 3.
NuRD-like complex components do not precipitate with the DRM complex. (A) Extracts from either WT or lin-37(n758) mutant animals were precipitated with antibodies against either HDA-1, LIN-37, or rabbit IgG. Immunoprecipitations were analyzed by using antibodies specific to the antigens indicated on the left. (B) WT embryo extracts were precipitated with antibodies that recognize LIN-9, LIN-52, or LIN-54 and were analyzed by Western blots with anti-HDA-1 antibodies. IN, 2% of the input. IP, 100% of the immunoprecipitate.
Mutations in NuRD-Like Complex Components Cause Vulval Developmental Defects Different from Those Caused by Animals Mutant for DRM Complex Components.
All of the proteins identified in the DRM complex, as well as HDA-1 and LET-418, are considered to be class B synMuv proteins (8, 9, 13, 14, 17, 24). Our data suggest that these proteins can be found in two distinct complexes. Interestingly, mutations that disrupt the two complexes have distinct phenotypic consequences. Whereas most class B synMuv mutants are completely non-Muv in the absence of mutations in class A or class C synMuv genes, hda-1 and let-418 single mutants display a weak Muv phenotype. Specifically, 20–31% hda-1 mutants are Muv (ref. 14 and data not shown), and 1.5% of let-418(n3536) mutants are Muv when cultured at 22.5°C (M.M.H., C.J.C. and H.R.H., unpublished results). In addition, in 7% of let-418(ar114) animals P8.p is induced (13); P8.p induction is a Muv phenotype (7). let-418 is unlike most class B synMuv genes in that the level of P8.p induction of let-418(ar114) animals is enhanced to 13–20% by a typical class B mutation (13), whereas a second class B mutation does not alter the vulval phenotype of most class B mutants. Together with our biochemical data, these genetic data suggest that members of the NuRD-like complex have functions distinct from those of the DRM complex in the regulation of vulval development.
DRM Complex Formation Is Sensitive to Mutations in lin-9, lin-53, and lin-54.
We investigated whether the DRM complex can form properly when specific components are either absent or mutated. We analyzed extracts from presumptive lin-35 and lin-37 null mutants, because these mutants can be maintained as homozygotes and therefore can be grown in sufficient quantities for biochemical analyses. For the genes lin-9, lin-52, lin-53, and lin-54, which have sterile null phenotypes (refs. 8, 17, and 24 and Supporting Materials and Methods in Supporting Text), we generated protein extracts from embryos with viable missense mutations that confer synMuv phenotypes.
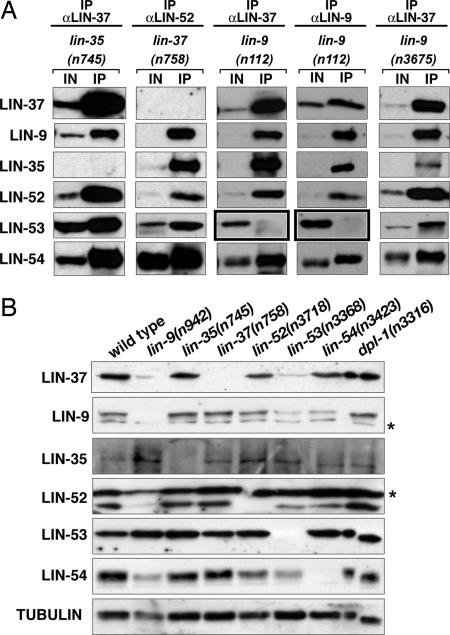
In the absence of either LIN-35 Rb or LIN-37, the remaining DRM complex components coprecipitate (Fig. 4A). The missense mutations in lin-52(n771), lin-53(n833), and lin-54(n2231) also did not affect the associations among complex members (data not shown). By contrast, a missense mutation in lin-9, n112, reduced the level of LIN-53 but not other DRM complex members in coimmunoprecipitations with either anti-LIN-37 antiserum or anti-LIN-9 antiserum (Fig. 4A, boxed immunoblots). The lin-9(n112) mutation affects a conserved residue (G341E) in a domain of LIN-9 that is conserved in the homologous human protein hLin9 and the homologous Drosophila proteins ALY and Mip130/TWIT (21, 36, 37). The only other identified missense mutation of lin-9 that causes a homozygous viable class B synMuv phenotype is n3675 (38). However, lin-9(n3675) did not reduce the levels of LIN-53 RbAp48 in the DRM complex (Fig. 4A). The lin-9(n3675) mutation (D305N) is in the same highly conserved region as the n112 mutation, but the altered residue is not conserved in Drosophila ALY or human Lin-9. These data suggest that an interaction between LIN-9 and LIN-53 might be important for the incorporation of LIN-53 into the DRM complex, but complex assembly cannot be the sole function of LIN-9 in vulval development. Specifically, lin-9(n3675) causes a synMuv phenotype but does not disrupt LIN-53 incorporation into the DRM complex, suggesting that another function of LIN-9 is modified in this mutant.
