Abstract
Legionella pneumophila, the causal agent of Legionnaires' disease, is an intracellular parasite and invades and proliferates within different eukaryotic cells, including human alveolar macrophages. After several 100-fold multiplication within host cells, the pathogens are released for new invasion by induction of apoptosis or necrosis. Here we report that L. pneumophila produces a glucosyltransferase, which selectively modifies an ≈50-kDa mammalian protein by using UDP-glucose as a cosubstrate. MS analysis identified the protein substrate as the mammalian elongation factor (EF)1A. Legionella glucosyltransferase modifies its eukaryotic protein substrate at serine-53, which is located in the GTPase domain of the EF. Glucosylation of EF1A results in inhibition of eukaryotic protein synthesis and death of target cells. Our findings show a mode of inhibition of protein synthesis by microbial pathogens and offer a perspective for understanding of the host-pathogen interaction of L. pneumophila.
Keywords: host–pathogen interaction, coralent modification, protein synthesis inhibition
Legionella pneumophila is the causative pathogen of legionellosis, also known as Legionnaires' disease, which is a human multisystem illness with severe pneumonia (1). Each year, 8,000–18,000 people are hospitalized with legionellosis in the United States with fatality rates of 5–30% (2). The pathogen is found in a wide variety of aquatic environments. L. pneumophila is a Gram-negative microorganism that invades and proliferates within different eukaryotic cells, including mammalian phagocytes (1). Natural hosts of the facultative intracellular parasites are free-living protozoa. Legionellae are able to induce alterations in the normal course of phagocytosis, including inhibition of acidification of phagosomes, prevention of phagosome–lysosome fusion, and suppression of oxidative burst (3–5). Moreover, legionellae alter phagocytic membrane biogenesis and trigger formation of “replicative vacuoles.” These organellae are associated with ribosome-studded membranes and support multiplication of the bacteria (6, 7). Eventually, host cells die because of induction of apoptosis or necrosis (8–10), and the pathogens are released to invade new pools of phagocytes of the host (10, 11). During recent years several genetic loci and bacterial factors have been identified that are suggested to be implicated in Legionella–host cell interactions (12). Among these are the type II and Dot/Icm type IV protein secretion systems (13–16). However, the precise functions of these factors and their host target proteins are largely still not well understood.
Recently, a 60-kDa glucosyltransferase from L. pneumophila was identified that modifies an ≈50-kDa protein in eukaryotic cells (17). The corresponding gene in L. pneumophila chromosome (accession no. NC_002942) has the identification number lpg1368 and is located in close proximity to coding sequences of the type II secretion system components. Here we studied the action of Legionella glucosyltransferase (termed Lgt1) on eukaryotic target cells. We report that Lgt1 glucosylates elongation factor (EF)1α (EF1A) at serine-53, a modification that blocks protein synthesis and causes death of target cells.
Results
DXD Motif and Cosubstrate Specificity of Lgt1.
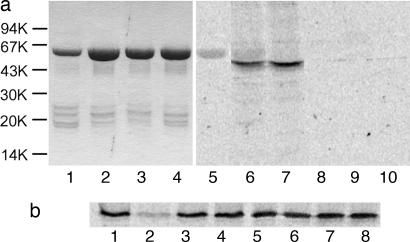
Comparison of the amino acid sequence of Lgt1 with those of large clostridial cytotoxins (e.g., Clostridium difficile toxins A and B), which are Rho GTPase-glucosylating bacterial protein toxins (18, 19), showed significant similarities in a region around the DXD motif (Table 1). The DXD motif is conserved in many prokaryotic and eukaryotic glucosyltransferases. It is involved in manganese coordination and cosubstrate binding and essential for enzyme activity of the toxins (20, 21). Change of the D246ID motif in Lgt1 to N246ID (Fig. 1a, lane 8) or N246IN248 (Fig. 1a, lane 9) inhibited glucosyltransferase activity. In contrast, substitution in the D198ID sequence of Lgt1 (data not shown in Table 1), which possesses no similarity with clostridial toxins, failed to affect substrate labeling (Fig. 1a, lane 7). To identify the cosubstrate specificity of Lgt1, different unlabeled activated sugars were added in 5-fold excess to the UDP-[14C]glucose-containing reaction mixtures. Only UDP-glucose significantly reduced modification of eukaryotic substrate, indicating that UDP-glucose was the specific cosubstrate of Lgt1 (Fig. 1b).
Table 1.
Partial amino acid alignment of Lgt1 and typical members of large clostridial toxins family
| Protein identification | GenBank accession no. | Partial amino acid sequence |
|---|---|---|
| L. pneumophila Lgt1 | NC_002942 (the ORF lpg1368) | G223NPAAASDLCRWIPELFNEGFYVDID-LPVDSSKIV257 |
| Clostridium sordellii lethal toxin | X82638 | W275NLAAASDILRISMLKEDGGVYLDVDILPGIQPDLF310 |
| Clostridium novyi α toxin | Z48636 | N264NLAAASDILRIAILKKYGGVYCDLDFLPGVNLSLF299 |
| C. difficile toxin A | M30307 | G262NLAAASDIVRLLALKNFGGVYLDVDMLPGIHSDLF297 |
| C. difficile toxin B | X53138 | W263NLAAASDILRISALKEIGGMYLDVDMLPGIQPDLF298 |
Amino acids that are identical in all five sequences appear in bold, and DXD motifs are underlined.
