Abstract
Cytotoxic T lymphocytes (CTL) can respond to a few viral peptide-MHC-I (pMHC-I) complexes among a myriad of virus-unrelated endogenous self pMHC-I complexes displayed on virus-infected cells. To elucidate the molecular recognition events on live CTL, we have utilized a self-assembled biosensor composed of semiconductor nanocrystals, quantum dots, carrying a controlled number of virus-derived (cognate) and other (noncognate) pMHC-I complexes and examined their recognition by antigen-specific T cell receptor (TCR) on anti-virus CD8+ T cells. The unique architecture of nanoscale quantum dot/pMHC-I conjugates revealed that unexpectedly strong multivalent CD8–MHC-I interactions underlie the cooperative contribution of noncognate pMHC-I to the recognition of cognate pMHC-I by TCR to augment T cell responses. The cooperative, CD8-dependent spread of signal from a few productively engaged TCR to many other TCR can explain the remarkable ability of CTL to respond to virus-infected cells that present few cognate pMHC-I complexes.
Keywords: CD8 coreceptor, peptide MHC clustering, sensitivity of T cell responses, antivirus immunity
Recognition of peptide-MHC (pMHC) complexes presented on the surface of infected cells by T cell receptors (TCR) initiates the T cell response against these cells. While TCR specifically recognizes peptide and polymorphic helices of MHC class I and class II proteins (1), the nonpolymorphic domain of these MHC proteins also interacts with the CD8 and CD4 coreceptors that mark distinct populations of T lymphocytes, i.e., CD8+ and CD4+ T cells. The MHC–coreceptor interactions occur regardless of the nature of MHC-bound peptide (2). Because MHC-I is expressed on every nucleated cell, CD8+ T cells (also known as cytotoxic T lymphocytes or CTL), can recognize and, if necessary, respond to virtually any infected cell in the body, and thus play a key role in anti-virus immunity.
Unraveling molecular recognition events as they occur at the cell membrane of T cells is essential for understanding mechanisms responsible for the unique sensitivity and selectivity of the T cell response. Soluble MHC proteins and their oligomers (3) have been extensively used for this purpose. However, the majority of these oligomers, MHC tetramers in particular, may not mimic the orientation and the typical distances between MHC monomers in their natural environment, and these limitations may influence the results of experiments in which these oligomers are used to explore TCR-mediated molecular events on the T cell surface. Indeed, the architecture of a multivalent ligand that binds to clustered receptors can strongly influence the ligand's biological activity (4).
Luminescent semiconductor nanocrystals (quantum dots, QDs) provide excellent nanoscaffolds to array several receptor molecules on their surfaces by metal-affinity-driven self-assembly, providing aggregate-free solutions of QD conjugates that specifically recognize their respective ligands (5–9).
In this report, we use self-assembled QD/pMHC-I conjugates (in which the MHC-I protein is HLA-A2) to evaluate mechanisms of antigen recognition by TCR on live CD8+ T cells. These QD/pHLA-A2 conjugates offer (i) a unique architecture with coherent orientation and spacing between proteins on the nanocrystal and (ii) control over the number of pHLA-A2 complexes immobilized on each QD. The binding of QDs carrying a predetermined number of cognate and noncognate pHLA-A2 complexes to live T cells shows that strong multivalent interactions between CD8 and MHC-I dominate the stability of TCR/pMHC-I/CD8 complexes and underlie striking noncognate pMHC-I cooperation with cognate pMHC-I that enhances T cell responses.
On the basis of these and other findings we suggest that strong, TCR-independent CD8–MHC-I interactions precede recognition of pMHC-I by TCR and facilitate rapid signal spreading from a few agonist-bound TCR to many other receptors within TCR clusters. This cooperative mechanism explains the highly sensitive response of CTL, which can respond to as few as perhaps a single cognate pMHC-I on a target cell (10, 11).
Results
The QD/pMHC Biosensor.
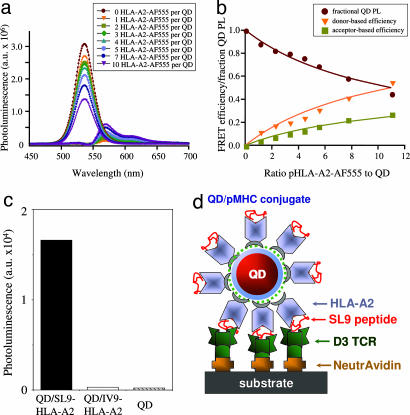
Nonradiative resonance energy transfer between a central QD and proximal dyes attached to the HLA-A2 protein was used to test conjugate formation and to determine the ratio of QD to pHLA-A2. Fig. 1a shows that changes in the deconvoluted photoluminescence (PL) signal of the solutions containing QD donors emitting at 530 nm and pHLA-A2 labeled with Alexa Fluor 555 acceptors are ratio dependent. These changes constitute a reliable indicator of protein immobilization on the QDs (5). Fig. 1b depicts the experimental values of the integrated PL signal as a function of dye-to-QD ratio. Because the dye was bound randomly to ε-amino groups on the protein surface, the data were simulated by using a variable donor–acceptor separation distances (with a normal distribution function). The best fit of the experimental data to theoretical curves was based on Förster formalism and was achieved by using a range of separation distances between donor and acceptor centered at 75 Å and a width of 40 Å. These values are consistent with the dimensions of the inorganic QD (core-shell) and the HLA-A2 protein molecule attached through the C terminus to the QD surface. The dependence of the PL signals on the ratio of labeled protein to QD also indicates that saturation of the surface with protein occurs at protein-to-QD ratios slightly exceeding 12, a value consistent with a maximum allowed by steric limitations of the protein packing around a nanocrystal (7).