Fig. 4.
Analysis of the DRM complex and its components in mutant backgrounds. (A) Anti-LIN-9, anti-LIN-37, or anti-LIN-52 antibodies were used to immunoprecipitate proteins from embryo extracts of the indicated genotype. Immunoprecipitations were analyzed by using the antibodies specific to the antigens listed on the left. IN, 2% of the input. IP, 100% of the immunoprecipitate. Black boxes denote places in which changes from WT are observed. (B) Protein from 100 L4 larvae of the genotype noted above each column was loaded per lane. The antibodies used for each blot are indicated on the left. Anti-tubulin antibodies were used to assess protein loading and transfer. Asterisks denote nonspecific immunoreactivity.
We could not obtain quantities of protein extract sufficient to analyze the association of DRM complex components by coimmunoprecipitation from sterile null mutants. Instead, we analyzed the total protein levels of DRM complex components when each member was removed by mutation to see whether the absence of one complex member affects the level of any other. In some cases, the absence of a core member of a protein complex can prevent the formation of the complex and can result in the degradation of other complex members. To compare sterile mutants to the WT, we examined L4 larvae, because adult WT hermaphrodites contain developing embryos that are absent in sterile mutants.
Null mutations in lin-9, lin-53, and lin-54 caused decreases in the protein levels of other DRM complex components, including LIN-9, LIN-37, LIN-52, and LIN-54 (Fig. 4B). In addition to lacking LIN-9, lin-9 null mutants also had decreased levels of LIN-37, LIN-52, and LIN-54 as compared with WT levels. Null mutants in lin-53 lacked LIN-53 and showed decreased levels of LIN-9, LIN-37, LIN-52, and LIN-54. In lin-54 null mutants, levels of LIN-9 and LIN-52 were decreased and, as expected, LIN-54 was absent. Loss of LIN-35, LIN-37, or DPL-1 proteins did not significantly affect the protein levels of any other DRM complex components. LIN-35 and LIN-53 levels remained unchanged in all genotypes examined, except as expected in lin-35 or lin-53 mutants, respectively (Fig. 4B). The mammalian homologs of these two proteins, Rb and RbAp48, are found in multiple complexes, and, thus, their overall levels might not depend on the presence of any single protein complex. Our data suggest that a complete loss of LIN-9, LIN-53, or LIN-54 can disrupt DRM complex formation and result in the degradation of other complex members. Alternatively, it is possible that the loss of any of these three proteins causes a decrease in the transcription or translation of some of the genes that encode DRM complex members.
Discussion
Here we describe the molecular identification and characterization of two class B synMuv genes, lin-37 and lin-54. We show that LIN-37 and LIN-54 form a multisubunit protein complex together with at least five other class B synMuv proteins: LIN-9, LIN-35 Rb, LIN-52, LIN-53 RbAp48, and DPL-1 DP. This DRM complex is biochemically and genetically distinct from a NuRD-like complex that includes HDA-1 HDAC1, LET-418 Mi2, and LIN-53 RbAp48. These findings suggest that LIN-35 Rb and DPL-1 DP likely have a function in vulval development distinct from recruitment of the NuRD complex.
The DRM Complex Likely Functions in Transcriptional Repression.