Fig. 1.
Glucosylation by Lgt1. (a) Glucosylation of target proteins in Caco-2 enterocyte lysate by Lgt1. Lanes 1–4 show SDS/PAGE of purified recombinant glucosyltransferases (2–4 μg per track, Coomassie staining). Lanes: 1, wild-type Lgt1; 2, D198N Lgt1; 3, D246N Lgt1; 4, D246N/D248N Lgt1. Lanes 5–10 show autoradiography of glucosylation caused by the glucosyltransferases. Caco-2 lysate was incubated with recombinant wild-type and mutant Lgt1 in the presence of UDP-[14C]glucose for 1 h. Lanes: 5, wild-type Lgt1 without Caco-2 cell extract (negative control); 6, wild-type Lgt1 plus Caco-2 cell extract; 7, D198N Lgt1 plus Caco-2 cell extract; 8, D246N Lgt1 plus Caco-2 cell extract; 9, D246N/D248N Lgt1 plus Caco-2 cell extract; 10, Caco-2 cell extract without Lgt1 (negative control). Molecular mass markers are indicated on the left. (b) Cosubstrate specificity of Lgt1. Glucosylation of Caco-2 cell lysates was performed with Lgt1 in the presence of 10 μM UDP-[14C]glucose and without (lane 1) or with (lanes 2–8) 50 μM each of unlabeled UDP-glucose (lane 2), UDP-galactose (lane 3), UDP-N-acetyl-galactosamine (lane 4), UDP-N-acetyl-glucosamine (lane 5), UDP-glucuronic acid (lane 6), GDP-mannose (lane 7), and glucose (lane 8).
Identification of the Eukaryotic Protein Substrate of Lgt1.
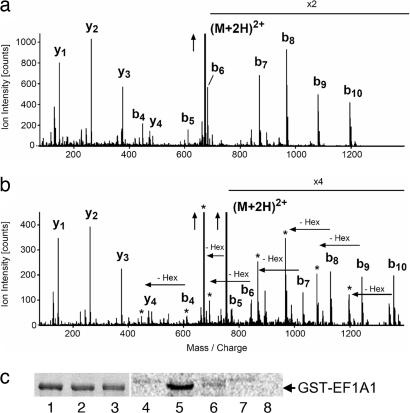
To analyze the eukaryotic protein substrate of the Legionella transferase, we glucosylated Caco-2 cell lysate with Lgt1 in the presence of UDP-glucose. Thereafter, proteins was separated by SDS/PAGE, cut from the gel, and digested by trypsin. The tryptic peptides were analyzed by capillary HPLC and tandem MS for peptide sequencing. In comparison to mock-treated lysate an additional peptide with the increase in molecular mass of exactly 162 Da could be found, suggesting glucosylation by the Legionella enzyme. Sequencing revealed that the peptide was glycine-52 through lysine-62 from eukaryotic EF1A (eEF1A) with mono-O-glucosylation at position serine-53 (Fig. 2a and b).
Fig. 2.
Analyses of target proteins of Lgt1 by MS and mutagenesis studies. (a and b) MS analysis of proteins in Caco-2 cell lysate glucosylated by Lgt1. (a) The fragment spectrum of the nonglucosylated peptide GSF(K-dimethyl)YAWVLDK of eEF1A1. (b) The fragment spectrum of the glucosylated peptides that were only found in samples exposed to Lgt1. The peaks labeled bn (b-ions) indicate N-terminal fragment ions, and the peaks labeled yn C-terminal fragment ions. The total mass of the glucosylated peptides is precisely 162 Da higher, consistent with a covalent modification by a hexose. The fragment spectrum in b demonstrates that all fragment ions that contain the N-terminal serine (the b-ions) are shifted by 162 Da in comparison to the nonglucosylated peptide fragment spectrum in a. Each of these b-ions is accompanied by a satellite fragment marked with an asterisk, representing a loss 162 Da corresponding to a hexose group. Such an additional fragment often is seen for covalent modifications by a single hexose group. The vertical arrows indicate that ion intensity was cut for adaptation to figure size; X2 and X4 indicate multiplication of ion intensity for better presentation of data and (M+2H)2+ indicates that the doubly charged peptide had been fragmented. (c) Glucosylation of recombinant GST-tagged wild-type and mutant eEF1A1 by Lgt1. Lanes 1–3 show SDS/PAGE of purified recombinant EF1A (Coomassie staining). Lanes: 1, wild-type eEF1A1-GST; 2, eEF1A1-GST with S53T mutation; 3, eEF1A1-GST with S53A mutation. Protein load is 3–4 μg per track. Lanes 4–8 show autoradiographic analysis of substrate activity of purified recombinant EF1A. Wild-type and mutant eEF1A1-GST fusion proteins were incubated with Lgt1 for 1 h in the presence of UDP-[14C]glucose. Lanes: 4, Lgt1 without eEF1A1-GST (negative control); 5, Lgt1 plus wild-type eEF1A1-GST; 6, Lgt1 plus eEF1A1-GST with S53T mutation; 7, Lgt1 plus eEF1A1-GST with S53A mutation; 8, wild-type eEF1A1-GST without Lgt1 (negative control). The position of GST-tagged EF1A is marked on the right. Please note the weak autoglucosylation activity of Lgt1 in lanes 4–7.
