Abstract
Programmed cell death (PCD) plays an important role during the life cycle of higher organisms. Although several regulatory mechanisms governing PCD are thought to be conserved in animals and plants, light-dependent cell death represents a form of PCD that is unique to plants. The light requirement of PCD has often been associated with the production of reactive oxygen species during photosynthesis. In support of this hypothesis, hydrogen peroxide and superoxide have been shown to be involved in triggering a PCD response. In the present work, we have used the conditional flu mutant of Arabidopsis to analyze the impact of another reactive oxygen species, singlet oxygen, on cell death. Unexpectedly, the light-dependent release of singlet oxygen alone is not sufficient to induce PCD of flu seedlings but has to act together with a second concurrent blue light reaction. This blue-light-specific trigger of PCD could not be attributed to a photosynthetic reaction or redox change within the chloroplast but to the activation of the blue light/UVA-specific photoreceptor cryptochrome. The singlet oxygen-mediated and cryptochrome-dependent cell death response differs in several ways from PCD triggered by hydrogen peroxide/superoxide.
Keywords: oxidative stress
Throughout the life cycle of plants, programmed cell death (PCD) is involved in a wide range of developmental processes (1, 2). PCD also is closely associated with defense reactions during plant–pathogen interactions or in response to abiotic stress. The onset of this PCD has been linked to the enhanced production of reactive oxygen species (ROS) (3, 4). In plants, chloroplasts are by far the most important site at which generation of ROS occurs. A variety of stress conditions that may trigger PCD limit the ability of plants to use light energy for photosynthesis and enhance the production of several ROS within the plastid compartment (5). Many studies have shown that the onset of PCD in plants is light-dependent (3, 6–10), suggesting a close link between the impairment of light-dependent photosynthetic electron transport under various stress conditions, the release of ROS, and the induction of PCD. For instance, generation of hydrogen peroxide and superoxide during photosynthesis has been implicated in the induction of the hypersensitive response, a hallmark of plant defenses during an incompatible plant/pathogen interaction (11, 12). We have used the conditional fluorescent (flu) mutant of Arabidopsis to analyze the impact of another ROS, singlet oxygen, that is generated within the plastid (13), on cell death. The singlet oxygen-induced cell death response of flu seedlings belongs to the class of light-dependent PCD. Unexpectedly, however, the light-dependent release of singlet oxygen alone is not sufficient to induce PCD of flu seedlings, but has to act together with a second concurrent blue light reaction. In plants two different families of blue light photoreceptors exist, the phototropins (14) and the cryptochromes (15–17). In the present work one of these photoreceptors, cryptochrome 1 (CRY1), is shown to be responsible for triggering the blue light-dependent, singlet oxygen-mediated PCD in Arabidopsis. The release of singlet oxygen within plastids of the flu mutant induces rapid changes in nuclear gene expression. The singlet oxygen-dependent up-regulation of a small subset of these genes is selectively suppressed in CRY1-deficient flu mutant plants. Previously the expression of these genes had been associated with various abiotic and biotic stress conditions, PCD, and oxidative stress but had not been shown to be under the control of CRY1. Our results suggest that singlet oxygen-mediated cell death is not confined to the flu mutant but may play a more general role also in wild type under various environmental stress conditions.
Results
In the dark, the conditional flu mutant of Arabidopsis accumulates the photosensitizer protochlorophyllide (Pchlide) that upon illumination transfers its excitation energy to ground state triplet oxygen, thereby elevating it to excited singlet oxygen (13, 18). The release of singlet oxygen in the flu mutant had been measured previously by using two different fluorescent sensors, that are both highly selective for singlet oxygen, dansyl-2,2,5,-tetramethyl-2,5-dihydro-1H-pyrrole (13, 19), and singlet oxygen sensor green (20). The generation of singlet oxygen starts within the first minute of illumination and has been shown to be confined to plastids (13, 21). Twenty-four hours after the dark-to-light shift, seedlings develop necrotic lesions (Fig. 1A). As had been shown previously, the majority of detectable singlet oxygen was seen to be released within the first 10 min of reillumination (13, 21). To examine the effect of light in a more specific manner on the death induced in flu, seedlings kept for 15 h in the dark were exposed to white light for 1 h at 100 μmol of photons per squared meter per second (henceforth, photons will be assumed, e.g., 100 μmol·m−2·s−1) to allow the production of singlet oxygen and were then either maintained under white light at the same fluence rate or transferred to the dark for the following 23 h. Under continuous illumination seedlings died (Fig. 1A), but remained alive and green, when transferred to the dark (Fig. 1B). This result indicates that the release of singlet oxygen during the first hour of reillumination is not sufficient to trigger the onset of PCD during a subsequent dark period. Similar to previous studies that had demonstrated the light-dependency of PCD (3, 6–10), the execution of PCD requires continuous illumination also in flu seedlings, suggesting that in addition to the initial light-dependent release of 1O2 a second light reaction is involved in triggering the cell death response.
