Abstract
Muscle activity contributes to muscle development and function largely by means of regulated gene expression. Many genes crucial to neuromuscular synapse formation, such as MuSK and nAChRs, are induced before muscle innervation or after muscle denervation, and this induction requires expression of the E-box binding, basic helix–loop–helix muscle-specific transcription factor, myogenin (Mgn). The mechanism by which muscle activity is coupled to gene expression is poorly defined. Here we report that inhibition of histone deacetylase (HDAC) activity attenuates the induction of activity-regulated genes in aneural myotubes and adult denervated muscle. The effect of HDAC inhibitors requires new protein synthesis, suggesting HDACs may regulate the expression of a Mgn transcriptional repressor. We identified Dach2 as a Mgn transcriptional repressor whose expression is dramatically reduced in an HDAC-dependent manner in developing aneural myotubes or adult denervated muscle. Dach2 overexpression in denervated muscle suppressed Mgn, nAChR, and MuSK gene induction, whereas Dach2 knockdown induced Mgn gene expression in innervated muscle and relieved Mgn promoter inhibition by HDAC inhibitors. Thus, a HDAC-Dach2-myogenin signaling pathway has been identified to decode nerve activity and control muscle gene expression in developing and adult skeletal muscle.
Keywords: acetylcholine receptor, depolarization, MuSK, synaptogenesis, transcription
Uncovering the mechanisms by which muscle activity controls gene expression is crucial to understanding how activity controls muscle development and function. One of the best studied systems documenting activity-dependent gene expression is the neuromuscular junction where muscle depolarization helps restrict expression of proteins necessary for nerve-muscle communication to the synapse (1). These proteins include nicotinic acetylcholine receptors (nAChRs) and the muscle-specific tyrosine kinase (MuSK). The mechanisms contributing to activity-dependent gene expression are just beginning to emerge. Early events in depolarization-dependent gene regulation include an activity-dependent increase in intracellular calcium (2–4) that activates a protein kinase C (5, 6) and/or calcium/calmodulin-dependent protein kinase (CaMK)II-dependent (7, 8) signal transduction cascade leading to suppression of nAChR gene expression. The mechanism by which these kinases lead to gene suppression is not known, although ultimately they must control the expression or function of transcription factors.
One set of transcription factors that can regulate the expression of genes involved in neuromuscular junction formation are bHLH MyoD family members, MyoD, myogenin (Mgn), myf5, and MRF4 (9–16). We recently showed that Mgn is necessary for nAChR gene induction in differentiated myotubes (17) and adult denervated skeletal muscle (15). Therefore, to further our understanding of activity-dependent control of gene expression, it is important to understand the mechanisms by which muscle activity controls Mgn gene expression.
Recent studies indicate that denervation-dependent gene induction is mediated by increased histone acetylation (18). Mgn and nAChR genes are hyperacetylated and induced after muscle denervation. The HDAC9 splice variant, MITR, is highly expressed in innervated muscle and contributes to Mgn gene repression by binding MEF2 and recruiting HDACs 1 and 3 to the Mgn promoter. However, MITR−/− mice still exhibit activity-dependent Mgn and nAChR gene regulation, suggesting additional mechanisms are involved.
Here we report the identification of a second mechanism for regulating gene expression by muscle activity. In apparent contradiction to the observation that muscle denervation is accompanied by chromatin hyperacetylation, we found HDAC activity is necessary for denervation-dependent gene induction. Interestingly, this regulation is mediated by the Mgn transcriptional repressor, Dach2. We show that HDAC activity is necessary for Dach2 gene suppression in denervated muscle and that Dach2 gene suppression is required for Mgn gene induction. Based on these results, along with previous data, we present a new model for activity-dependent gene expression in skeletal muscle.
Results
Mgn Gene Induction in Inactive Muscle Requires HDAC Activity.
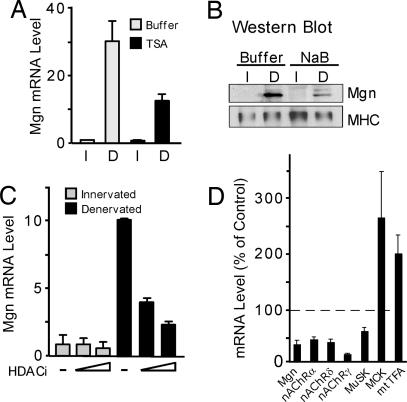
Activity-dependent suppression of Mgn and nAChR gene expression in innervated muscle is associated with histone hypoacetylation, presumably because of increased HDAC activity (18). To determine whether HDAC activity is necessary for Mgn gene suppression, we exposed innervated and denervated muscle to HDAC inhibitors (Fig. 1). Both long-term (14 days; Fig. 1 A and B) and short-term (3 days; Fig. 1C) denervated sternomastoid muscles were examined. The HDAC inhibitor, trichostatin A (TSA), was applied locally to sternomastoid muscles, in vivo, for 12 h just before harvesting muscles for Mgn RNA analysis by real-time quantitative PCR (Fig. 1 A and C) or protein analysis on Western blots (Fig. 1B). Surprisingly, HDAC inhibition did not relieve Mgn gene suppression in innervated muscle, but, instead, suppressed Mgn gene and protein induction after muscle denervation (Fig. 1 A–C). The inability to completely reverse Mgn gene expression (80% maximal inhibition; Fig. 1C) with TSA probably reflects: (i) its inability to completely penetrate the muscle, (ii) the relatively short time of application (last 12 h of a 3-day or 14-day denervation), and (iii) the presence of myogenin gene activators, such as MEF2, in denervated muscle. In contrast, myosin (Fig. 1B) and γ-actin RNA (data not shown) expression were not affected by muscle denervation or HDAC inhibition. To ascertain whether these results were unique to the sternomastoid muscle, we examined the effect of HDAC inhibition on diaphragms explanted to organ culture (Fig. 1D) (19, 20). Consistent with the results obtained in vivo by using the sternomastoid muscle, gene induction in denervated diaphragm is also attenuated by HDAC inhibition (Fig. 1D).