To confirm MS data, eEF1A1 was expressed in Escherichia coli and purified as a GST fusion protein (Fig. 2c, lanes 1–3). When purified GST-eEF1A1 was used in glucosylation assay, labeling of an ≈80-kDa protein corresponding to the molecular mass of the fusion protein was observed (Fig. 2c, lane 5). The results were essentially the same when eEF1A2, which is highly similar to eEF1A1, was used as a protein substrate (data not shown). To verify further the identification of eEF1A as a target of the Legionella enzyme, we changed the acceptor amino acid serine-53 of eEF1A1 to threonine and alanine. Whereas glucosylation of S53T-eEF1A was reduced, S53A-eEF1A was not at all modified by Lgt1 (Fig. 2c, lanes 6 and 7). Altogether, these data confirmed the MS data and showed that eEF1A is modified by Lgt1 at serine-53.
Cytotoxic Activity of Lgt1 Enzyme.
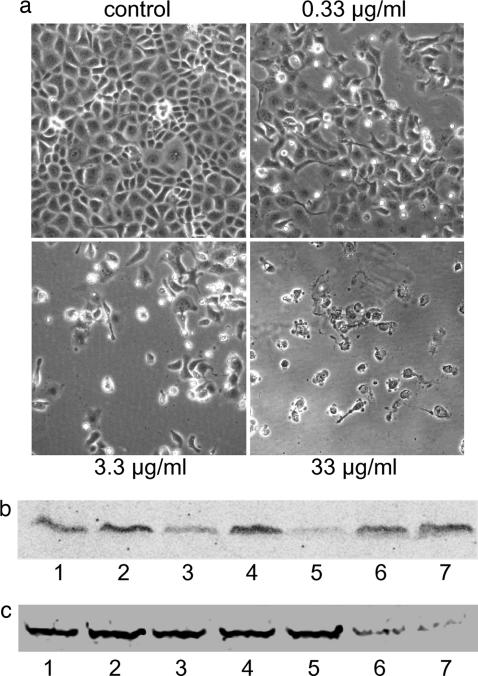
EF1A plays a pivotal role in protein synthesis (22). In addition, eEF1A appears to be involved in several other cellular processes (23, 24), including regulation of the actin cytoskeleton and cell morphology (25). Therefore, we studied the potential cytopathic activity of Lgt1 in various eukaryotic cells. When Lgt1 was directly added to medium with cultivated HeLa epithelial cells, Caco-2 enterocytes or embryonic bovine lung (EBL) cells, we did not detect any effects (data not shown), most likely because of failure of the protein up-take. However, when recombinant L. pneumophila protein was introduced intracellularly by electroporation, the enzyme caused major morphological alterations. Approximately 15–18 h after electroporation, Lgt1 induced rounding up of cells and started destruction of the cell monolayer. At 40–48 h after electroporation with Lgt1 (3.3 μg/ml), almost total destruction of the cell monolayer occurred (Fig. 3a). In contrast, introduction of the double D246N/D248N mutant by electroporation at high concentrations (33 μg/ml) (Fig. 3, control) or electroporation without Lgt1 (data not shown) did not produce any cytotoxic effects.
Fig. 3.
Effects of Lgt1 on host cells. (a) Morphological changes in EBL cells induced by Lgt1. EBL cells were electroporated in the presence of Lgt1 at concentrations of 33, 3.3, and 0.33 μg/ml, with double D246N/D248N Lgt1 mutant at 33 μg/ml (control). Thereafter the cells were replated. After 48 h of incubation, microscopic pictures of cells were taken. (b) Back-glucosylation of the protein substrate of Lgt1 in lysate of EBL cells previously electroporated with Lgt1. Shown are samples from cell lysates electroporated with Lgt1 (3.3 μg/ml; lanes 1, 3, and 5), double D246N/D248N Lgt1 mutant (3.3 μg/ml; lanes 2, 4, and 6), or without Legionella proteins (lane 7). EBL cells were collected 30 min (lanes 1 and 2), 1.5 h (lane 3 and 4), and 4.5 h (lane 5, 6, and 7) after electroporation and used in the glucosylation assay with Lgt1 and UDP-[14C]glucose. Labeled proteins were analyzed by SDS/PAGE and phosphorimaging (shown). (c) Back-glucosylation of the protein substrate of Lgt1 in lysate of A549 cells, previously infected with L. pneumophila. A549 cells were infected with L. pneumophila at a multiplicity of infection of 10. The cells were harvested 1, 2, 3, 4, 8, and 24 h after start of the infection and lysed as described in Materials and Methods. Cell lysate was glucosylated with Lgt1 (4 μg) in the presence of UDP-[14C]glucose. Labeled proteins were analyzed by SDS/PAGE (see Fig. 6, which is published as supporting information on the PNAS web site) and phosphorimaging for [14C]glucosylation (shown). Lanes: 1, noninfected cells; 2–7, infected cells 1, 2, 3, 4, 8 and 24 h after start of the infection, respectively. Shown is a representative experiment that was repeated several times with similar results.