Fig. 1.
Formation of functional QD/pHLA-A2 conjugates. (a) Deconvoluted photoluminescence (PL) spectra of QD and Alexa Fluor 555 (AF555)-labeled pHLA-A2 for different dye-to-protein ratios are shown. a.u., atomic units. (b) Changes of the fluorescence intensity versus the dye-to-QD ratio show that up to 12 pHLA-A2 complexes can be attached to each QD. FRET, Förster resonance energy transfer. (c) Immobilized D3 TCR specifically recognize cognate (QD/SL9-HLA-A2) but not noncognate (QD/IV9-HLA-A2) conjugates at 150 nM. Data shown are representative of four independent experiments. (d) Schematic drawing of QD/SL9-HLA-A2 conjugate bound to TCR immobilized on a plastic surface.
Thus, on the surface of the QD the pMHC complexes are in virtual contact with each other and with the TCR-binding surface facing away into the surrounding solution. This configuration may be contrasted with pMHC-I tetramers, in which the distance between pMHC arms is determined by the geometry of streptavidin binding sites. The pMHC–pMHC distance in the tetramer is ≈80 Å (12), which is higher than the optimal distance between pMHC arms (≈30 Å) required for effective binding to the T cell surface (13, 14). Although QD/pMHC may not mimic precisely the well documented cell surface MHC clusters (15), the spacing and orientation of pMHC on QDs would appear to be a closer approximation than tetramers to the clustered arrangement of MHC proteins on the cell surface.
To further characterize the QD/pMHC conjugates we tested their ability to specifically bind to surface-immobilized soluble TCR from CTL clone D3, which is specific for the HIV-derived peptide SLYNTVATL (SL9) in association with HLA-A2 (16). As evident from Fig. 1c, immobilized D3 TCR captures cognate QD/SL9-HLA-A2 conjugate but not QD/pHLA-A2 carrying an HIV-derived irrelevant peptide, ILKEPVHGV (IV9) (17), or unconjugated QDs.
QD/pMHC Binding to the Surface of Live T Cells.
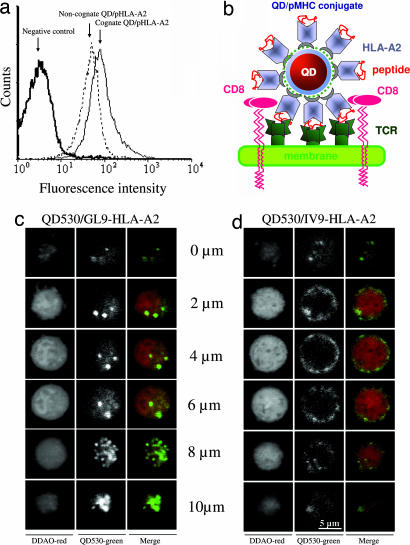
We next examined the ability of QD/pHLA-A2 conjugates to bind TCR in its natural environment, i.e., on the surface of live CTL. Flow cytometric analysis showed that both cognate QD/pHLA-A2 conjugate containing the influenza-virus-derived peptide GILGFVFTL (GL9) and noncognate QD/IV9-HLA-A2 conjugate strongly bind to the surface of CER43 CTL (18) (Fig. 2a). Strong binding of noncognate QD/pMHC to the surface of live CTL was observed at various conjugate concentrations, but binding of cognate QD/pMHC was always stronger than that of noncognate conjugates (Fig. 5, which is published as supporting information on the PNAS web site). The specificity of the QD/GL9-HLA-A2 binding was also evident from the analysis of fluorescence images of CER43 CTL incubated with both QD-pHLA-A2 conjugates at 37°C. Cognate conjugates produced punctate surface and intracellular staining, indicative of QD/GL9-HLA-A2 clustering at the cell surface and subsequent uptake (Fig. 2c). In contrast, noncognate conjugates produced more homogeneous staining limited to the cell surface (Fig. 2d and Figs. 6 and 7, which are published as supporting information on the PNAS web site). Similar strong binding to CER43 CTL was also observed for four different noncognate QD/pHLA-A2 conjugates containing irrelevant HLA-A2-restricted peptides and for various noncognate conjugates to another CTL clone (68A62)(data not shown). These data suggest that efficient binding of noncognate QD/pHLA-A2 to the surface of live CTL does not depend on the nature of HLA-A2-bound peptide and is likely mediated by the HLA-A2 interactions with CD8 coreceptor.
Fig. 2.
Binding of QD/pHLA-A2 conjugates to the surface of live CTL. (a) Although cognate QD/GL9-HLA-A2 conjugate binds specifically to CER43 CTL, noncognate QD/IV9-HLA-A2 conjugate shows high TCR-independent binding to the CTL. QD concentration was 1 μM; the binding was performed for 30 min at 4°C. (b) Schematic drawing of QD/HLA-A2 and peptide complexed with a TCR bound to the membrane of a CD8+ cell. (c and d) Binding of cognate (QD/GL9-HLA-A2) (c) but not noncognate (QD/IV9-HLA-A2) (d) conjugates to CER43 CTL leads to internalization of the QD conjugates. Images of the distribution of QD conjugates (green) in various z sections of CTL are shown.