Fig. 1.
HDAC inhibition attenuates denervation-dependent Mgn gene induction. (A and C) Relative Mgn RNA level in innervated and 14-day (A) or 3-day (C) denervated sternomastoid muscles bathed, in vivo, in buffer or HDAC inhibitors (HDACi) for 12 h before tissue harvest (n = 2, for each individual experiment). Error bars are standard deviation. (B) Western blot analysis of Mgn protein levels in innervated and 14-day denervated sternomastoid muscle bathed, in vivo, in buffer or NaB for 12 h before tissue harvest. (D) RNA levels in adult mouse diaphragm muscles placed in organ culture with TSA or vehicle for 48 h (n = 4). Error bars are standard error of the mean. All RNA levels were assayed by real-time PCR and normalized to γ-actin, which did not change after muscle denervation or HDAC inhibition. I, innervated; D, denervated.
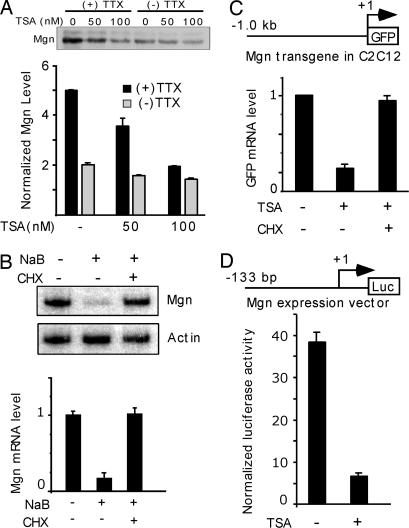
To determine whether HDAC activity also participates in gene induction in differentiated myotubes, we treated C2C12 myotubes with HDAC inhibitors and assayed Mgn gene expression (Fig. 2). Without influencing myotube morphology, HDAC inhibition dramatically suppressed Mgn protein levels preferentially in inactive myotubes (+TTX) (Fig. 2A). This suppression is not a result of increased Mgn protein degradation because Mgn mRNA levels also decreased drastically after HDAC inhibition (Fig. 2B). Interestingly, blocking new protein synthesis with cycloheximide (CHX) completely eliminated the suppressive effect of HDAC inhibitors on Mgn expression (Fig. 2 B and C), suggesting this effect is mediated by a Mgn transcriptional repressor. We confirmed that Mgn transcription was suppressed by HDAC inhibition (Fig. 2C) and mapped this effect to a 133-bp region of the Mgn promoter (Fig. 2D).
Fig. 2.
HDAC inhibitors require new protein synthesis to inhibit Mgn gene expression. Differentiated myotubes were used to investigate the effect of HDAC inhibition on Mgn gene expression. (A) Western blot analysis of Mgn protein levels in active (−TTX) and inactive (+TTX) myotubes treated with either buffer or TSA for 6 h. Graph below the blot shows quantification of Mgn protein levels shown on Western blot after normalization to total protein applied to the gel (n = 3). (B) New protein synthesis is required for Mgn RNA suppression by HDAC inhibitors. C2C12 myotubes were treated with buffer or NaB in the presence or absence of cycloheximide (CHX) for 6 h before isolating RNA for radioactive RT PCR and analysis on polyacrylamide gels (autoradiogram shown, Upper). (B Lower) Graph shows quantification of data shown in autoradiogram (n = 3). (C and D) Mgn promoter activity is suppressed by HDAC inhibitors. (C) C2C12 myotubes harboring an integrated 1-kb Mgn promoter driving GFP gene expression were treated with and without TSA and CHX for 12 h before quantifying GFP and actin RNA levels by real-time PCR (n = 3). GFP RNA level is normalized to actin RNA. (D) C2C12 cells were transfected with a 133-bp Mgn promoter-luciferase expression vector along with CMV-CAT for normalization. Transfected myotubes were treated with and without TSA for 12 h before cell harvest for luciferase and CAT assays (n = 3). Error bars are standard deviation.
Dach2 Is a HDAC-Regulated Gene That Contributes to Mgn Repression in Innervated Muscle.
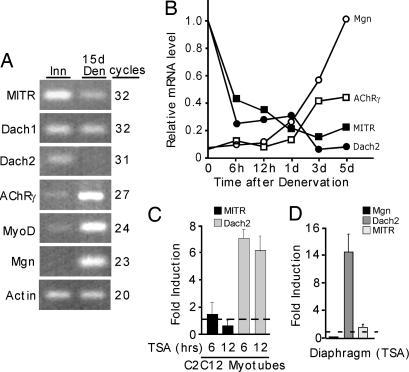
The above data suggests that a transcriptional repressor mediating Mgn gene suppression in innervated muscle is inhibited in an HDAC-dependent manner after muscle denervation. Candidate repressors, previously reported to inhibit Mgn transcription, include MITR (18) and Dach proteins (21). There are two Dach genes expressed in mammals, Dach1 and Dach2 (22, 23). Interestingly, RT-PCR reveals that Dach2 and MITR, but not Dach1, are dramatically suppressed after muscle denervation (Fig. 3A). Shortly after muscle denervation, we observed a very rapid drop in Dach2 and MITR expression that preceded Mgn and nAChR gene induction (Fig. 3B). Thus MITR and Dach2 emerge as good candidates for inhibiting Mgn expression in innervated muscle. However, only Dach2 is regulated in a HDAC-dependent manner (Fig. 3 C and D).