To confirm that morphological changes in target cells were conveyed by glucosylation of eEF1A, we performed “back-glucosylation.” We reasoned that any EF1A molecules that had already been glucosylated in live cells would not be available as a substrate in the subsequent in vitro reaction; therefore, enzyme activity in vivo could be deduced by the failure to incorporate [14C]glucose in vitro. To this end, EBL cells were electroporated in the presence of Lgt1 (3.3 μg/ml) and lysed by sonication after the time intervals indicated in Fig. 3b and subsequently used in the [14C]glucosylation assay. As shown in Fig. 3b, samples collected 30 min after electroporation exhibited a slight reduction of [14C]glucose labeling (Fig. 3b, lane 1). Weak labeling still could be observed after 1.5 h (Fig. 3b, lane 3), and almost no glucosylation was detected with cells after 4.5 h of incubation (Fig. 3b, lane 5), suggesting a causative relationship of glucosylation of eEF1A and morphological changes of EBL cells induced by Lgt1.
In another set of experiments, we studied whether the modification of host EF1A takes place during intracellular proliferation of L. pneumophila. For this purpose, human alveolar basal epithelial (A549) cells were infected with virulent bacteria. Cells were harvested after different time periods (1–24 h), and cell extracts were prepared. These extracts were tested in the back-glucosylation assay with UDP-[14C]glucose as a cosubstrate similar to the assay described above for intoxication studies. As shown on Fig. 3c, coculturing of bacterial and A549 host cells for >8 h resulted in inhibition of subsequent in vitro [14C]glucosylation. Similar results were obtained with EBL cells (data not shown). Taken together, these data suggested that Lgt1 is produced during the infection cycle, gains access to the cytosol, and modifies eEF1A.
Inhibition of Eukaryotic Protein Synthesis by Lgt1.
One of the main functions of eEF1A is participation in protein synthesis. Therefore, we studied the consequences of glucosylation of eEF1A on protein synthesis by in vitro transcription/translation and in vivo methionine incorporation assays.
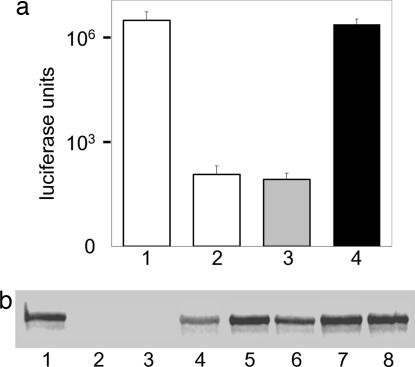
In vitro transcription and translation of the luciferase gene with reticulocyte lysate in the presence of [35S]methionine resulted in production of the corresponding protein. This could be demonstrated either by luminescence assay (Fig. 4a) or by autoradiography (Fig. 4b). Increasing amounts of wild-type Lgt1 but not of the DXD mutant enzyme inhibited synthesis of luciferase (Fig. 4). Essentially the same inhibition of protein synthesis was observed by autoradiography with an actin-coding plasmid or with luciferase-coding mRNA (data not shown). These data suggested that Lgt1 did affect protein synthesis in vitro. Next, [35S]methionine incorporation into newly synthesized proteins was studied in intact cells. To this end, EBL cells were electroporated in the presence of increasing concentrations of wild-type or mutant Lgt1. After 2 h of intoxication, [35S]methionine was added to the medium. After further incubation for 3 h, incorporation of radioactivity was studied. As shown in Fig. 5, intoxication of EBL cells with wild-type Lgt1 but not with the mutant enzyme reduced incorporation of radioactivity in trichloroacetic acid-precipitated proteins. As determined by trypan blue assay, >95% of cells were viable after this time of incubation (data not shown). A similar inhibition of incorporation of radioactivity was observed when EBL cells were treated with Lgt1 for 1 h before they were pulsed with [35S]methionine for only 1 h (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 4.
Inhibition of transcription/translation by Lgt1. (a) Luminescence assay. A transcription/translation reaction was performed in the absence (columns 1 and 2) or presence of recombinant wild-type (column 3) or D246N-mutated (column 4) Lgt1 (each at 2.4 μg/ml). In these experiments, matrix DNA was luciferase gene-containing plasmid (columns 1, 3, and 4). Column 2 is a negative control without matrix DNA. All measures were done in triplicate. Error bars indicate standard deviations. Differences in the values of columns 1 and 2 with those of columns 3 and 4 are statistically significant (P < 0.01). White columns show control results, the gray column shows wild-type Lgt1, and the black column shows mutated Lgt1. The y axis represents logarithmic scale. (b) Autoradiographic assay. A transcription/translation reaction has been performed in the absence (lanes 1 and 2) or presence (lanes 3–8) of recombinant wild-type (lanes 3–5) or D246N-mutated (lanes 6–8) Lgt1. Concentrations of L. pneumophila proteins were 2.4 μg/ml in lanes 3 and 6; 240 ng/ml in lanes 4 and 7; and 24 ng/ml in lanes 5 and 8. Matrix DNA present in lanes 1 and 3–8 was luciferase gene-containing plasmid coding for an ≈60-kDa protein. Lane 2 is a negative control without matrix DNA.
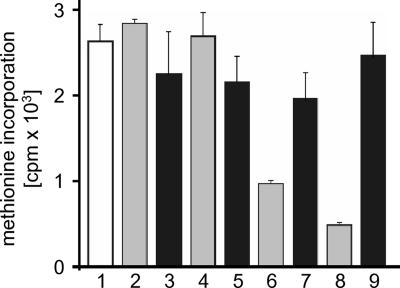
Fig. 5.