Role of CD8–HLA-A2 Interactions in Recognition of QD/pHLA-A2.
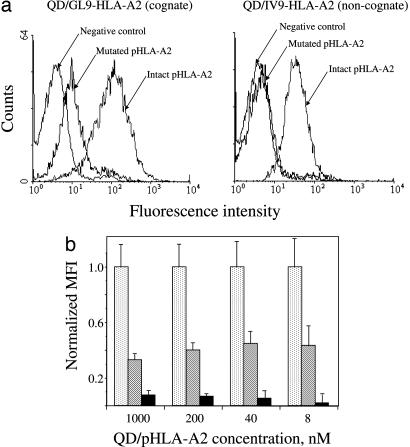
To evaluate the role of CD8–MHC-I interactions in the binding of QD/pMHC to the surface of live CTL, we used soluble HLA-A2 containing a single mutation in a nonpolymorphic domain (A245V). This mutation disrupts CD8 binding to MHC-I and leads to a 5-fold decrease in the intrinsic binding affinity of CD8αα to MHC-I (19).
As evident from Fig. 3a, the binding of noncognate conjugate (QD/IV9-HLA-A2) with mutated HLA-A2 to CER43 cells was indistinguishable from that of unconjugated QDs. The mutation in cognate pHLA-A2 significantly decreased the binding of QD/GL9-HLA-A2 conjugate to live CTL as well. These experiments clearly show a significant contribution of CD8–MHC-I interactions to the binding of both cognate and noncognate QD/pHLA-A2 conjugates to the surface of live CTL. Mutation of HLA-A2 did not affect recognition of pHLA-A2 by the TCR as evident from the binding of cognate pHLA-A2 tetramers to immobilized TCR or the binding of soluble TCR tetramers to immobilized peptide–HLA-A2 in a cell-free system (see Figs. 8 and 9, which are published as supporting information on the PNAS web site).
Fig. 3.
Binding of noncognate QD/pHLA-A2 conjugates to live CTL is CD8-dependent. (a) A single mutation in HLA-A2 that disrupts HLA-A2–CD8 interactions abolishes binding of noncognate QD conjugates (Right) and significantly reduced binding of cognate QD conjugates (Left) to live CTL. (b) The dependence of normalized mean fluorescence intensity (MFI) of CER43 CTL incubated with cognate QD/GL9-HLA-A2 (stippled bars), noncognate QD/IV9-HLA-A2 (hatched bars), and cognate QD/GL9-HLA-A2 with A245V mutation (solid bars) upon concentration of the QD/pHLA-A2 conjugates is shown. MFIs measured with either cognate QD/pHLA-A2 or noncognate QD/pHLA-A2 or cognate QD/mutated pHLA-A2 were divided by MFI measured with cognate QD/pHLA-A2 for every given concentration to determine normalized MFI values. Data represent mean ± SD.
That an optimal positioning and spacing of pMHC on QDs facilitates unexpectedly strong CD8–MHC-I interactions is evident from the comparison of the binding of noncognate pHLA/tetramer and QD/pHLA-A2 to live CTL. The binding of noncognate pHLA-A2/tetramers was very weak (Fig. 10, which is published as supporting information on the PNAS web site). In addition, the effect of A245V mutation on the binding of the pHLA-A2/tetramer was less pronounced (Fig. 10). Thus, close “packaging” of pHLA-A2 on QDs apparently favors optimal multivalent CD8–pHLA-A2 interactions, whereas assembly of pMHC into the tetramer imposes constrains on their ability to get engaged in optimal multivalent interactions with cell surface CD8.
To estimate the relative contributions of the CD8 and TCR interactions to the formation of TCR/pMHC-I/CD8 ternary complexes, we compared normalized MFI of CTL incubated with cognate QD/pHLA-A2 conjugates containing either intact or mutated HLA-A2 and noncognate conjugates bearing intact pHLA-A2 over a wide range of conjugate concentrations (shown in Fig. 3b). Normalized cell staining with the noncognate conjugate was independent of conjugate concentration as long as the binding was still measurable and was significantly higher than normalized staining produced with the mutated cognate pHLA-A2 conjugate; the latter progressively decreased at lower conjugate concentrations. In marked contrast, the binding of noncognate pHLA-A2/tetramer to live cells was barely detectable, whereas normalized binding of the cognate tetramer containing mutated HLA-A2 was stronger, indicating that the tetramer binding was mainly dependent on the TCR–pMHC interactions and was less depended on CD8 (Fig. 11, which is published as supporting information on the PNAS web site). These data demonstrate that the strength of multivalent CD8–MHC-I interactions (avidity) observed with QD/pHLA-A2 conjugates is substantially higher than that of the TCR–pMHC interactions; this also suggests that TCR and CD8 on the T cell cooperate in the recognition of cognate pMHC (see Fig. 3b and below).
The significance of CD8–MHC-I interactions has been confirmed by examining the binding of noncognate QD/pHLA-A2 to a CD8-negative CTL line developed in our laboratory (20) and by analyzing the effect of anti-CD8 antibody on the binding of cognate and noncognate QD/pHLA-A2 to CD8+ CTL (Figs. 12 and 13, which are published as supporting information on the PNAS web site).