The DRM complex is similar to two recently described and highly similar complexes that contain several Drosophila homologs of class B synMuv proteins (20, 28). The Myb–MuvB complex was purified by immunoprecipitation of the LIN-54 homolog Mip120 or the LIN-9 homolog Mip130 from Drosophila tissue-culture cells and coimmunoprecipitating proteins were identified by mass spectrometry. The Myb–MuvB complex contains stoichiometric levels of Mip130, RBF, Mip40, Mip120, p55, dDP, dE2F2, and dLin52, which are homologs of LIN-9, LIN-35, LIN-37, LIN-54, LIN-53, DPL-1, EFL-1, and LIN-52, respectively. This complex also contains substoichiometric amounts of Rpd3, the fly homolog of HDA-1, and L(3)MBT, a protein similar to the class B synMuv protein LIN-61 (28). The dREAM complex was identified by biochemical purification of Drosophila Rb-containing complexes from embryo extracts followed by mass spectrometry and Western blot analyses. The dREAM complex contains all of the proteins identified in the Myb–MuvB complex at stoichiometric levels except for dLin52 (20). The differences between the dREAM and Myb–MuvB complexes might be a consequence of the methods used for purification or might reflect the existence in different tissues or during different developmental stages of multiple subcomplexes with overlapping components. Both the dREAM and Myb–MuvB complexes can mediate transcriptional repression of many E2F-target genes (20, 28).
The similarity between the C. elegans DRM complex and the Drosophila dREAM and Myb–MuvB complexes indicates that the DRM complex likely also acts in transcriptional repression. Given the broad expression patterns of the synMuv genes and the multiple phenotypic abnormalities caused by the loss of individual synMuv proteins, we propose that, similar to their Drosophila counterparts, the DRM complex proteins are involved in the repression of many targets important for diverse biological functions.
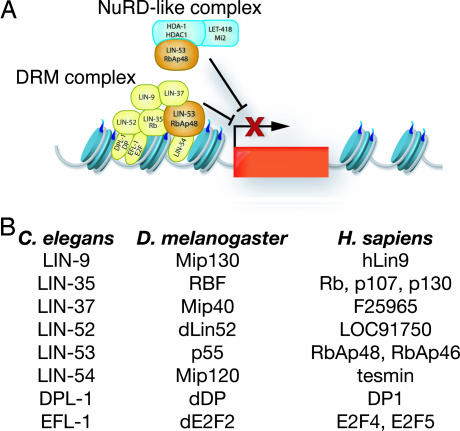
The DRM complex differs slightly from both the dREAM and the Myb–MuvB complexes. Unlike the dREAM complex (20), the DRM complex contains a LIN-52 dLin52-like protein. Unlike the Myb–MuvB complex (28), the DRM complex does not contain HDA-1 Rpd3 or LIN-61 L(3)MBT (M.M.H. and H.R.H., unpublished results). The similarities of the DRM, dREAM, and Myb–MuvB complexes suggest that there is a core complex consisting of LIN-35 RBF, EFL-1 E2F2, DPL-1 DP, LIN-9 Mip130, LIN-37 Mip40, LIN-52 dLin-52, LIN-53 p55, and LIN-54 Mip120 and that this complex might associate with other proteins during specific stages of development or in certain cell types (Fig. 5A).
Fig. 5.
A model of the DRM and NuRD-like complexes. (A) The DRM and NuRD-like complexes are distinct and act to repress the transcription of genes that promote the expression of vulval cell fates (indicated by the red box). It is not known whether the DRM and NuRD-like complexes act on the same set of target genes. LIN-53 RbAp48 is a component of both the NuRD-like and the DRM complexes. See text for details. (B) A list of DRM complex members and the homologs found in D. melanogaster and Homo sapiens.
The dREAM and Myb–MuvB complexes both contain the DNA-binding protein Myb (20, 28). There is no clear Myb homolog in C. elegans (20, 39). It is possible that the C. elegans DRM complex does not contain a Myb ortholog or that the functional ortholog of the Drosophila Myb protein found in the dREAM and Myb–MuvB complexes might not be readily identifiable by comparisons of primary sequence.