Fig. 3.
Dach2 expression is regulated in an activity- and HDAC-dependent manner. (A) Gene expression in innervated and 15-day denervated TA muscle. RT-PCR was used to evaluate gene expression, and reaction products were visualized on agarose gels stained with ethidium bromide. The number of PCR cycles is indicated. (B) Time course of gene expression after muscle denervation. At the indicated times, innervated and denervated TA muscles were harvested for RNA analysis by real-time PCR. Experiment was repeated twice with similar results. (C) Dach2 RNA is specifically induced by HDAC inhibition. C2C12 myotubes were treated with TSA or buffer for 6 and 12 h before isolating RNA and assaying Dach2 and MITR RNA levels by real-time PCR (n = 3). (D) Diaphragms in organ culture induce Dach2 expression. Diaphragms were cultured for 48 h in the presence of TSA or buffer (n = 3). RNA was assayed by real-time PCR and normalized to actin RNA levels. Error bars are standard deviation.
We confirmed Dach2 is a Mgn transcriptional repressor that mediates HDAC-dependent regulation by (i) overexpressing Dach2 in myotubes harboring the 133-bp Mgn promoter (Fig. 4A) and (ii) rescuing TSA-mediated Mgn repression by Dach2 knockdown (Fig. 4B). It is interesting that TSA-mediated suppression was somewhat more robust at inhibiting Mgn promoter activity than Dach2-mediated suppression, perhaps suggesting that HDAC inhibitors are suppressing Mgn expression in both a Dach2-dependent and independent fashion. This latter idea is consistent with our finding that Dach2 knockdown does not completely rescue Mgn promoter activity in TSA-treated cells (Fig. 4B).
Fig. 4.
Dach2 inhibits Mgn promoter activity and Dach2 knockdown abrogates TSA-mediated Mgn promoter inhibition. (A) Dach2 overexpression inhibits Mgn promoter activity. C2C12 cells were cotransfected with the 133-bp Mgn promoter-luciferase expression vector, CMV-CAT for normalization, and either pCS2GFP or pCDNA3Dach2. Cells were differentiated, and 4 days posttransfection, myotubes were harvested for luciferase and CAT assays (n = 3). (B) Dach2 knockdown rescues Mgn gene expression in myotubes treated with TSA. C2C12 cultures were transfected with the 133-bp Mgn promoter driving luciferase expression along with CMV-CAT, and with either a control or two different Dach2-targeted siRNAs (1# and 2#). Transfected and differentiated myotubes were treated with either TSA or vehicle for 12 h before harvesting for luciferase and CAT assays (n = 3). (C and D) The MEF3 element of the Mgn promoter confers Dach2-dependent suppression on the minimal enkephalin promoter (MEK). HEK cells (C) (n = 3) or adult denervated muscle (D) (n = 4) were transfected or electroporated, respectively, with the indicated vectors. Three days later, cells or tissue were harvested for luciferase and CAT assays. Error bars are standard deviation.
Dach proteins are thought to regulate Mgn promoter activity via their interaction with Six/Eya proteins bound to the Mgn promoter's MEF3 site (21, 24). Consistent with this idea is our finding that the Mgn promoter with a mutated MEF3 site was barely suppressed by Dach2 overexpression (Fig. 4A) and 3xMEF3 sites ligated upstream of the minimal enkephalin promoter (MEK) confers Dach2-dependent suppression when transfected into HEK cells (Fig. 4C) or electroporated into adult denervated muscle (Fig. 4D).
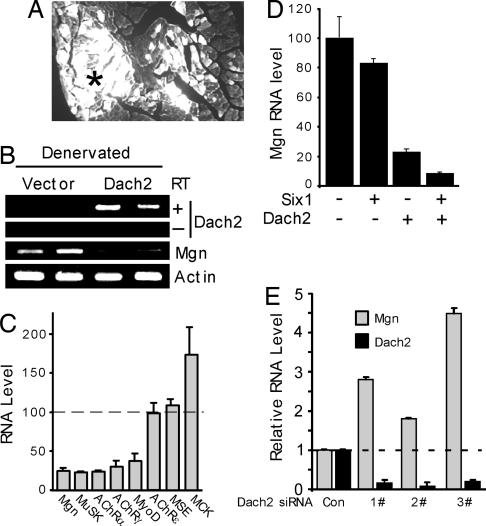
To test whether Dach2 suppression is necessary for denervation-dependent gene induction, we overexpressed Dach2 in adult denervated tibialis anterior (TA) muscle (Fig. 5). For these experiments, adult innervated TA muscle was electroporated with a GFP expression vector along with either a control or Dach2 expression plasmid. Twelve days later muscles were denervated for 3 days before harvesting transfected muscle fibers by using a dissecting microscope equipped with fluorescence optics (Fig. 5A). RNA isolated from electroporated fibers was assayed by using PCR, followed by gel electrophoresis to visualize amplified products (Fig. 5B), and real-time PCR (Fig. 5C) to obtain more quantitative results. Increased expression of Dach2 in denervated muscle was verified and was accompanied by a dramatic reduction of Mgn expression (Fig. 5B). In addition, we found that genes normally induced by muscle denervation such as Mgn, MyoD, nAChR α-, γ- and δ-subunits, and MuSK, were suppressed by Dach2 expression (to ≈25% of their control values; Fig. 5C), whereas muscles electroporated with the empty vector expressed these genes at a high level (100% in Fig. 5C). The specificity of this effect is demonstrated by genes encoding the nAChR ε-subunit (nAChRε), muscle-specific creatine kinase (MCK), and muscle-specific enolase (MSE), which were unaffected or slightly increased by Dach2 over-expression (Fig. 5C).