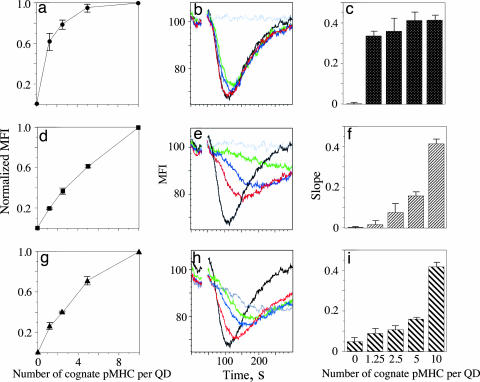
Noncognate pMHC-I Enhances Binding of Cognate pMHC-I in Cooperative CD8-Dependent Manner.
Studies indicate that MHC proteins on the surface of target and antigen-presenting cells are organized in clusters (21, 22). Because clusters on virus-infected cells may contain pMHC complexes presenting both viral and self peptides, we sought to mimic the clusters with QDs bearing a controlled number of cognate and noncognate pMHC and to determine how the presence of noncognate pHLA-A2 influences the binding of the QD conjugates to CTL. We examined the binding of QD conjugates presenting cognate and noncognate pHLA-A2 at various ratios, keeping the total number of pHLA-A2 complexes fixed, i.e., 10 complexes per QD. The concentration of QD conjugates was 200 nM. The binding was first performed at 4°C to minimize TCR-mediated uptake of the cell-surface-bound QD/pHLA-A2. We have found that the dependence of MFI on the number of cognate pHLA-A2 per QD is exponential, indicating that recognition of noncognate pHLA-A2 is enhanced by the presence of cognate pHLA-A2 (Fig. 4a). This effect was dramatically reduced when noncognate pHLA-A2 with a mutation disrupting CD8–MHC-I interactions was used (Fig. 4d). Similar experiments with mutated cognate pHLA-A2 resulted in almost linear dependence (Fig. 4g), suggesting that the effect of enhancement is mostly determined by CD8–pMHC-I interactions. The cooperative effect of the binding of QDs carrying cognate and noncognate pHLA-A2 was also evident at a lower concentration of QD conjugates, i.e., 40 nM (Fig. 14, which is published as supporting information on the PNAS web site) as well as at 37°C (Fig. 15, which is published as supporting information on the PNAS web site). These data clearly demonstrate cooperative, CD8-dependent contribution of noncognate pMHC-I to recognition of cognate pMHC-I ligands by CD8+ T cells.
Fig. 4.
CD8-dependent cooperation of noncognate and cognate pHLA-A2 recognized by CTL. QDs were loaded with a mixture of intact cognate and noncognate (a–c), intact cognate and mutated (A245V) noncognate (d–f), and intact and mutated (A245V) cognate (g–i) pHLA-A2 complexes. The total number of pHLA-A2 molecules (10 per QD) was fixed, whereas the number of intact cognate pHLA-A2 molecules per QD was 0 (gray), 1.25 (green), 2.5 (blue), 5 (red), and 10 (black). Effectiveness of the binding of QD/pHLA-A2 to live CTL (a, d, and g) was characterized by normalized MFI. Slope (c, f, and i) of the Ca2+ flux was determined from the initial decay of the kinetic curve in each experiment (b, e, and h). Results of representative experiments are shown.
CD8-Dependent Cooperation Between Noncognate and Cognate pMHC Augments T Cell Response.
Having shown cooperative contribution of noncognate pMHC-I to the binding of QD conjugates to live T cells, we then asked whether a similar cooperation appears in stimulation of the T cell. To answer the question, we used QDs that display cognate and noncognate pMHC at various ratios to stimulate T cells with known specificity and monitored intracellular Ca2+ flux kinetics to measure stimulation. The experiments were performed in Ca2+-free medium containing 1 mM EGTA to preclude outside Ca2+ influx and to focus on Ca2+ mobilization from intracellular stores to measure the T cell response. By preserving the total number of pMHC per QD (10 pMHC per QD), we systematically replaced cognate pMHC with noncognate pMHC; we found no significant effect on the kinetics and magnitude of Ca2+ mobilization (Fig. 4 b and c), as long as the QDs contained some cognate pMHC. Even at the ratio of noncognate-to-cognate pMHC ≈9:1 the parameters of the Ca2+ flux were very similar to those measured with QDs presenting only cognate pMHC (10 complexes per QD) (Fig. 4 b and c). Importantly, QD carrying only noncognate pMHC did not induce detectable Ca2+ flux (Fig. 4b), indicating that the CD8–MHC-I interactions, which are important for binding, did not themselves provide a stimulatory signal. The ability of noncognate pMHC with the mutation disrupting CD8–MHC-I interactions to facilitate the T cell response to cognate pMHC was severely impaired (Fig. 4e). Stimulation of T cells with QDs bearing intact cognate and mutated noncognate pMHC resulted in significantly slower Ca2+ mobilization and a lower maximum concentration of intracellular Ca2+; both parameters were strongly dependent on the number of intact cognate pMHC per QD (Fig. 4 e and f). We also tested the role of CD8–MHC-I interactions in recognition of cognate pMHC by varying the ratio of the mutated and intact cognate pMHC on QDs. Again, the replacement of the intact pMHC-I with increasing numbers of the mutated pMHC-I led to a slower kinetics and decrease of the maximum concentration of Ca2+ mobilization in T cells stimulated with the QD conjugates (Fig. 4 h and i). These results reveal an essential CD8-dependent cooperation between cognate pMHC and noncognate pMHC-I that augments T cell response.