Inhibition of [35S]methionine incorporation by Lgt1. EBL cells were electroporated with wild-type (columns 2, 4, 6, and 8) and double D246N/D248N Lgt1 mutant (columns 3, 5, 7, and 9) in methionine-free MEM. Two hours after intoxication, cells were pulsed with 0.5 μCi [35S]methionine for a further 3 h, lysed, and assayed for incorporation of radioactivity into proteins. Columns: 1, control cells electroporated without Legionella proteins; 2–9, cells were electroporated with Legionella proteins at 33 ng/ml (columns 2 and 3), 0.33 μg/ml (columns 4 and 5), 3.3 μg/ml (columns 6 and 7), or 33 μg/ml (columns 8 and 9). All measures have been done in triplicate. Error bars indicate standard deviations. Differences between the values in columns 6 and 7 with those in columns 8 and 9 are statistically significant (P < 0.01). The white column represents the control experiment, the gray columns represent wild-type Lgt1, and black columns represent mutant Lgt1.
Discussion
Here we show that eEF1A is mono-O-glucosylated by Lgt1 at serine-53, a modification that blocks protein synthesis. eEF1A is an ≈50-kDa GTP-binding protein possessing GTPase activity, which is necessary for recruitment of aminoacyl-tRNA to the A-site of ribosomes. The GTPase activity of eEF1A is stimulated by interaction of the mRNA codon with the anticodon of the aminoacyl-tRNA at the decoding center of the 30S subunit of the ribosome and allows dissociation of eEF1A from the tRNA (26, 27). No direct structure of mammalian eEF1A is available; however, recently, the very similar EF1A from Saccharomyces cerevisiae was crystallized in a complex with the EF1Bα (28). eEF1A consists of three domains: domain 1 (or G-domain) harbors the nucleotide binding site and has a typical Ras GTPase-like structure (29); domains II and III have β-barrel structure and are directly involved in aminoacyl-tRNA binding (28).
Serine-53 of eEF1A, which is modified by Lgt1, is located in the G-domain near the switch-1 region of the GTPase. Switch regions show major conformational changes dependent on GDP or GTP-binding (30). In the functionally equivalent prokaryotic EF-Tu, the switch-1 region (in EF-Tu amino acids 53–59) undergoes a dramatic change from a β-structure in the GDP form to a α-structure in the GTP-bound form (31). However, the switch-1 region is not well defined in eEF1A, because two additional helices (A* and A′) are inserted in this region of the EFs. So far no functional changes in the switch-1 region have been reported for eEF1A. In addition, serine-53 of eEF1A has to be compared with alanine-42 in E. coli EF-Tu and no major structure change occurs with this residue upon nucleotide exchange in EF-Tu (31). However, our findings show that the attachment of a glucose moiety at serine-53 inhibits the function of eEF1A and blocks protein synthesis. At least two explanations are possible. First, glucosylation of serine-53 prevents a conformational change in the switch-1 region, which is essential for the function of the EF. Second, attachment of glucose onto serine-53, although not directly influencing the structure of the switch-1 region, prevents protein–protein interaction by sterical hindrance. The name “effector region,” which is used for the switch-1 region of many GTPases, illustrates that this part of the protein often is important for GTPase-effector interaction. The large clostridial cytotoxins like C. difficile toxin A and B, which glucosylate threonine-37 in the switch-1 region of the GTPase RhoA, block the conformational change required for activation and prevent interaction of the GTPase with effectors by sterical hindrance (32, 33).
Thus, although we cannot precisely explain how glucosylation of eEF1A inhibits protein synthesis, our findings indicate that the region around serine-53 is important for the function of eEF1A in protein synthesis. Notably, eEF1A is posttranslationally modified by various mechanisms, including phosphorylation (34), methylation of lysine residues, attachment of glycerylphosphorylethanolamine (35), and carboxymethylation (36). Of special interest is the fact that methylation occurs at lysine-55 (35), which is in the vicinity of the glucosylation site. So far, the functional consequences of lysine methylation are not clear.
Invasion of phagocytic cells by Legionella and its intracellular replication depend on the ability of bacteria to control numerous functions of the host. Recent studies indicate that manipulation of vesicular traffic of eukaryotic cells is an important property of many invasive microorganisms (37). Thus, phagosome remodeling and defects in endocytic fusion events observed during intracellular proliferation of L. pneumophila have been shown to be altered by the so called Dot/Icm type IV secretion system (13–15, 38). Such a secretion apparatus is responsible for export of a set of bacterial effector proteins with an ultimate aim to promote development of a replicative vacuole, which is a specialized organelle permissive for intracellular growth of Legionella (15).
Another critically important target, attacked by various pathogenic bacteria, is the eukaryotic protein synthesis machinery. Inhibition of mRNA translation on ribosomes is known to be accomplished by several mechanisms. Shiga- and Shiga-like toxins act as rRNA N-glycosidases (39), whereas diphtheria toxin and Pseudomonas aeruginosa exotoxin A inhibit protein synthesis by ADP-ribosylation of eukaryotic EF2 (40). Here we describe modification of eEF1A by Lgt1 as a mode of inhibition of translation by bacterial effectors and demonstrate that such glucosylation takes place during proliferation of the bacteria in target cells. It should be noted that the inhibition of protein synthesis induced by L. pneumophila was observed 15 years ago (41), although its molecular mechanism was not investigated.