Fig. 5.
Dach2 overexpression suppresses denervation-dependent gene induction and Dach2 knockdown induces Mgn gene expression in innervated muscle. (A) Muscle electroporation with a GFP expression plasmid facilitates identification of electroporated muscle fibers. TA muscle was electroporated with a GFP expression vector (5 μg), and, 14 days later, TA muscle was isolated and GFP fluorescence was examined in muscle cross sections. White areas are GFP expression, and the asterisk in the center represents the region where we isolate GFP+ fibers for RNA analysis. (B) Dach2 overexpression in denervated muscle suppresses Mgn RNA expression. Represented is an agarose gel showing PCR amplification of Dach2, Mgn, and γ-actin RNA isolated from denervated muscle fibers electroporated with either a control or Dach2-expressing vector. Dach2 overexpression induced high levels of Dach2 RNA. Dach2 RNA was amplified in the presence (+) of reverse transcriptase (RT) but not in its absence (−). Note that, when Dach2 expression is high, Mgn expression is low, whereas actin RNA levels are unaffected. (C) Real-time PCR analysis of RNA from 3-day denervated muscle electroporated with control or Dach2 expression vector. Dach2 electroporated muscle RNA levels are normalized to vector electroporated muscle RNA values that were set to 100% (n = 4). (D) Six1 synergizes with Dach2 to mediate Mgn gene suppression. Real-time PCR analysis of RNA from 3-day denervated muscle electroporated with the indicated plasmids. RNA values are normalized to vector electroporated muscle RNA values that are set to 100% (n = 4). (E) siRNA-mediated Dach2 knockdown in innervated muscle induces Mgn expression. Innervated TA muscle was electroporated with control or one of three different Dach2 targeted siRNAs (1#, 2#, and 3#). Fourteen days postelectroporation, TA muscles were isolated, and Mgn and Dach2 RNA levels were quantified by real-time PCR (n = 4). Error bars are standard error of the mean.
Because Dach-mediated gene repression is thought to be via interactions with Six proteins bound to the Mgn promoters MEF3 site (21, 24), we investigated whether Six proteins may enhance Dach2-mediated gene repression in denervated muscle. Although Six1 overexpression had only a modest effect on Mgn RNA levels, it did enhance Dach2-mediated suppression, resulting in a 90% reduction in Mgn gene expression (Fig. 5D). Interestingly, we found that Dach2 expression in 14-day denervated muscle also resulted in ≈90% reduction in myogenin RNA levels (Fig. 7, which is published as supporting information on the PNAS web site), perhaps indicating Six protein levels or activity may increase during the later stages of muscle denervation.
To investigate whether Dach2 is sufficient for mediating Mgn gene repression in innervated muscle, we knocked down Dach2 expression with siRNAs. For these experiments we electroporated innervated TA muscle with one of three different Dach2-targeted siRNAs or a control siRNA, along with a GFP expression vector to identify electroporated fibers. Previous studies have demonstrated prolonged gene silencing in skeletal muscle harboring gene-specific siRNAs (15, 25). Fourteen days postelectroporation, GFP+ fibers were isolated and assayed for Dach2 and Mgn RNA. Consistent with Dach2 functioning as a repressor of Mgn gene expression, we found that Dach2 knockdown in innervated muscle increased Mgn RNA levels (Fig. 5D). However, this increased Mgn expression is still much less than what is observed in denervated muscle, suggesting additional regulatory mechanisms contribute to the very high Mgn levels in denervated muscle.
Discussion
Although activity-dependent gene regulation has been studied for decades, little is known about the mechanisms underlying this regulation. Unraveling these regulatory mechanisms may not only contribute to a better understanding of how muscle activity contributes to muscle function but also may suggest novel therapies for improving muscle function after injury or disease. We used Mgn, MyoD, MuSK, and nAChR gene expression as probes for studying activity-dependent gene regulation in muscle. We previously demonstrated that MuSK and nAChR gene induction after muscle denervation requires activation of Mgn gene expression (15). Here we report that (i) HDAC activity is necessary for gene induction in inactive muscle (aneural myotubes and denervated adult muscle), (ii) Dach2 is an activity- and HDAC-regulated Mgn transcriptional repressor, (iii) Dach2 mediates its effects on gene expression via the Mgn promoter's MEF3 site, and (iv) Dach2 suppression after muscle denervation is necessary for Mgn, MyoD, nAChR, and MuSK gene induction. This HDAC-Dach2-Mgn signaling pathway represents a signal transduction cascade by which muscle decodes neural activity to control gene expression.
HDAC Activity Is Required for Mgn, nAChR, and MuSK Gene Induction in Inactive Muscle.