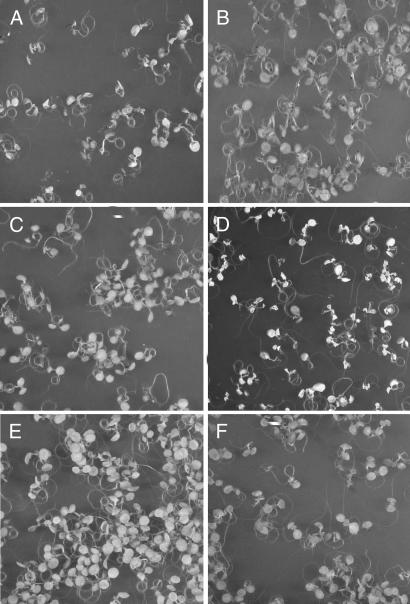
Fig. 1.
Blue-light dependency of the cell death response induced in seedlings of the flu mutant. Seedlings of flu were kept under continuous light for 5 days, followed by 15 h of darkness and exposure to white light for 1 h at 100 μmol·m−2·s−1 to allow singlet oxygen production. The seedlings were subsequently placed for 23 h in white light at 100 μmol·m−2·s−1 (A), in darkness (B), or in red (C) or blue (D) light, both at 10 μmol·m−2·s−1. (E and F) Control seedlings that were kept for 24 h under red (E) or blue (F) light at 10 μmol·m−2·s−1 without the 1-h white-light treatment.
The proposed additional light reaction was investigated by testing different light qualities for their ability to induce PCD in flu. After the 15 h dark period and 1 h of reillumination with white light, seedlings were transferred to either blue or red light for the remaining 23 h. The light intensities during this period were reduced from 100 μmol·m−2·s−1 for the white light treatment to lower fluence rates for the red and blue light to minimize a possible oxidative stress caused by the release of singlet oxygen during the 23-h illumination period. When exposed to fluence rates of <5 μmol·m−2·s−1 seedlings did not show a cell death response (data not shown). However, at fluence rates of 10 μmol·m−2·s−1, flu seedlings may show a similar response to blue light as those kept for 24 h at 100 μmol·m−2·s−1 of white light (Fig. 1D), whereas under red light no cell death response could be detected (Fig. 1C). Seedlings exposed for 24 h to either red or blue light at 10 μmol·m−2·s−1 without the initial 1 h white light treatment at 100 μmol·m−2·s−1 did not show the cell death response (Fig. 1 E and F). The lower light intensity was still sufficient to sustain growth of the plants (A.D., unpublished results).
These results obtained with intact seedlings were confirmed and extended by using a protoplast system that had been established previously to characterize and quantify the death response in flu induced by the release of 1O2 (22). Protoplasts from wild-type and flu seedlings were isolated after the 15 h dark period and were exposed for 1 h to white light. The protoplast suspension was then divided into four aliquots which were either maintained under white light or transferred to the dark, or to blue or red light, respectively. Dead cells were detected by staining protoplasts with Evans blue dye (8, 22) 24 h after the initial release of singlet oxygen. Almost all of the flu protoplasts kept under white light or transferred to blue light died during this incubation, whereas those transferred to the dark or to red light were protected from PCD (Fig. 2A).
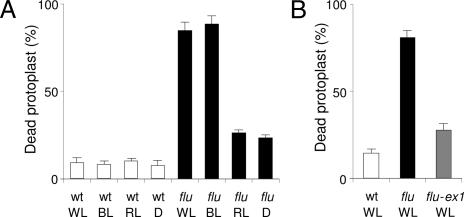
Fig. 2.