Termination of peptide synthesis apparently leads to the death of intoxicated cells. However, to what extent is cell death favorable for an intracellular pathogen? At least three consequences have to be considered. First, the rate of cell death due to protein synthesis inhibition is relatively slow (days), compared with death induced by cytoskeleton-targeting (hours) or cytolytic membrane-targeting (minutes) toxins. This delayed rate of cell death could favor unrestricted proliferation of pathogens within replicative vacuoles of a “defenseless” host. Second, at the final stages of the intracellular life cycle, legionellae have to escape the eukaryotic cell, which can no longer support proliferation of hundreds of bacteria. Induction of apoptotic processes at late phases of infection has been suggested to be involved in this process (10, 42). In this connection, termination of protein synthesis, induced by Lgt1, could trigger programmed cell death, thus supporting release of Legionella to the extracellular milieu. Finally, it cannot be excluded that reduction of protein synthesis is necessary for fine tuning host cell physiology and synchronizing host cell metabolism toward parasite requirements.
Materials and Methods
Construction, Expression, and Purification of Recombinant Proteins.
The nucleotide sequence coding for Lgt1 (accession no. NC_002942; denoted lpg1368) was PCR-amplified by FideliTaq DNA polymerase (Amersham Biosciences, Moscow, Russia) with the primers LUT1 and LUT2 (Table 2, which is published as supporting information on the PNAS web site). As a matrix we used chromosomal DNA isolated from L. pneumophila Philadelphia I cells. PCR product was cut with EcoRI/SalI restriction endonucleases and ligated into pGEX-4T3 vector. The resulting plasmid p214–4T3 coded for Lgt1 fused N-terminally to GST tag. Expression and purification of recombinant proteins using glutathione-Sepharose were done as suggested by the manufacturer (Amersham Biosciences) and used as GST-tagged or cleaved proteins. Site-directed mutagenesis was performed with QuikChange (Stratagene, La Jolla, CA). The primers used are listed in Table 2.
Glucosyltransferase Assay.
Eukaryotic cell extracts (proteins at 7–10 mg/ml) used as substrates in the reaction were prepared by sonication of Caco-2 or EBL cells (17, 28). Glucosylation reaction was carried out in 20 μl of a mixture consisting of 20 mM Tris·HCl (pH 7.5), 150 mM NaCl, 1 mM MnCl2, 2–5 μg of recombinant Legionella enzyme, and 50–70 μg of crude cell extract or 2–4 μg of purified recombinant eEF1A1 or eEF1A2 (the EF-coding plasmids were gifts from Charlotte R. Knudsen, University of Aarhus, Aarhus, Denmark) and 10 μM of UDP-[14C]glucose (American Radiolabeled Chemicals, St. Louis, MO). The mixture was incubated at 37°C for 1 h. The reaction was stopped by the addition of Laemmli sample buffer and heating at 100°C for 5 min. The samples were then subjected to SDS/PAGE and scanned on a PhosphorImager.
MS Identification of Proteins Modified by Lgt1.
Caco-2 cell lysate was exposed to UDP-glucose in the absence or presence of Lgt1. An aliquot was also UDP-[14C]glucosylated to define the position of the labeled eukaryotic band. Proteins were analyzed by SDS/PAGE. Lgt1-treated and untreated protein bands, which corresponded in size to the [14C]glucosylated substrate, were cut off from the SDS gel, reduced, alkylated, and digested overnight with trypsin as described (43). The extracted peptide mixtures were dried, taken up in 3 μl of 33% formic acid, their pH adjusted with 3 μl of 25% NH3, and injected onto a C18 precolumn (300 μm wide, 2 cm long) for desalting. Afterward, desalted preparations were separated with a capillary HPLC (Ultimate; Dionex, Idstein, Germany) on a 75-μm-wide, 11-cm-long column packed with C18 material (C-18 ODS AQ; YMC Europe, Dinslaken, Germany) and analyzed online with a quadrupole time of flight instrument (Micromass Q-TOF I, Waters, Elstree, United Kingdom). Fragment spectra of peptides were preprocessed with our own software package (44) before they were submitted to a database search to identify the underlying proteins (Mascot; Matrix Science, London, United Kingdom) (45).
Intoxication of Eukaryotic Cells by Electroporation.
EBL cells were grown in an atmosphere of 5% CO2/95% air at 37°C until confluency on a 9-cm Petri dish in MEM (46). Cells from one Petri dish were trypsinized and resuspended in 2–3 ml of fresh MEM (the resulting density was 2 × 106 to 3 × 106 cells per milliliter), to which different amounts of recombinant L. pneumophila proteins were added. Electroporation settings were 200 V and 950 μF for 4-mm standard electroporation cuvette (GenePulser; Bio-Rad, Hercules, CA). Immediately after the 15–25 msec pulse, cells were seeded into 24-well cell culture plates or 10-cm Petri dishes and incubated for 1 h in a CO2 incubator. Afterward, cells were washed with TBS and incubation continued for up to 3 days in fresh MEM. At daily time points, cells were subjected to phase-contrast microscopy.