We used pharmacological inhibition of HDAC's in C2C12 myotubes and adult muscle fibers to show HDAC activity is necessary for Mgn, nAChR, and MuSK gene induction in inactive muscle. Although HDAC inhibitors have previously been used to study myogenesis (26) and have been shown to inhibit Mgn expression in differentiated myotubes (27, 28), their effect on adult skeletal muscle has not been reported. Our finding that both mature myotubes and adult denervated muscle require HDAC activity and Dach2 repression for Mgn and Mgn-dependent gene induction suggests a common underlying mechanism. The observation that innervated muscle gene expression was relatively unperturbed by HDAC inhibition and that denervated muscle HDAC inhibition specifically affected genes whose expression is normally induced after muscle denervation, highlights the specificity of the drug treatments.
Because Mgn gene suppression is accompanied by chromatin hypoacetylation in innervated muscle (18), we were surprised to find that HDAC activity is not required for this suppression and suggests redundant modes of repression. Indeed MITR binds the Mgn transcriptional activator MEF2 (29, 30), and may provide a HDAC activity-independent mode of Mgn gene repression. In addition, posttranslational control of MEF2 activity (ref. 31 and references therein) and a dearth of Mgn transcriptional activators are also likely to contribute to its expression level.
Our finding that HDAC activity was required for Mgn and Mgn-dependent gene induction in inactive muscle suggests HDACs inhibit expression of a transcriptional repressor that is normally expressed in active muscle. Experiments employing cycloheximide confirmed this idea and suggested that the Mgn promoter was a possible target for this repression.
HDACs Couple Muscle Depolarization with Gene Expression.
The observation that HDAC activity is necessary for denervation-dependent gene induction, along with previous reports indicating class II HDACs nuclear export is regulated by calcium (32) and muscle activity (33), suggests a mechanism by which changes in intracellular calcium are coupled to gene expression. Although we have not identified the specific HDACs that mediate Dach2 suppression, we suspect that class II HDACs (HDACs 4, 5, 6, or 7) will be involved. Class II HDACs are calcium responsive HDACs whose nuclear-cytoplasmic distribution is regulated by calcium (32). These particular HDACs are often excluded from the nucleus when calcium levels are high. Recently HDAC4 nuclear-cytoplasmic shuttling was shown to be regulated by muscle depolarization (33). Therefore, HDAC4 represents a good candidate for mediating the effects of muscle denervation on Dach2 expression.
Calcium-dependent nuclear export of class II HDACs is mediated by CaMK-dependent HDAC phosphorylation (32, 33). Interestingly, we previously reported that a calcium/CaMK-dependent signal transduction cascade also mediates Mgn and nAChR gene suppression by muscle activity (7, 8). Thus, calcium-dependent nuclear localization of class II HDACs seem to play a key role in coupling muscle activity to the genome.
Dach2 Is a HDAC-Regulated Mgn Gene Suppressor.
Dach2 is the vertebrate homolog of the Drosophila Dachshund (Dac) gene. Dac along with eyeless (eye), eyes absent (eya), and sine oculis (so) are components of compound eye formation (34). In vertebrates these genes also participate in muscle formation and Six/sine oculus proteins regulate Mgn expression via its MEF3 site (24). Six:Dach complexes have been reported to inhibit Mgn promoter activity (21) and Eya, which is a Dach interacting protein (35), may convert the function of this complex from gene suppression to gene activation (21). Consistent with this data are our observations that Dach2-mediated Mgn repression is via the Mgn promoter's MEF3 site, and this repression is enhanced by Six protein overexpression.
Dach2 overexpression in denervated TA muscle suppressed Mgn, MyoD, nAChR, and MuSK gene expression, whereas Dach2 siRNA-mediated knockdown induced Mgn expression in innervated muscle. Our data suggest that Dach2 suppresses Mgn promoter activity via its MEF3 site, leading to nAChR and MuSK suppression. The mechanism by which Dach2 leads to MyoD suppression is not clear, although it is possible that in adult denervated muscle Mgn and MyoD regulate each other's expression.
The 2- to 4-fold increase in Mgn levels after Dach2 knockdown in innervated muscle does not approach the induction of Mgn observed after muscle denervation (>30-fold induction) and likely reflects additional mechanisms of Mgn suppression. Indeed, MITR is a Mgn repressor (18) that does not seem to be regulated by the HDAC-Dach2 signaling pathway. In addition, siRNA-mediated Dach2 knockdown is not complete, and residual Dach2 expression may be sufficient to maintain low levels of Mgn gene expression. Recently, a Dach2 knockout mouse was generated and found to be viable with no gross defects in eye or brain development (36). Although skeletal muscle was not specifically examined, limbs were reported to appear normal. Whether the expression of genes induced by muscle inactivity is perturbed in the Dach2 knockout animal is not known; however, if Dach2 represents just one of multiple pathways controlling activity-dependent gene expression, an obvious phenotype may be hard to discern.
Model for Activity-Dependent Control of Skeletal Muscle Gene Expression.
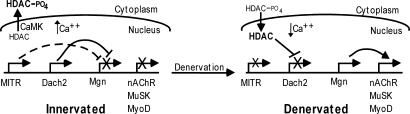
The results reported here, along with previous studies, allow us to formulate a model for muscle activity-dependent gene expression (Fig. 6). It is proposed that specific class II HDACs are exported from the nucleus in a calcium/CaMKII-dependent manner in active muscle and translocate to the nucleus on silencing of muscle activity (32, 33), providing a link between changes in muscle calcium and gene expression. In innervated muscle, MITR and Dach2 are highly expressed. MITR mediates Mgn gene repression (dotted line) by binding MEF2 and perhaps recruiting HDACs to the Mgn promoter (18). In contrast, Dach2 suppresses Mgn transcription via its interaction with Six proteins bound to the Mgn promoters MEF3 site (21, 24). On muscle denervation, specific class II HDACs are imported into the nucleus and suppress Dach2 expression and perhaps other genes that are down-regulated after muscle denervation. The reduced expression of Dach2 and MITR in denervated muscle allows for Mgn gene induction, which participates in the activation of other genes such as MuSK and nAChRs.