Quantification of the blue-light-dependent cell death response induced in protoplasts of the flu mutant. (A) After 15 h of darkness, protoplasts from wild type (wt) and flu were prepared and exposed to white light (WL) for 1 h at 100 μmol·m−2·s−1 to allow singlet oxygen production. Protoplasts were subsequently placed in white light at 100 μmol·m−2·s−1, blue light (BL) at 10 μmol·m−2·s−1, red light (RL) at 10 μmol·m−2·s−1, or in darkness (D) for 23 h, and the percentage of dead protoplasts was calculated. (B) After a 15-h dark period, protoplasts from wild type, flu and the double mutant flu–executer1 were transferred to white light at 100 μmol·m−2·s−1. The percentage of dead protoplasts was calculated 24 h after the light/dark/light treatment. The percentage of dead protoplasts was calculated as described in ref. 22. The values represent the mean and standard deviations of four experiments with protoplasts extracted from 400–500 seedlings for each sample. A minimum of 100 protoplasts were counted per sample. ex1, executer1.
Enhanced levels of singlet oxygen trigger cell death responses that may range from necrotic reactions resulting from photooxidative damage to the activation of a genetically controlled PCD (21). The extent of cell damage caused by the cytotoxicity of singlet oxygen was determined by analyzing protoplasts prepared from seedlings of flu and flu-executer1. The executer1 mutation has been shown to abrogate the singlet oxygen-mediated collapse of seedlings and defines a genetically controlled cell death response (21). When protoplasts of flu-executer1 were exposed for 24 h to white light (100 μmol·m−2·s−1) after singlet oxygen production, the majority of them survived similar to flu protoplasts shifted to the dark or to red light after 1 h of white light treatment (Fig. 2A). The percentage of dead cells was only slightly higher than in protoplast suspensions isolated from control wild-type seedlings, suggesting that during the first hour of white light exposure only a small fraction of protoplasts was physically damaged because of the toxicity of singlet oxygen (Fig. 2).
Light-dependent cell death in plants has often been associated with photosynthetic electron transport that under excessive light may generate elevated levels of hydrogen peroxide/superoxide. These ROS have been implicated with triggering various cell death reactions (3, 6–10, 23). The possible involvement of light-driven electron transport in mediating the blue light-dependent cell death response was tested by incubating wild-type and flu protoplasts in the presence of 3-(3,4-dichlorphenyl)-1-1-dimethylurea that is known to block forward electron transport by binding to the secondary quinone electron acceptor of photosystem II (PSII), QB. Cell death in flu protoplasts treated with 3-(3,4-dichlorphenyl)-1–1-dimethylurea was not suppressed (Fig. 5, which is published as supporting information on the PNAS web site). Hence, the proposed additional light reaction needed for the execution of singlet oxygen-mediated cell death cannot be attributed to a light-dependent signal derived from photosynthetic electron transport.
In plants two families of photoreceptors are responsible for the perception of blue light. The blue light-sensing cryptochromes control several photomorphogenic responses in plants including hypocotyl elongation, the setting of the circadian clock, and the control of flowering time (15–17), whereas the phototropins control various light responses such as phototropism, plastid relocation, and stomatal opening (14). In Arabidopsis, two cryptochromes, CRY1 (15) and CRY2 (17), and two phototropins, PHOT1 and PHOT2 (14) are involved in mediating these responses.
The possible involvement of cryptochrome in mediating the blue light dependency of PCD induced by singlet oxygen was tested by crossing the flu mutant with cry1 and cry2 mutants and with the double mutant cry1–cry2. As cryptochromes are able to influence the expression of plastidic genes (24, 25) that could in turn modify the production of the photosensitizer Pchlide in plastids and in this way affect the severity of the singlet oxygen-dependent cell death response, the concentration of Pchlide in the double and triple mutants was determined and compared with that in the original flu mutant. flu mutants kept in the dark accumulate ≈10 times more Pchlide than the wild type, consistent with previous reports (18), and the double mutants flu–cry1 and flu–cry2 as well as the triple mutant flu–cry1–cry2 accumulated similar amounts of Pchlide as flu (Fig. 6, which is published as supporting information on the PNAS web site). Although it is conceivable that inactivation of CRY1 may affect the level of other photosensitizing molecules we found no experimental evidence to support this notion (data not shown).