Infection of Epithelial Cells by L. pneumophila.
The L. pneumophila serogroup 1 strain Philadelphia I was grown on buffered charcoal-yeast extract agar for 2 days at 37°C before use. A549 cells (American Type Culture Collection, Manassas, VA) were cultured in HAM's F-12 medium, plated in 10-cm dishes, and infected with L. pneumophila at a multiplicity of infection of 10 (i.e., 10 bacteria per cell) in culture medium without FCS. After the time intervals indicated in Fig. 3, A549 cells were washed with ice-cold PBS, scraped in PBS containing 10% glycerol, and stored at −20°C. Immediately before the glucosyltransferase reaction, cells were lysed by 1% Nonidet P-40 detergent on ice for 5 min, and clarified by centrifugation at 14,000 rpm (Eppendorf centrifuge 5402) for 10 min. EBL cells were cultured as described above and were infected at a multiplicity of infection in the range of 0.1–100. After up to 24 h of incubation, infected EBL cells were washed with TBS, suspended by trypsin treatment, washed again, lysed, processed as described for A549, and used immediately in the reaction.
In Vitro Transcription/Translation Assay.
For the in vitro transcription/translation assay, rabbit reticulocyte lysate system L4610 was used as suggested by the manufacturer (Promega, Mannheim, Germany). As a target DNA for transcription/translation experiments, luciferase gene-contained plasmid or Drosophila melanogaster actin act88F gene-contained plasmid (gift from John C. Sparrow, University of York, York, United Kingdom) were used. In translation experiments, luciferase mRNA was used. To investigate the influence of Lgt1 on protein synthesis, different amounts of recombinant active enzyme or site-mutated proteins were added. The mixture was incubated for 90 min at 30°C. A portion of the reaction mixture (1 microliter per measurement) was read in a luminometer. For autoradiography experiments, 5 μl of the reaction mixture was subjected to SDS/PAGE and scanned on a PhosphorImager.
Methionine Incorporation Assay.
For the methionine incorporation assay, EBL cells were harvested into MEM without l-methionine (MEM-M) and were intoxicated by electroporation (as described above) with wild-type or site-mutated recombinant L. pneumophila proteins. After 1 or 2 h of incubation at 37°C, cells were washed and pulsed with [35S]methionine (0.5 μCi per well of a 24-well plate) for 1 or 3 h in MEM-M. After this time, cells were washed with TBS and lysed by 0.1% SDS supplemented with 0.2 mg/ml BSA. Proteins were precipitated by 10% trichloroacetic acid and the amounts of incorporated [35S]methionine were measured by filter assay and liquid scintillation counting (41). At the time points indicated in Fig. 5, >95% of cells were viable as determined by trypan blue assay, and the differences in cell numbers per well were insignificant, as determined by cell counting.
Supplementary Material
Acknowledgments
The work was supported by the Bundesministerium für Bildung und Forschung, International Association for the Promotion of Cooperation with Scientists from the New Independent States of the Former Soviet Union Project 05-1000004-7756 (to Y.B. and K.A.), and the Deutsche Forschungsgemeinschaft (K.A.).
Abbreviations
- EF
elongation factor
- eEF1A
eukaryotic EF1A
- EBL
embryonic bovine lung.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Fields BS, Benson RF, Besser RE. Clin Microbiol Rev. 2002;15:506–526. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marston BJ, Plouffe JF, File TM, Jr, Hackman BA, Salstrom SJ, Lipman HB, Kolczak MS, Breiman RF. Arch Intern Med. 1997;157:1709–1718. [PubMed] [Google Scholar]
- 3.Horwitz MA. J Exp Med. 1983;158:2108–2126. doi: 10.1084/jem.158.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horwitz MA, Maxfield FR. J Cell Biol. 1984;99:1936–1943. doi: 10.1083/jcb.99.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Summersgill JT, Raff MJ, Miller RD. Microb Pathog. 1988;5:41–47. doi: 10.1016/0882-4010(88)90079-4. [DOI] [PubMed] [Google Scholar]
- 6.Horwitz MA. J Exp Med. 1983;158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coers J, Monahan C, Roy CR. Nat Cell Biol. 1999;1:451–453. doi: 10.1038/15687. [DOI] [PubMed] [Google Scholar]
- 8.Gao LY, Abu KY. Infect Immun. 1999;67:862–870. doi: 10.1128/iai.67.2.862-870.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrne B, Swanson MS. Infect Immun. 1998;66:3029–3034. doi: 10.1128/iai.66.7.3029-3034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao LY, Kwaik YA. Environ Microbiol. 2000;2:79–90. doi: 10.1046/j.1462-2920.2000.00076.x. [DOI] [PubMed] [Google Scholar]