We propose that the DRM complex could be recruited to DNA by multiple DNA-binding factors, including LIN-54 and the heterodimeric transcription factor formed by EFL-1 and DPL-1 (Fig. 5A). The DRM complex could then act with the NuRD-like complex to repress transcription. Alternatively, the DRM complex and the NuRD-like complex could act sequentially. The NURD-like complex could deacetylate the N-terminal tails of histones, and the DRM subsequently could bind these unmodified histone tails, preventing their acetylation. The dREAM complex previously has been shown to bind unmodified histone H4 tails, supporting this model (20). This binding might be mediated by LIN-53, because the mammalian homolog RbAp48 binds histone H4 (35). Deacetylated histones are associated with transcriptionally repressed areas of the genome. Thus, by protecting histone tails from future acetylation, the DRM complex could act to maintain transcriptional repression of nearby genes.
The DRM Complex and a NuRD-Like Complex Have Separable Functions During Vulval Development.
Although neither the DRM nor the dREAM complexes contains known chromatin-modifying or chromatin-remodeling enzymes, these complexes might require the activity of a histone deacetylase to mediate transcriptional repression, as noted above. Mutations in genes encoding components of either the DRM or the NuRD-like complex require an additional class A or class C synMuv mutation to produce a highly penetrant Muv phenotype. However, we found that mutations affecting two of the NuRD-like complex components, HDA-1 and LET-418, alone can cause low penetrance Muv phenotypes, suggesting that the chromatin-remodeling and chromatin-modifying activities of this complex might be required more broadly for the transcriptional repression of genes necessary for proper vulval development than is the activity of the DRM complex. Perhaps other class B synMuv proteins not associated with the DRM complex, for example, HPL-2, LIN-36, or LIN-61, act with the DRM complex to maintain the repressed state formed by the activity of the NuRD-like complex.
Does Rb Function as a Component of a DRM-Like Complex to Control Development in Mammals?
The high degree of conservation shared by the DRM/Myb-MuvB/dREAM complexes in C. elegans and Drosophila and the important roles that the components of DRM complex play in C. elegans development suggest that a similar complex exists in other organisms, including humans. The core components of these complexes have homologs in humans (Fig. 5B), and the human homolog of LIN-9, hLin-9, can associate with Rb to specifically promote differentiation (but not to inhibit cell-cycle progression) (37). Perhaps Rb or other Rb-family proteins act within the context of a DRM-like complex to control differentiation. Rb could act as a tumor suppressor through such DRM-mediated regulation of differentiation in addition to its role in cell-cycle regulation. Further biochemical and genetic studies of nematodes, insects, and mammals should elucidate the role that this conserved protein complex plays in development and in carcinogenesis.
Materials and Methods
Culture Conditions and Strains.
All strains were cultured at 20°C, unless otherwise specified, on NGM agar seeded with E. coli strain OP50 (40). N2 (Bristol) was the WT strain. Mutant alleles used are described in Supporting Material and Methods.
Transgenic Strains.
Germ-line transformations were performed as described in ref. 41. For details, see Supporting Materials and Methods.
Antibody Preparation, Immunocytochemistry, and Western Blots.
For details of antibody preparation and specificity, see Supporting Materials and Methods and Fig. 9, which is published as supporting information on the PNAS web site. All antibodies were used at a 1:1,000 dilution for Western blots, except for anti-LIN-53, anti-LIN-54, and anti-DPL-1, which were used at 1:500. Larvae and adults for immunostaining were fixed as described in ref. 42. Embryos were fixed as described in ref. 43. Antibodies were used at a 1:100 dilution for immunocytochemistry.
Embryo Lysates.
Embryos were harvested from liquid cultures, resuspended in 1 ml of lysis buffer (Supporting Materials and Methods) for each gram of embryos and frozen in liquid nitrogen. The embryos were thawed and sonicated. Lysates were clarified, and the protein concentration was determined. Lysates were diluted to 5–10 mg/ml and were used immediately or stored at −80°C.
Immunoprecipitation Experiments.