Protoplasts from wild-type, flu, flu–cry1, flu–cry2, and flu–cry1–cry2 seedlings were isolated after the 15 h dark period and were kept under white light for 1 h to allow singlet oxygen production to occur and were subsequently transferred to blue light. The percentage of dead protoplasts was calculated 1, 4, 8, and 24 h after the light/dark/light treatment. As shown in Fig. 3A, the number of protoplasts of flu that had died during the incubation rapidly increased and reached ≈50% after 8 h and 80% after 24 h, whereas the number of dead protoplasts of wild-type seedlings remained at an almost constant low level. In contrast to protoplasts of flu, those extracted from flu–cry1 seedlings exhibited a strong suppression of the singlet oxygen-induced cell death response (Fig. 3A) down to the level of flu control protoplasts shifted to the dark (Fig. 2A). In protoplasts of the double mutant flu–cry2, however, only a minor reduction of the percentage of dead protoplasts was detected (Fig. 3A). The preponderance of the role played by CRY1 over that of CRY2 during the induction of PCD was confirmed by results obtained with the triple mutant flu–cry1–cry2. Protoplasts obtained from this mutant line showed the same extent of suppression of singlet oxygen-mediated PCD as the flu–cry1 double mutant (Fig. 3A), consistent with CRY2 not being essential for mediating the blue light-dependent induction of PCD.
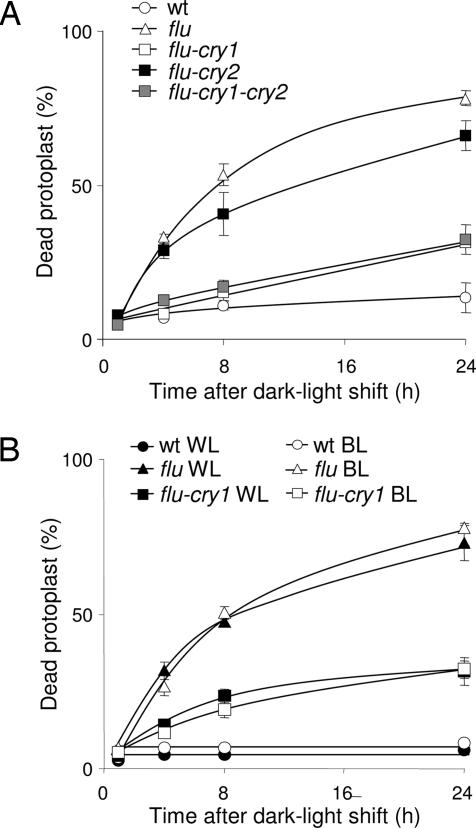
Fig. 3.
The effect of light quality and the cryptochrome mutations on the extent of cell death in flu. (A) After a 15-h dark period, protoplasts from wild type, flu, the double mutants flu–cry1 and flu–cry2, and the triple mutant flu–cry1–cry2 were kept under white light (WL) at 100 μmol·m−2·s−1 for 1 h to allow singlet oxygen production and then transferred to blue light (BL) at 10 μmol·m−2·s−1. The percentage of dead protoplasts was calculated 1, 4, 8, and 24 h after the light/dark/light treatment. (B) After 15 h of darkness, protoplasts from wild type, flu, and the double mutant flu–cry1 were exposed to white light for 1 h at 100 μmol·m−2·s−1 to allow singlet oxygen production. Protoplasts were subsequently placed in white light at 100 μmol·m−2·s−1 or blue light at 10 μmol·m−2·s−1 and the percentage of dead protoplasts was calculated 1, 4, 8, and 24 h after the dark/light treatment. The percentage of dead protoplasts was calculated as described in ref. 22. The values represent the mean and standard deviations of four experiments with protoplasts extracted from 400–500 seedlings each. A minimum of 100 protoplasts were counted per sample.
Similar experiments were also done with protoplasts of flu–phot1, flu–phot2, and flu–phot1–phot2 seedlings. Inactivation of either PHOT1 or PHOT2 or both phototropins did not affect the rapid increase of the singlet oxygen-induced cell death response of flu protoplasts (results not shown). Thus, 1O2-mediated cell death responses in flu seem to be controlled by blue light and CRY1. To confirm that CRY1 and blue light are indeed the main factors in mediating the light dependency of the PCD induced in flu, the blue light- and white light-dependent inductions of PCD were compared in protoplasts extracted from wild-type, flu, and flu–cry1 seedlings. As shown in Figs. 2 and 3B, no obvious difference in the percentage of dead cells was detected in protoplast suspensions obtained from flu seedlings after they were exposed to either white light or blue light after the initial 1 h white light treatment. This suggests that blue light is sufficient and necessary for the execution of PCD in flu. Moreover, no obvious difference in the suppression of cell death of protoplasts extracted from flu–cry1 seedlings was detected after they were transferred to blue or white light (Fig. 3B).