Discussion
Three principal findings emerged from this study. First, using self-assembled QD/pMHC conjugates that mimic small clusters of tightly packed pMHC proteins on the cell membrane, we have found that antigen-independent multivalent CD8–MHC-I interactions at the surface of live T cells are much stronger than expected. Inasmuch as the number of noncognate pMHC-I molecules greatly exceeds the number of cognate pMHC molecules on target cells (10, 11) and the on-rate of CD8 binding to MHC-I is at least as fast as TCR binding to cognate pMHC (23), it is likely that CD8–MHC-I interactions occur independently of, and precede, cognate pMHC-I recognition by TCR, a view that is consistent with previous findings showing that CD8 recruitment is not peptide specific (24). Second, these interactions are responsible for the cooperative binding of cognate and noncognate pMHC to TCR that leads to augmented T cell response, as evident from the rapid and extensive Ca2+ mobilization from intracellular stores. Third, QDs carrying only noncognate pMHC complexes did not stimulate T cells, although they bind almost as well as cognate QD/pMHC conjugates to T cells.
While these findings are in line with a previously published report showing an essential role of noncognate pMHC-II in antigen recognition by CD4+ T cells (25), they also reveal a profound difference in the role of CD4 and CD8 coreceptors in activation of CD4+ and CD8+ T cells, respectively. Mutation of the CD4-binding site of noncognate pMHC-II does not affect its ability to facilitate a response of CD4+ T cells induced by cognate pMHC-II and also does not impair stimulatory capacity of a strong pMHC-II agonist (25). In contrast, we have shown here that CD8–MHC-I interactions are critical for a CD8+ T cell response that is stimulated by cognate and noncognate pMHC-I complexes functioning in concert.
Although engagement of self pMHC complexes appears to be essential for triggering responses of both CD8+ and CD4+ T cells, the mechanisms by which self pMHC contribute to antigen recognition are different for these T cell subsets. The difference is largely determined by the disparate contributions of CD4 and CD8 coreceptors to the stability of the TCR/pMHC/coreceptor complex. The affinity of CD4 for MHC-II protein is barely measurable (i.e., ≈5 × 103 M−1) and is lower than that of TCR interactions with pMHC ligands (26). In contrast, CD8 binds to MHC-I with higher affinity, i.e., 104 to 105 M−1, which is comparable to that of many TCR–pMHC interactions (27). Thus, additional TCR recruitment to the agonist pMHC-II–TCR complex on CD4+ T cells seems to be more driven by recognition of self pMHC-II by TCR than by CD4 coreceptor engagement (28). In contrast, we propose that the CD8-MHC-I interactions play a key role facilitating the contribution of self pMHC-I to cognate pMHC-I binding and stimulating CTL.
On the basis of these considerations, we propose a model, according to which CD8–MHC-I interactions mediate activation of many TCR in the vicinity of a few TCR engaged with cognate pMHC. Although precise molecular mechanisms by which the activation process spreads from a few to many TCR are not entirely clear, we envision that they are initiated by the formation of a TCR dimer, in which one TCR molecule binds to agonist pMHC and another one associates with self pMHC (25). This initial triggering event may then be further propagated, leading to the formation of a higher order of TCR/pMHC-I/CD8 assemblies (12), in which TCR bound to cognate and noncognate pMHC become activated. How precisely engagement of TCR with cognate pMHC leads to activation of TCR bound to noncognate pMHC remains to be understood. Nevertheless, this CD8-dependent activation of many TCR seems to be a rapid sequential process resembling a chain reaction. Such a process is sensitive to termination at any step even with a stochiometrically minute amount of an inhibitor. In accord with this model, very little soluble CD8, amounting to only ≈2% of cell surface MHC-I molecules, blocked CTL activation, apparently through obstructing CD3ζ chain phosphorylation (29). This great dependence on CD8 may explain why CD8–MHC-I interactions can increase the sensitivity of CTL responses by one millionfold (30).
Effectiveness of the signal spread is expected to depend strongly on the close proximity between pMHC proteins. Indeed, at least a fraction of MHC molecules on the cell surface are concentrated in clusters, and disruption of these clusters impairs the effectiveness of antigen presentation to T cells (31, 32). A tight packaging of pMHC on QDs that closely mimics MHC clustering on the cell membrane appears to be critical for cooperative recognition of cognate and noncognate pMHC leading to the signal spread. In fact, our attempts to observe cooperation between cognate and noncognate pMHC on a glass-supported bilayer, where individual pMHC molecules can freely diffuse and not form clusters, have been unsuccessful (K. Somersalo, N.A., Y.S., and M.L.D., unpublished observation).
A similar model has been previously proposed for activation of CD4+ T cells, according to which the activation is initiated by the formation of TCR–pMHC class II dimers containing an agonist and self peptides, and the CD4 coreceptor with associated p56lck, which is recruited to the dimer, phosphorylates and, thereby, activates both TCR molecules (28). TCR bound to self pMHC-II can then be serially exchanged with many other TCR molecules that become activated, spreading the signal. In this model, unlike ours, TCR recognition of self pMHC-II, but not CD4–MHC-II interactions, primarily drives the signal spread.