Fig. 6.
Model of activity-dependent gene regulation. MITR and Dach2 are Mgn gene repressors. In innervated muscle, calcium levels are high, and CaMK-mediated phosphorylation of specific HDACs leads to their nuclear export. MITR and Dach2 expression is high in innervated muscle. MITR inhibits Mgn gene expression (dotted line) by sequestering MEF2 from the Mgn promoter. Dach2 inhibits Mgn gene expression (solid line) by means of its interaction with Six proteins residing at the Mgn promoter's MEF3 site. On muscle denervation, HDACs are imported into the nucleus and suppress Dach2 gene expression. MITR expression is also inhibited by an unknown mechanism. The resulting relief of Mgn repression is necessary for denervation-dependent gene induction.
Materials and Methods
Cell Culture, Plasmids, and Antibodies.
Primary muscle and C2C12 cells were cultured as described (7). Tetrodotoxin (TTX) (5 μM) was used to keep myotubes inactive. Cells were treated with 5–20 mM sodium butyrate (NaB, dissolved in H2O) or 50–100 nM Trichostatin A (TSA, dissolved in DMSO). Cycloheximide (dissolved in ethanol) was used at 50 μg/ml. The 1-kb Mgn promoter spanning nucleotides −1001 to +1 (translation initiation) and driving EGFP expression was stably integrated into C2C12 cells by using G418 (400 μg/ml) selection. Mgn G133-Luc harbors the 133-bp Mgn promoter (−133 to +1) driving luciferase expression. 3xMEF3MEKpXP2 harbors the minimal enkephalin (MEK) promoter with 3× MEF3 sites (GGGGCTCAGGTTTCTGTG) upstream and drives luciferase expression. CMV–chloramphenicol acetyltransferase (CAT) has been described (7). Mgn (F5D) and myosin (MF20) antibody were purchased from the Iowa Developmental Studies Hybridoma bank. Protein expression was examined by Western blot.
Animal Model, Diaphragm Culture, and HDAC Inhibition.
The sternomastoid muscles of adult mice were exposed and unilaterally denervated by removing a 2-mm section of the nerve. On day 2.5 or 13.5 postdenervation, innervated and denervated muscles were bathed, in vivo, in 250 mM NaB, 5 μM TSA or 15 μM TSA and 250 mM NaB for 12 h while the mice were anesthetized. Control animal received vehicle (0.9% NaCl with or without 0.1% DMSO). After drug treatment, both innervated and denervated sternomastoid muscles were collected for RNA analysis. The lower hind limb muscles of adult mice were denervated as described (10). For organ culture of hemidiaphragms, mice were killed by cervical dislocation, and the diaphragm was dissected and immersed in 10% FBS, DMEM. Connective tissue and fat were removed, and diaphragms were cut into halves, placed in fresh 10% FBS, DMEM with 0.1%DMSO or 5 μM TSA, and incubated with periodic rocking for 48 h in 8% CO2 at 37°C.
In Vivo Muscle Electroporation.
For Dach2 overexpression experiments, 7.5 μg pCS2EGFP plasmid and either 15 μg of Dach2 expression vector or control vector were injected into innervated TA muscle of anesthetized adult mice. For Dach2 and Six overexpression experiments, 2 μg pCS2EGFP, ±3 μg of Dach2, ±3 μg of Six1, ±3 μg of control expression vector were injected into innervated TA muscle. For Dach2 knockdown, 320 picomoles Stealth siRNAs targeting Dach2 (Invitrogen, Carlsbad, CA) or control siRNA (similar GC content; Invitrogen) were injected into innervated TA muscle. Nucleic acid uptake was facilitated by placing electrodes, coated with ultrasound transmission gel, on each side of the leg and by using a BTX840 square wave electroporator to deliver six pulses of 140 V/cm of 60-ms duration with an interval of 100 ms. For 3-day denervation experiments, animals were allowed to recover for 12 days before muscle denervation. A stereomicroscope (Leica) equipped with fluorescence optics was used to isolate electroporated/GFP+ muscle fibers. GFP+ fibers were isolated in ice-cold PBS and then immediately lysed in TRIzol (Invitrogen). Stealth modified Dach2 siRNA targeted the following sequences: Dach2 siRNA 1#, AUAAAUGGGAGAUCCAGACGAUUGC; Dach2 siRNA 2#, UAAACAGUAAUAGUUACCUCCUUGC; Dach2 siRNA 3#, UAUCUUGCAAUACUCCCAAAGAACG.
RNA Isolation and RT-PCR.