So far little is known about how CRY1 might interfere with the singlet oxygen-dependent signaling of PCD in Arabidopsis. CRY1 is localized within the nucleus (16), whereas singlet oxygen is generated within the plastid compartment (13). Shortly after the release of singlet oxygen massive changes in the expression of nuclear genes are induced (13, 22). Because of the short half-life of singlet oxygen, it seems unlikely that this ROS leaves the plastid compartment and directly controls nuclear gene activities (26, 27). Instead, singlet oxygen-derived second messengers may be expected to be involved in retrograde control of nuclear gene expression. As a first step toward identifying nuclear genes that are under the control of both singlet oxygen and CRY1 and that may be involved in activating the cell death response of flu seedlings, total RNA extracted from flu, flu–cry1, cry1, and wild-type seedlings reilluminated for six hours was analyzed. Because the release of singlet oxygen during the first hour of reillumination of flu seedlings was necessary, but not sufficient for the induction of the cell death response, a blue light-dependent modulation of singlet oxygen-induced gene expression changes may be required for the ultimate execution of PCD during the subsequent reillumination period. First, the total number of genes activated differentially in seedlings by singlet oxygen after six hours of reillumination was assessed by comparing global changes in gene expression of flu seedlings with those of wild-type seedlings. Total RNA was first transcribed into cDNAs and then into biotinylated complementary RNAs that were hybridized to Affymetrix chips representing ≈24,000 genes or >95% of the total genome of Arabidopsis. Genes with a 3-fold or greater differential expression that were either up-regulated or down-regulated in flu relative to wild type were selected. A total of 972 genes belonging to this group were identified. 588 genes were up-regulated, and 384 were down-regulated. The possible impact of cryptochrome on the expression of these genes was determined by comparing transcript levels in flu and flu–cry1. None of the genes that were down-regulated in flu relative to wild type after six hours of reillumination were significantly altered in their expression level after the inactivation of CRY 1. On the other hand, among the 588 genes that were up-regulated in flu relative to wild type, the expression of only a small subset of 34 genes was selectively repressed in flu–cry 1 seedlings (Table 1, which is published as supporting information on the PNAS web site). The identification of these genes reveals two findings. First, in contrast to previous expression studies of cryptochrome-deficient mutants that had found a large proportion of blue light-activated genes to encode plastid proteins (28), only 3 of the 34 genes up-regulated in flu and repressed in flu–cry 1 seedlings were predicted to encode plastid proteins. Second, the expression of all of the 34 genes has been associated previously with various abiotic and biotic stress conditions, PCD, and oxidative stress caused by norflurazon treatment, but had not been shown to be under the control of cryptochrome (www.genevestigator.ethz.ch/at) (29). None of these genes are rapidly up-regulated during the first 30 min of reillumination. Thus, CRY1-dependent gene activation seems to be a secondary step affecting genes that are distinct from those directly controlled by the initial singlet oxygen-mediated retrograde signaling. Two of the Cry1-dependent genes, At1g61120 and At4g21830, that encode a putative S-linalool synthase and a methionine sulfoxide reductase domain-containing protein, respectively, were selected for a more detailed quantitative analysis of transcript changes after various lengths of reillumination.
The plastidic methionine sulfoxide reductase had recently been linked to cold and oxidative stress (30–32).