Antibodies were cross-linked to Protein A Dynabeads (Invitrogen, Carlsbad, CA) (Supporting Material and Methods). Five hundred microliters of precleared lysate were incubated with antibody-bound beads at 4°C for 1–2 h and then were washed three times for 5 min each at 4°C in lysis buffer. The beads were resuspended in 20 μl of 2× protein sample buffer and separated by SDS/PAGE. In most cases, HRP-conjugated protein A (Bio-Rad, Hercules, CA) was used for detection of protein by Western blot after coimmunoprecipitation experiments.
Gel Filtration Chromatography.
Embryo extract in buffer [15 mM Hepes pH 7.6/0.1 M KCl/3 mM MgCl2/0.1 mM EDTA/10% glycerol with Complete EDTA-free protease inhibitors (Roche Applied Sciences, Mannheim, Germany)] was precipitated with solid ammonium sulfate added to a final concentration of 20%. The supernatant from this precipitation then was brought to a concentration of 30% ammonium sulfate, and protein pellets were resuspended in elution buffer (EB) (25 mM Hepes pH 7.6/150 mM KCl/50 mM MgCl2/1 mM DTT/0.1 mM EDTA/0.1% Nonidet P-40/10% glycerol) combined, and 50 μl were loaded onto a preequilibrated Superose 6 PC 3.2/30 column (GE Healthcare, Piscataway, NJ). Samples were eluted by using EB and collected in 75-μl fractions. Proteins of known molecular masses (GE Healthcare, Piscataway, NJ) were used as standards.
Supplementary Material
Acknowledgments
We thank our colleagues for helpful comments and discussion about the manuscript, B. Castor for DNA sequence determination, N. An for strain management, members of the H.R.H. laboratory for construction of the deletion library, J. Bowers and S. Bell for assistance with gel filtration chromatography, A. Coulson for cosmid clones, Y. Kohara for cDNA clones, the C. elegans Genome Sequencing Consortium for genomic sequence, and the C. elegans Genetics Center for some of the strains used in this work. This work was supported by National Institutes of Health Grant GM24663. M.M.H. was a Howard Hughes Predoctoral Fellow. C.J.C. was a David H. Koch Fellow. H.R.H. is the David H. Koch Professor of Biology at Massachusetts Institute of Technology and an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- synMuv
synthetic multivulva
- NuRD
Nucleosome Remodeling and Deacetylase
- DRM
DP/Rb/MuvB
Footnotes
The authors declare no conflict of interest.
References
- 1.Becker PB, Horz W. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 2.Ayer DE. Trends Cell Biol. 1999;9:193–198. doi: 10.1016/s0962-8924(99)01536-6. [DOI] [PubMed] [Google Scholar]
- 3.Sulston JE, Horvitz HR. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 4.Sulston JE, White JG. Dev Biol. 1980;78:577–597. doi: 10.1016/0012-1606(80)90353-x. [DOI] [PubMed] [Google Scholar]
- 5.Horvitz HR, Sulston JE. Genetics. 1980;96:435–454. doi: 10.1093/genetics/96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferguson EL, Horvitz HR. Genetics. 1989;123:109–121. doi: 10.1093/genetics/123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceol CJ, Horvitz HR. Dev Cell. 2004;6:563–576. doi: 10.1016/s1534-5807(04)00065-6. [DOI] [PubMed] [Google Scholar]
- 8.Ceol CJ, Horvitz HR. Mol Cell. 2001;7:461–473. doi: 10.1016/s1097-2765(01)00194-0. [DOI] [PubMed] [Google Scholar]
- 9.Lu X, Horvitz HR. Cell. 1998;95:981–991. doi: 10.1016/s0092-8674(00)81722-5. [DOI] [PubMed] [Google Scholar]
- 10.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Zelewsky T, Palladino F, Brunschwig K, Tobler H, Hajnal A, Muller F. Development (Cambridge, UK) 2000;127:5277–5284. doi: 10.1242/dev.127.24.5277. [DOI] [PubMed] [Google Scholar]
- 14.Dufourcq P, Victor M, Gay F, Calvo D, Hodgkin J, Shi Y. Mol Cell Biol. 2002;22:3024–3034. doi: 10.1128/MCB.22.9.3024-3034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Unhavaithaya Y, Shin TH, Miliaras N, Lee J, Oyama T, Mello CC. Cell. 2002;111:991–1002. doi: 10.1016/s0092-8674(02)01202-3. [DOI] [PubMed] [Google Scholar]