Whereas responses of most CD8 T cells are strongly influenced by the CD8 coreceptor, CD8 T cell recognition of allogeneic and other strong agonist pMHC-I may be CD8-independent (33). Likewise, CD8-dependence of the CTL response is less evident at a high density of cognate pMHC-I on target cells. Under these circumstances, many TCR can be activated directly without CD8-mediated signal spread. Thus, the role of CD8 becomes critical when CTL attack target cells that present a very low density of cognate pMHC as in chronically infected and cancer cells (34, 35).
Materials and Methods
Reagents.
We have used the human CTL clone CER43, which recognizes influenza virus peptide GL9 (18), and human CTL clone 68A62 specific for HIV-derived peptide IV9 (17).
An HLA-A2 gene with point mutation (alanine-245 to valine, A245V) was produced as described in ref. 36. Soluble mutated and nonmutated pHLA-A2 and pHLA-A2 tetramers were produced as described in refs. 16 and 37. HLA-A2 was labeled with Alexa Fluor 555 (Molecular Probes, Eugene, OR).
Soluble D3 TCR specific for SL9–HLA-A2 was produced as described in ref. 37 with some modifications. A sequence of the biotin-receptive site GLNDIFEAQKIEWHE was introduced at the C terminus of the D3 TCR α-chain.
Peptide IV9 was a generous gift from Herman Eisen (Massachusetts Institute of Technology). GL9, SL9, and Tax peptides were synthesized by Research Genetics (Huntsville, AL).
The nanocrystals were prepared by using a previously reported stepwise approach (38, 39). Chemical functionalities were added to the QDs with dihydrolipoic acid (DHLA) by exchanging the native capping shell with DHLA as described in ref. 5. These steps produce stable and aggregate-free QDs in neutral to basic buffer solutions.
QD/pMHC conjugate formation was driven by self-assembly between His6-terminated proteins and QDs at the desired QD-to-pHLA-A2 molar ratios in 10 mM sodium tetraborate buffer/25 mM NaCl, pH 8.0, for 30 min at 22–25°C. This procedure produces stable solutions of QD/pHLA-A2 conjugates with the desired protein-to-QD ratio.
Förster Resonance Energy Transfer.
The conjugates were characterized by steady-state measurements of Förster resonance energy transfer between a central QD donor and the proximal dye-labeled pHLA-A2 proteins as previously described (7, 8). Proportional amounts of dye-labeled and unlabeled pHLA-A2 were combined with 0.5 μM QDs in 100 μl of 10 mM sodium tetraborate buffer (pH 9.5), and the mixture was incubated for 20 min at room temperature. Molar ratios of labeled pHLA-A2 to QDs varied from 0 to 14, whereas the total amount of protein per QD in each conjugate remained constant. Emission spectra were collected by using a SPEX Fluorolog-3 fluorimeter (Jobin Yvon/SPEX, Edison, NJ). Spectral deconvolution of the PL spectra was performed by using a custom algorithm in MATLAB (7, 40). For other details see Supporting Methods, which is published as supporting information on the PNAS web site.
QD/pMHC Binding to TCR in a Cell-Free System.
Biotinylated D3 TCR was immobilized on 96-well plates coated with NeutrAvidin (Pierce, Rockford, IL). QD/SL9-HLA-A2 conjugates at 0.15 μM in 10 mM sodium tetraborate buffer/0.25 mM NaCl, pH 8.0, containing 4% nonfat dry powdered milk were added to immobilized D3 TCR. QD solution (0.15 μM) with no attached pHLA-A2 or QDs bearing irrelevant pMHC were used as negative controls. After incubation for 1 h the fluorescence signal was collected by using a Tecan Safire plate reader (Tecan, Durham, NC).
Flow Cytometry Analysis.
QDs emitting at 530 nm (QD-530) were used in these experiments. CTL were incubated with QD conjugates at various concentrations for 30 min at 4°C, washed free of unreacted QD conjugates, and analyzed on an Epics XL-MCL flow cytometer (Beckman Coulter, Fullerton, CA). To block CD8-MHC-I interactions the QD conjugates were incubated with the CTL in the presence of anti-CD8 antibodies (kindly provided by Bice Perussia, Thomas Jefferson University) that inhibit TCR-mediated lysis of target cells by CTL.
In some experiments QD-530 were loaded with mixture of cognate (GL9-HLA-A2) and noncognate (IV9-HLA-A2) pMHC. The number of the cognate pMHC molecules per dot was 10, 5, 2.5, 1.25, and 0, and the total number of pMHC molecules per QD was always 10. Both cognate and noncognate pHLA-A2 proteins were either intact or contained a mutation in the nonpolymorphic domain of HLA-A2 (A245V) that disrupts HLA-A2–CD8 interaction. Loading and staining buffers contained excess of noncognate peptide (3 × 10−4 M). Staining of CER43 CTL with the QD conjugates was performed at either 4°C or 37°C for 30 min. Effectiveness of the binding of QD/pHLA-A2 to live CTL was characterized by normalized MFI; the latter was calculated as follows: (MFIn − MFI0)/(MFI10 − MFI0), where MFIn, MFI0, and MFI10 are measured with QD bearing predetermined number (0 < n < 10) of cognate pHLA-A2, zero cognate pHLA-A2, and ten cognate pHLA-A2 complexes, correspondingly.
Ca2+ Flux Measurements.