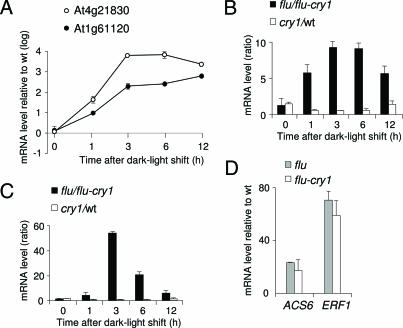
Transcript concentrations of the two singlet oxygen- and blue light-responsive genes At1g61120 and At4g21830 increased in flu during the first three hours of reillumination (Fig. 4A). Transcripts of At1g61120 continued to accumulate in flu during the following 9 h of reillumination, whereas those of At4g21830 reached a maximum between 3 and 6 h of reillumination and declined afterward (Fig. 4A). The singlet oxygen-mediated induction of both genes was repressed in flu–cry1 seedlings (Fig. 4 B and C). In cry1 seedlings without the flu mutation transcripts reached similar levels as wild type, suggesting that in the absence of the initial release of singlet oxygen cryptochrome-dependent regulation of these genes plays only a minor role (Fig. 4 B and C). The flu and flu–cry1 seedlings released similar amounts of singlet oxygen shortly after the beginning of reillumination as indicated by the rapid and drastic up-regulation of ERF1 and ACS6 in both mutant lines after 1 h of light exposure (Fig. 4D).
Fig. 4.
Activation of two selected genes in the flu mutant and their suppression in the flu–cry1 double mutant. (A–C) Two of the genes shown in Table 1 and identified by cluster analysis as being up-regulated in flu and down-regulated in flu–cry1 6 h after a dark/light shift (At1g61120, putative S-linalool synthase; At4g21830, methionine sulfoxide reductase domain-containing protein) were selected for an independent determination of their transcript levels by real-time quantitative PCR. Results shown are mean values ± SD of four measurements from two independent experiments. Five-day-old seedlings of wild type, cry1, flu, and flu–cry1 grown under continuous light at 100 μmol·m−2·s−1 were shifted to the dark for 15 h and reexposed for various lengths of time to white light. Total RNAs were extracted, and transcript levels were determined as described in Materials and Methods. (A) Relative expression levels of these two genes in flu versus wild-type plants after the dark/light shift. (B and C) The effect of the inactivation of CRY1 on the relative expression levels of At1g61120 (B) and At4g21830 (C) in flu (flu/flu–cry1) and in wild-type (cry1/wt) plants after the dark/light shift. (D) Rapid induction of the two singlet oxygen-dependent genes ACS6 and ERF1 (28) in flu and flu–cry1 1 h after the dark/light shift.
Discussion
The light requirement of PCD has often been associated with the production of ROS during photosynthesis. The initiation of PCD during an incompatible plant-pathogen interaction or in response to elicitor treatment may occur in the dark (23) but the subsequent execution of lesion formation has been shown to require light and to depend on the generation of hydrogen peroxide/superoxide during photosynthesis (3, 11, 23). Plants under light stress may generate these two ROS by using photorespiration or the reduction of molecular oxygen by photosystem I to keep the acceptors of PSII in a partially oxidized state, thus minimizing the risk of photoinhibition (33, 34). Once the plant's capacity to quench excess light energy is no longer sufficient to avoid photoinhibition, enhanced levels of singlet oxygen may be produced by PSII. Because conditions favoring the release of enhanced levels of hydrogen peroxide/superoxide during photosynthesis differ from those that stimulate the generation of singlet oxygen, perception of these different ROS by the plant may be expected to evoke different stress responses. Even though both groups of ROS may trigger a cell death response, singlet oxygen-mediated cell death indeed differs from hydrogen peroxide/superoxide-dependent cell death responses in several ways. During the pathogen-induced initiation of PCD in the dark a rapid activation of the 9-lipoxygenase (LOX) pathway has been shown to endorse the rapid enzymatic production of oxylipins within the cytosol that seem to play an essential role in establishing the host's resistance in at least some incompatible plant/pathogen interactions (35, 36). During the light-dependent spreading of necrotic lesions, hydrogen peroxide/superoxide supports the massive nonenzymatic release of various lipid peroxidation products and at the same time suppresses the activity of the 9-LOX pathway (23). Singlet oxygen, however, activates the 13-LOX pathway within the plastid compartment, but not the 9-LOX pathway, and leads to the rapid enzymatic formation of a subset of oxylipins that are distinct from those generated during plant-pathogen interaction (13). In addition to its dependence on the release of hydrogen peroxide/superoxide the spreading of cell death during an incompatible interaction is further enhanced by a second light reaction that is mediated by phytochrome (37). As shown in the present work singlet oxygen is necessary, but not sufficient to trigger the cell death response of the flu mutant and requires also a second light-dependent reaction. However, in contrast to the light-dependent stimulation of PCD during plant-pathogen interaction, this additional light reaction requires the activation of the UVA/blue light-absorbing photoreceptor CRY1.