- 16.Solari F, Ahringer J. Curr Biol. 2000;10:223–226. doi: 10.1016/s0960-9822(00)00343-2. [DOI] [PubMed] [Google Scholar]
- 17.Thomas JH, Ceol CJ, Schwartz HT, Horvitz HR. Genetics. 2003;164:135–151. doi: 10.1093/genetics/164.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Owen AB, Stuart J, Mach K, Villeneuve AM, Kim S. Genome Res. 2003;13:1828–1837. doi: 10.1101/gr.1125403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poulin G, Dong Y, Fraser AG, Hopper NA, Ahringer J. EMBO J. 2005;24:2613–2623. doi: 10.1038/sj.emboj.7600726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korenjak M, Taylor-Harding B, Binne UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A. Cell. 2004;119:181–193. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 21.Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR. Nature. 2002;420:833–837. doi: 10.1038/nature01228. [DOI] [PubMed] [Google Scholar]
- 22.Thomas JH, Horvitz HR. Development (Cambridge, UK) 1999;126:3449–3459. doi: 10.1242/dev.126.15.3449. [DOI] [PubMed] [Google Scholar]
- 23.Melendez A, Greenwald I. Genetics. 2000;155:1127–1137. doi: 10.1093/genetics/155.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beitel GJ, Lambie EJ, Horvitz HR. Gene. 2000;254:253–263. doi: 10.1016/s0378-1119(00)00296-1. [DOI] [PubMed] [Google Scholar]
- 25.Cvitanich C, Pallisgaard N, Nielsen KA, Hansen AC, Larsen K, Pihakaski-Maunsbach K, Marcker KA, Jensen EO. Proc Natl Acad Sci USA. 2000;97:8163–8168. doi: 10.1073/pnas.090468497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deplancke B, Mukhopadhyay A, Ao W, Elewa AM, Grove CA, Martinez NJ, Sequerra R, Doucette-Stamm L, Reece-Hoyes JS, Hope IA, et al. Cell. 2006;125:1193–1205. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 27.Beall EL, Bell M, Georlette D, Botchan MR. Genes Dev. 2004;18:1667–1680. doi: 10.1101/gad.1206604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis PW, Beall EL, Fleischer TC, Georlette D, Link AJ, Botchan MR. Genes Dev. 2004;18:2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walhout AJ, Sordella R, Lu X, Hartley JL, Temple GF, Brasch MA, Thierry-Mieg N, Vidal M. Science. 2000;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- 30.Lai JS, Herr W. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Page BD, Guedes S, Waring D, Priess JR. Mol Cell. 2001;7:451–460. doi: 10.1016/s1097-2765(01)00193-9. [DOI] [PubMed] [Google Scholar]
- 32.Dyson N. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 33.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Sun ZW, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- 35.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 36.White-Cooper H, Leroy D, MacQueen A, Fuller MT. Development (Cambridge, UK) 2000;127:5463–5473. doi: 10.1242/dev.127.24.5463. [DOI] [PubMed] [Google Scholar]
- 37.Gagrica S, Hauser S, Kolfschoten I, Osterloh L, Agami R, Gaubatz S. EMBO J. 2004;23:4627–4638. doi: 10.1038/sj.emboj.7600470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ceol CJ, Stegmeier F, Harrison MM, Horvitz HR. Genetics. 2006;173:709–726. doi: 10.1534/genetics.106.056465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lipsick JS. Genes Dev. 2004;18:2837–2844. doi: 10.1101/gad.1274804. [DOI] [PubMed] [Google Scholar]
- 40.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mello CC, Kramer JM, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finney M, Ruvkun G. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- 43.Guenther C, Garriga G. Development (Cambridge, UK) 1996;122:3509–3518. doi: 10.1242/dev.122.11.3509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.