Total RNA was isolated by using TRIzol and quantified spectrophotometrically. One to two micrograms of total RNA was used to generate cDNA with oligo(dT) primers and Superscipt II reverse transcriptase. DNA was amplified by using Taq polymerase and 0.2 mM dNTPs spiked, in some instances, with 1 μCi α-32P-dCTP (10 mCi/ml). The amplification profile was 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min. Amplified samples were analyzed on 1.5% agarose gels and stained with ethidium bromide or 6% acrylamide gels and subjected to autoradiography. γ-actin was used for normalization in all PCRs. For more quantitative data, real-time PCR was performed with iQ SYBR green Supermix on iCycler iQ real-time detection system. The procedure followed the manufacturer's instruction, and the reaction was at 20 μl scale. The specificity and efficiency of the primers used in real-time PCRs were first verified by PAGE or agarose gel electrophoresis. PCR cycles are 95°C for 30 s, 58°C for 30 s, and 72°C for 40 s. All real-time PCR results were normalized to γ-actin. The primers used are Mgn forward 5′-CTCAGCTTAGCACCGGAAGCCCGA, reverse 5′-ATTGCCCCACTCCGGAGCGCAGGAG; nAChR α-subunit forward 5′-CGTCTGGTGGCAAAGCT, reverse 5′-CCGCTCTCCATGAAGTT; nAChR γ-subunit forward 5′-ACGGTTGTATCTACTGGCTG, reverse 5′-GATCCACTCAATGGCTTGC; nAChR δ-subunit forward 5′-GTGATCTGTGTCATCGTACT, reverse 5′-GCTTCTCAAACATGAGGTCA; nAChR ε-subunit forward: 5′-AGACCTACAATGCTGAGGAGG, reverse 5′-GGATGATGAGCGTATAGATGA; MuSK forward 5′-CTCGTCCTCCCATTAATGTAAAAA, reverse 5′-TCCAGCTTCACCAGTTTGGAGTAA; muscle-specific creatine kinase forward 5′-GCCGGCGATGAGGAGTCCTACA, reverse 5′-GCAGTGCGGAGGCAGAGTGTAA; γ-actin forward 5′-ACCCAGGCATTGCTGACAGGATGC, reverse 5′-CCATCTAGAAGCATTTGCGGTGGACG; muscle-specific enolase forward 5′-AAAAGTTGACAAATTATGATTGA, reverse 5′-AATCATGAACTCCTGCATGG; Dach1 forward 5′-CATGGACACCACTACCTCC, reverse 5′-GTTGGACCTGCTTTTCTTGA; Dach2 forward 5′-ACTGAAAGTGGCTTTGGATAA, reverse 5′-TTCAGACGCTTTTGCATTGTA; HDAC9 (MITR) forward 5′-TCAGAGGTTCCTATGGGCCTG, reverse 5′-TGGAGACGTTCCACTGAGGG; mitochondrial transcription factor A, forward 5′-GAGCTTGTAAATGAGGCTTG, reverse 5′-CACTTCGACGGATGAGATC; MyoD, forward 5′-TACCCAAGGTGGAGATCCT, reverse 5′-GCGGTGTCGTAGCCATTCT; GFP, forward 5′-GACGACGGCAACTACAAGAC, reverse 5′-GGTCACGAACTCCAGCAGGAC.
Supplementary Material
Acknowledgments
We thank Drs. C. Tabin (Harvard University, Cambridge, MA), X. Li and M. Rosenfeld (both from University of California at San Diego, La Jolla, CA), and G. Mardon (Baylor University, Waco, TX) for providing Dach expression plasmids; Dr. Z. Wu (Hong Kong University of Science and Technology, Hong Kong, China) for wild-type and MEF3-mutated G133 Mgn promoter constructs; Dr. P. Maire (Université Paris V, Paris, France) for MEF3 plasmid; and Drs. M. Akaaboune and I. Valenzuela (University of Michigan) for advice on sternomastoid muscle denervation. We thank Dr. J. Parent (University of Michigan) for sharing his BTX electroporator. We also thank M. Marvin and X. Zhang for technical assistance and members of the Goldman laboratory for helpful discussions. This work was supported by National Institutes of Health Grant 5 R01 NS025153 (to D.G.).
Abbreviations
- CaMK
calcium/calmodulin-dependent protein kinase
- CAT
chloramphenicol acetyltransferase
- HDAC
histone deacetylase
- Mgn
myogenin
- MuSK
muscle-specific tyrosin kinase
- NaB
sodium butyrate
- nAChR
nicotinic acetylcholine receptor
- TA
tibialis anterior
- TSA
trichostatin A.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Sunesen M, Changeux J-P. J Neurocyt. 2003;32:677–684. doi: 10.1023/B:NEUR.0000020616.53664.80. [DOI] [PubMed] [Google Scholar]
- 2.Adams L, Goldman D. J Neurobiol. 1998;35:245–257. doi: 10.1002/(sici)1097-4695(19980605)35:3<245::aid-neu2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 3.Klarsfeld A, Changeaux J-P. Proc Natl Acad Sci USA. 1985;82:4558–4562. doi: 10.1073/pnas.82.13.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C-F, Schmidt J. FEBS Lett. 1994;338:277–280. doi: 10.1016/0014-5793(94)80283-1. [DOI] [PubMed] [Google Scholar]
- 5.Huang C-F, Tong J, Schmidt J. Neuron. 1992;9:671–678. doi: 10.1016/0896-6273(92)90030-h. [DOI] [PubMed] [Google Scholar]
- 6.Klarsfeld A, Laufer R, Fontaine B, Devillers-Thiery A, Dubreuil C, Changeaux J-P. Neuron. 1989;2:1229–1236. doi: 10.1016/0896-6273(89)90307-3. [DOI] [PubMed] [Google Scholar]
- 7.Tang H, Sun Z, Goldman D. J Biol Chem. 2001;276:26057–26065. doi: 10.1074/jbc.M101670200. [DOI] [PubMed] [Google Scholar]