CTL (107 per ml) were loaded with 5 μM Fura Red AM (Molecular Probes) mixed with 20% pluronic F-127 in complete medium containing 4 mM probenecid at 37°C for 30 min. After the first wash, the cells were further incubated for 30 min at 37°C to allow de-esterification of the dye and then were washed twice and resuspended in the assay buffer (Dulbecco's PBS containing 0.1 mM MgCl2, 5 mM glucose, and 0.025% BSA) at 106 per ml. EGTA was then added to the cell suspension to the final concentration of 1 mM. Freshly prepared QD conjugates were promptly added to 1 ml of the CTL suspension at 10 or 2 nM, and the samples were analyzed on an Epics XL-MCL flow cytometer. Typically, the data collection was initiated 20 s after the QD conjugates were combined with the CTL after the background measurements. The data were analyzed with FlowJo software (Tree Star, Ashland, OR).
Microscopy.
CTL were loaded with CellTrace Far Red DDAO dye (Molecular Probes) and were incubated with QD-530/pHLA-A2 conjugates (200–400 nM) for 20 min at 37°C and fixed with 3.7% formaldehyde. z-sections imaging of the cells was performed on an LSM 510 laser scanning confocal microscope (Carl Zeiss Microimaging, Thornwood, NY). Images were processed by Photoshop (Adobe Systems, San Jose, CA).
Supplementary Material
Acknowledgments
We thank members of the Sykulev, Dustin, and Mattoussi laboratories for support and stimulating discussions. This work was supported by National Institutes of Health grants (to Y.S. and M.L.D.). We thank A. Ervin and L. Chrisey at the Office of Naval Research for supporting this project. We also thank A. Krishnan at the Defense Advanced Research Projects Agency for financial support. A.R.C. was supported by a National Research Council Fellowship.
Abbreviations
- CTL
cytotoxic T lymphocyte(s)
- MFI
mean fluorescence intensity
- PL
photoluminescence
- pMHC
peptide-MHC
- QD
quantum dot(s)
- TCR
T cell receptor(s).
Footnotes
The authors declare no conflict of interest.
References
- 1.Rudolph MG, Wilson IA. Curr Opin Immunol. 2002;14:52–65. doi: 10.1016/s0952-7915(01)00298-9. [DOI] [PubMed] [Google Scholar]
- 2.Gao GF, Rao Z, Bell JI. Trends Immunol. 2002;23:408–413. doi: 10.1016/s1471-4906(02)02282-2. [DOI] [PubMed] [Google Scholar]
- 3.Hugues S, Malherbe L, Filippi C, Glaichenhaus N. J Immunol Methods. 2002;268:83–92. doi: 10.1016/s0022-1759(02)00202-8. [DOI] [PubMed] [Google Scholar]
- 4.Gestwicki JE, Cairo CW, Strong LE, Oetjen KA, Kiessling LL. J Am Chem Soc. 2002;124:14922–14933. doi: 10.1021/ja027184x. [DOI] [PubMed] [Google Scholar]
- 5.Mattoussi H, Mauro JM, Goldman ER, Anderson GP, Sundar VC, Mikulec FV, Bawendi MG. J Am Chem Soc. 2000;122:12142–12150. [Google Scholar]
- 6.Medintz IL, Clapp AR, Mattoussi H, Goldman ER, Fisher B, Mauro JM. Nat Mater. 2003;2:630–638. doi: 10.1038/nmat961. [DOI] [PubMed] [Google Scholar]
- 7.Clapp AR, Medintz IL, Mauro JM, Fisher BR, Bawendi MG, Mattoussi H. J Am Chem Soc. 2004;126:301–310. doi: 10.1021/ja037088b. [DOI] [PubMed] [Google Scholar]
- 8.Goldman ER, Medintz IL, Whitley JL, Hayhurst A, Clapp AR, Uyeda HT, Deschamps JR, Lassman ME, Mattoussi H. J Am Chem Soc. 2005;127:6744–6751. doi: 10.1021/ja043677l. [DOI] [PubMed] [Google Scholar]
- 9.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 10.Sykulev Y, Joo M, Vturina I, Tsomides TJ, Eisen HN. Immunity. 1996;4:565–571. doi: 10.1016/s1074-7613(00)80483-5. [DOI] [PubMed] [Google Scholar]
- 11.Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Nature. 2002;419:845–849. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Miguel G, Alarcon B, Iglesias A, Bluethmann H, Alvarez-Mon M, Sanz E, de la Hera A. Proc Natl Acad Sci USA. 1999;96:1547–1552. doi: 10.1073/pnas.96.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cochran JR, Cameron TO, Stone JD, Lubetsky JB, Stern LJ. J Biol Chem. 2001;276:28068–28074. doi: 10.1074/jbc.M103280200. [DOI] [PubMed] [Google Scholar]
- 14.Cebecauer M, Guillaume P, Mark S, Michielin O, Boucheron N, Bezard M, Meyer BH, Segura JM, Vogel H, Luescher IF. J Biol Chem. 2005;280:23820–23828. doi: 10.1074/jbc.M500654200. [DOI] [PubMed] [Google Scholar]