- 11.Molmeret M, Bitar DM, Han L, Kwaik YA. Infect Immun. 2004;72:4040–4051. doi: 10.1128/IAI.72.7.4040-4051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrews HL, Vogel JP, Isberg RR. Infect Immun. 1998;66:950–958. doi: 10.1128/iai.66.3.950-958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brand BC, Sadosky AB, Shuman HA. Mol Microbiol. 1994;14:797–808. doi: 10.1111/j.1365-2958.1994.tb01316.x. [DOI] [PubMed] [Google Scholar]
- 14.Berger KH, Isberg RR. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 15.Segal G, Feldman M, Zusman T. FEMS Microbiol Rev. 2005;29:65–81. doi: 10.1016/j.femsre.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Cianciotto NP. Trends Microbiol. 2005;13:581–588. doi: 10.1016/j.tim.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Belyi I, Popoff MR, Cianciotto NP. Infect Immun. 2003;71:181–186. doi: 10.1128/IAI.71.1.181-186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Just I, Gerhard R. Rev Physiol Biochem Pharmacol. 2004;152:23–47. doi: 10.1007/s10254-004-0033-5. [DOI] [PubMed] [Google Scholar]
- 19.Just I, Selzer J, Wilm M, Von Eichel-Streiber C, Mann M, Aktories K. Nature. 1995;375:500–503. doi: 10.1038/375500a0. [DOI] [PubMed] [Google Scholar]
- 20.Busch C, Hofmann F, Selzer J, Munro J, Jeckel D, Aktories K. J Biol Chem. 1998;273:19566–19572. doi: 10.1074/jbc.273.31.19566. [DOI] [PubMed] [Google Scholar]
- 21.Reinert DJ, Jank T, Aktories K, Schulz GE. J Mol Biol. 2005;351:973–981. doi: 10.1016/j.jmb.2005.06.071. [DOI] [PubMed] [Google Scholar]
- 22.Kapp LD, Lorsch JR. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 23.Chuang SM, Chen L, Lambertson D, Anand M, Kinzy TG, Madura K. Mol Cell Biol. 2005;25:403–413. doi: 10.1128/MCB.25.1.403-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hotokezaka Y, Tobben U, Hotokezaka H, Van LK, Beatrix B, Smith DH, Nakamura T, Wiedmann M. J Biol Chem. 2002;277:18545–18551. doi: 10.1074/jbc.M201022200. [DOI] [PubMed] [Google Scholar]
- 25.Gross SR, Kinzy TG. Nat Struct Mol Biol. 2005;12:772–778. doi: 10.1038/nsmb979. [DOI] [PubMed] [Google Scholar]
- 26.Ogle JM, Ramakrishnan V. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 27.Ogle JM, Carter AP, Ramakrishnan V. Trends Biochem Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 28.Andersen GR, Pedersen L, Valente L, Chatterjee I, Kinzy TG, Kjeldgaard M, Nyborg J. Mol Cell. 2000;6:1261–1266. doi: 10.1016/s1097-2765(00)00122-2. [DOI] [PubMed] [Google Scholar]
- 29.Kjeldgaard M, Nyborg J, Clark BFC. FASEB J. 1996;10:1347–1368. [PubMed] [Google Scholar]
- 30.Vetter IR, Wittinghofer A. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 31.Abel K, Yoder MD, Hilgenfeld R, Jurnak F. Structure (London) 1996;4:1153–1159. doi: 10.1016/s0969-2126(96)00123-2. [DOI] [PubMed] [Google Scholar]
- 32.Sehr P, Joseph G, Genth H, Just I, Pick E, Aktories K. Biochemistry. 1998;37:5296–5304. doi: 10.1021/bi972592c. [DOI] [PubMed] [Google Scholar]
- 33.Geyer M, Wilde C, Selzer J, Aktories K, Kalbitzer HR. Biochemistry. 2003;42:11951–11959. doi: 10.1021/bi034529v. [DOI] [PubMed] [Google Scholar]
- 34.Venema RC, Peters HI, Traugh JA. J Biol Chem. 1991;266:12574–12580. [PubMed] [Google Scholar]
- 35.Dever TE, Costello CE, Owens CL, Rosenberry TL, Merrick WC. J Biol Chem. 1989;264:20518–20525. [PubMed] [Google Scholar]
- 36.Zobel-Thropp P, Yang MC, Machado L, Clarke S. J Biol Chem. 2000;275:37150–37158. doi: 10.1074/jbc.M001005200. [DOI] [PubMed] [Google Scholar]
- 37.Salcedo SP, Holden DW. Curr Opin Microbiol. 2005;8:92–98. doi: 10.1016/j.mib.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 38.Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR. Nat Cell Biol. 2006;8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- 39.Sandvig K. Toxicon. 2001;39:1629–1635. doi: 10.1016/s0041-0101(01)00150-7. [DOI] [PubMed] [Google Scholar]
- 40.Wilson BA, Collier RJ. Curr Top Microbiol Immunol. 1992;175:27–42. doi: 10.1007/978-3-642-76966-5_2. [DOI] [PubMed] [Google Scholar]
- 41.McCusker KT, Braaten BA, Cho MW, Low DA. Infect Immun. 1991;59:240–246. doi: 10.1128/iai.59.1.240-246.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.bu-Zant A, Santic M, Molmeret M, Jones S, Helbig J, Abu KY. Infect Immun. 2005;73:5339–5349. doi: 10.1128/IAI.73.9.5339-5349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 44.Gentzel M, Kocher T, Ponnusamy S, Wilm M. Proteomics. 2003;3:1597–1610. doi: 10.1002/pmic.200300486. [DOI] [PubMed] [Google Scholar]
- 45.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 46.Busch C, Orth J, Djouder N, Aktories K. Infect Immun. 2001;69:3628–3634. doi: 10.1128/IAI.69.6.3628-3634.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.