- 8.Macpherson P, Kostrominova T, Tang H, Goldman D. J Biol Chem. 2002;277:15638–15646. doi: 10.1074/jbc.M109864200. [DOI] [PubMed] [Google Scholar]
- 9.Tang S, Jo SA, Burden SJ. Development (Cambridge, U.K.) 1994;120:1799–1804. doi: 10.1242/dev.120.7.1799. [DOI] [PubMed] [Google Scholar]
- 10.Walke W, Xiao G, Goldman D. J Neurosci. 1996;16:3641–3651. doi: 10.1523/JNEUROSCI.16-11-03641.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bessereau J-L, Stratford-Perricaudet LD, Piette J, Poupon CL, Changeux J-P. Proc Natl Acad Sci USA. 1994;91:1304–1308. doi: 10.1073/pnas.91.4.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilmore BP, Goldman D, Chahine KG, Gardner PD. Dev Biol. 1995;168:416–428. doi: 10.1006/dbio.1995.1091. [DOI] [PubMed] [Google Scholar]
- 13.Merlie JP, Mudd J, Cheng T-C, Olson EN. J Biol Chem. 1994;269:2461–2467. [PubMed] [Google Scholar]
- 14.Su C-T, Huang C-F, Schmidt J. FEBS Lett. 1995;366:131–136. doi: 10.1016/0014-5793(95)00496-v. [DOI] [PubMed] [Google Scholar]
- 15.Tang H, Veldman MB, Goldman D. J Biol Chem. 2006;281:3943–3953. doi: 10.1074/jbc.M511317200. [DOI] [PubMed] [Google Scholar]
- 16.Liu S, Sinner DS, Schmidt MM, Danielson JA, Wang S, Schmidt J. J Biol Chem. 2000;275:41364–41368. doi: 10.1074/jbc.M004172200. [DOI] [PubMed] [Google Scholar]
- 17.Tang H, Macpherson P, Argetsinger LS, Cieslak D, Suhr ST, Carter-Su C, Goldman D. Cell Signalling. 2004;16:551–563. doi: 10.1016/j.cellsig.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Mejat A, Ramond F, Bassel-Duby R, Khochbin S, Olson EN, Schaeffer L. Nat Neurosci. 2005;8:313–321. doi: 10.1038/nn1408. [DOI] [PubMed] [Google Scholar]
- 19.Brockes JP, Hall ZW. Proc Natl Acad Sci USA. 1975;72:1368–1372. doi: 10.1073/pnas.72.4.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bevan S, Steinbach JH. J Physiol (London) 1983;336:159–177. doi: 10.1113/jphysiol.1983.sp014574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Ohhi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, Rosenfeld MG. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 22.Caubit X, Thangarajah R, Theil T, Wirth J, Nothwang HG, Ruther U, Krauss S. Dev Dyn. 1999;214:66–80. doi: 10.1002/(SICI)1097-0177(199901)214:1<66::AID-DVDY7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 23.Davis RJ, Shen W, Sandler YI, Heanue TA, Mardon G. Mech Dev. 2001;102:169–179. doi: 10.1016/s0925-4773(01)00307-0. [DOI] [PubMed] [Google Scholar]
- 24.Spitz F, Demignon J, Porteu A, Kahn A, Condordet J-P, Daegelen D, Maire P. Proc Natl Acad Sci USA. 1998;95:14220–14225. doi: 10.1073/pnas.95.24.14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golzio M, Mazzolini L, Moller P, Rols MP, Theissie J. Gene Ther. 2005;12:246–251. doi: 10.1038/sj.gt.3302405. [DOI] [PubMed] [Google Scholar]
- 26.Lezzi S, Cossu G, Nervi C, Sartorelli V, Puri PL. Proc Natl Acad Sci USA. 2002;99:7757–7762. doi: 10.1073/pnas.112218599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnston LA, Tapscott SJ, Eisen H. Mol Cell Biol. 1992;12:5123–5130. doi: 10.1128/mcb.12.11.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ugarte G, Brandan E. J Biol Chem. 2006;281:18473–18481. doi: 10.1074/jbc.M600918200. [DOI] [PubMed] [Google Scholar]
- 29.Zhou X, Richon VM, Rifkind RA, Marks PA. Proc Natl Acad Sci USA. 2000;97:1056–1061. doi: 10.1073/pnas.97.3.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X, Marks PA, Rifkinid RA, Richon VM. Proc Natl Acad Sci USA. 2001;98:10572–10577. doi: 10.1073/pnas.191375098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma K, Chan JKL, Zhu G, Wu Z. Mol Cell Biol. 2005;25:3575–3582. doi: 10.1128/MCB.25.9.3575-3582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKinsey TA, Zhang CL, Olson EN. Mol Cell Biol. 2001;21:6312–6321. doi: 10.1128/MCB.21.18.6312-6321.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Randall WR, Schneider MF. J Cell Biol. 2005;168:887–897. doi: 10.1083/jcb.200408128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silver S, Rebay I. Development (Cambridge, U.K.) 2005;132:3–13. doi: 10.1242/dev.01539. [DOI] [PubMed] [Google Scholar]
- 35.Heanue TA, Reshef R, Davis RJ, Mardon G, Olivere G, Tomarev S, Lassaar AB, Tabin CJ. Genes Dev. 1999;13:3231–3243. doi: 10.1101/gad.13.24.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis RJ, Pesah YI, Harding M, Paylor R, Mardon G. Genesis. 2006;44:84–92. doi: 10.1002/gene.20188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.