- 15.Matko J, Bushkin Y, Wei T, Edidin M. J Immunol. 1994;152:3353–3360. [PubMed] [Google Scholar]
- 16.Anikeeva N, Lebedeva T, Krogsgaard M, Tetin SY, Martinez-Hackert E, Kalams SA, Davis MM, Sykulev Y. Biochemistry. 2003;42:4709–4716. doi: 10.1021/bi026864+. [DOI] [PubMed] [Google Scholar]
- 17.Tsomides TJ, Walker BD, Eisen HN. Proc Natl Acad Sci USA. 1991;88:11276–11280. doi: 10.1073/pnas.88.24.11276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valitutti S, Muller S, Dessing M, Lanzavecchia A. J Exp Med. 1996;183:1917–1921. doi: 10.1084/jem.183.4.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao GF, Willcox BE, Wyer JR, Boulter JM, O'Callaghan CA, Maenaka K, Stuart DI, Jones EY, Van Der Merwe PA, Bell JI, Jakobsen BK. J Biol Chem. 2000;275:15232–15238. doi: 10.1074/jbc.275.20.15232. [DOI] [PubMed] [Google Scholar]
- 20.Somersalo K, Anikeeva N, Sims TN, Thomas VK, Strong RK, Spies T, Lebedeva T, Sykulev Y, Dustin ML. J Clin Invest. 2004;113:49–57. doi: 10.1172/JCI200419337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Q, Edidin M. Biophys J. 2001;81:196–203. doi: 10.1016/S0006-3495(01)75691-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lebedeva T, Dustin ML, Sykulev Y. Curr Opin Immunol. 2005;17:251–258. doi: 10.1016/j.coi.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 23.Wyer J, Willcox B, Gao G, Gerth U, Davis S, Bell J, van der Merwe P, Jakobsen B. Immunity. 1999;10:219–225. doi: 10.1016/s1074-7613(00)80022-9. [DOI] [PubMed] [Google Scholar]
- 24.Yachi PP, Ampudia J, Gascoigne NR, Zal T. Nat Immunol. 2005;6:785–792. doi: 10.1038/ni1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krogsgaard M, Li QJ, Sumen C, Huppa JB, Huse M, Davis MM. Nature. 2005;434:238–243. doi: 10.1038/nature03391. [DOI] [PubMed] [Google Scholar]
- 26.Xiong Y, Kern P, Chang H, Reinherz E. J Biol Chem. 2001;276:5659–5667. doi: 10.1074/jbc.M009580200. [DOI] [PubMed] [Google Scholar]
- 27.Gao GF, Jakobsen BK. Immunol Today. 2000;21:630–636. doi: 10.1016/s0167-5699(00)01750-3. [DOI] [PubMed] [Google Scholar]
- 28.Li QJ, Dinner AR, Qi S, Irvine DJ, Huppa JB, Davis MM, Chakraborty AK. Nat Immunol. 2004;5:791–799. doi: 10.1038/ni1095. [DOI] [PubMed] [Google Scholar]
- 29.Sewell AK, Gerth UC, Price DA, Purbhoo MA, Boulter JM, Gao GF, Bell JI, Phillips RE, Jakobsen BK. Nat Med. 1999;5:399–404. doi: 10.1038/7398. [DOI] [PubMed] [Google Scholar]
- 30.Holler PD, Kranz DM. Immunity. 2003;18:255–264. doi: 10.1016/s1074-7613(03)00019-0. [DOI] [PubMed] [Google Scholar]
- 31.Anderson HA, Hiltbold EM, Roche PA. Nat Immunol. 2000;1:156–162. doi: 10.1038/77842. [DOI] [PubMed] [Google Scholar]
- 32.Lebedeva T, Anikeeva N, Kalams SA, Walker BD, Gaidarov I, Keen JH, Sykulev Y. Immunology. 2004;113:460–471. doi: 10.1111/j.1365-2567.2004.01985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho BK, Lian KC, Lee P, Brunmark A, McKinley C, Chen J, Kranz DM, Eisen HN. Proc Natl Acad Sci USA. 2001;98:1723–1727. doi: 10.1073/pnas.98.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsomides TJ, Aldovini A, Johnson RP, Walker BD, Young RA, Eisen HN. J Exp Med. 1994;180:1283–1293. doi: 10.1084/jem.180.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cox AL, Skipper J, Chen Y, Henderson RA, Darrow TL, Shabanowitz J, Engelhard VH, Hunt DF, Slingluff CL., Jr Science. 1994;264:716–719. doi: 10.1126/science.7513441. [DOI] [PubMed] [Google Scholar]
- 36.Bodinier M, Peyrat MA, Tournay C, Davodeau F, Romagne F, Bonneville M, Lang F. Nat Med. 2000;6:707–710. doi: 10.1038/76292. [DOI] [PubMed] [Google Scholar]
- 37.Anikeeva N, Lebedeva T, Sumaroka M, Kalams SA, Sykulev Y. J Immunol Methods. 2003;277:75–86. doi: 10.1016/s0022-1759(03)00179-0. [DOI] [PubMed] [Google Scholar]
- 38.Murray CB, Norris DJ, Bawendi MG. J Am Chem Soc. 1993;115:8706–8715. [Google Scholar]
- 39.Dabbousi BO, Rodriguez-Viejo J, Mikulec FV, Heine JR, Mattoussi H, Ober R, Jensen KF, Bawendi MG. J Phys Chem. 1997;101:9463–9475. [Google Scholar]
- 40.Clapp AR, Medintz IL, Fisher BR, Anderson GP, Mattoussi H. J Am Chem Soc. 2005;127:1242–1250. doi: 10.1021/ja045676z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.