Thus far, mainly two different modes of blue light-dependent control of stress responses have been analyzed. CRY1 has been shown to control the phenylpropanoid pathway by regulating its key enzyme, chalcone synthase that stimulates the synthesis of various protectants such as anthocyanins in UVA/blue light-treated plants (38). In the flu mutant the release of singlet oxygen does not enhance the level of chalcone synthase transcripts and does not stimulate anthocyanin accumulation.
A second blue light-dependent activity that may conceivably affect the singlet oxygen-mediated cell death response is the blue light-dependent turnover and synthesis of the D1- and D2-proteins encoded by the plastid DNA (39, 40). Both proteins form part of the core of PSII and have been implicated with the detoxification of singlet oxygen and the photoprotection of PSII (41, 42). The blue light-dependent synthesis of D2 has been attributed to the CRY1-dependent transcription of nuclear genes encoding sigma factors that within the plastid form part of the plastid-encoded RNA polymerase and are thought to confer different promoter specificities to the plastid-encoded RNA polymerase during chloroplast gene transcription in response to various developmental and environmental cues (43, 44). Sigma 5 in particular has been shown to be required for the transcription of the D2 protein. Inactivation of CRY1 impedes the blue-light dependent expression of SIGMA5, blocks the transcription of the D2 protein gene and thus would be expected to impair the photoprotection of PSII and enhance singlet oxygen-mediated stress responses. However, contrary to this predicted cell death-promoting consequence of CRY1 inactivation, the singlet oxygen-mediated cell death response as shown in the present work is suppressed in the flu–cry1 double mutant and thus is unlikely to be caused by a reduced turnover rate of the D2 protein. This conclusion is supported by the transcript concentrations of PsbD in flu, flu–cry1, and wild type, that reached similar levels in all three lines (N.S.C., unpublished results).
Upon the release of singlet oxygen rapid and drastic changes in nuclear gene expression have been shown to affect ≈5% of the genome of Arabidopsis thaliana (13). At least some of these gene expression changes seem to be linked to visible singlet oxygen-mediated stress responses such as seedling lethality and growth inhibition and cell death responses of mature plants (13, 21). Because generation of singlet oxygen is restricted to the plastid compartment, its effect on nuclear gene expression implies retrograde control of nuclear genes by plastid-derived signals. Activation of a very small subset of these nuclear genes was found to depend on the presence of blue light. CRY1 has been shown to be localized within the nucleus (16), and it seems to confer the blue light-dependent control to the 34 singlet oxygen-responsive genes identified in the present study. Interestingly, these genes were not found among the nuclear genes that were rapidly and transiently up-regulated in the flu mutant right after the dark-to-light shift (13). Thus, the CRY1-dependent genes represent a distinct and separate set of genes whose activation followed the initial singlet oxygen-mediated signaling and reached its maximum only after 6–12 h of reillumination. This result is in close agreement with our findings that the initial release of singlet oxygen is mandatory but not sufficient to trigger the cell death response and that the subsequent blue light-dependent activation of CRY1 is required for the execution of cell death. All of the singlet oxygen-up-regulated genes that were repressed in cry1–flu had previously been associated with either responses to various abiotic and/or biotic stresses that may invoke cell death or directly with the activation or suppression of PCD.
Based on these results it is tempting to suggest that the singlet oxygen-mediated cell death program is not restricted to the flu mutant grown under nonpermissive light/dark cycles but may play a more general role during various adverse environmental conditions that hitherto has remained unnoticed.
Materials and Methods
Plant Material and Growth Conditions.
Seeds were sterilized and sown under aseptic conditions on agar plates Gamborg B5 supplemented with 0.5% sucrose. Seedlings were grown for 5 days at 20°C in continuous light (100 μmol·m−2·s−1), transferred to the dark for 15 h and reilluminated for various lengths of time under white light (100 μmol·m−2·s−1) blue light (10 μmol·m−2·s−1) or red light (10 μmol·m−2·s−1). Seedlings were subjected to light from commercially available light sources: Sylvania LUXLINE plus F18/860 for white light, Philips TLD Blue 18W/18 for blue light, and Philips TLD Red 18W/15 for red light. Mutants used in this work were flu1–3 (18), executer1 (21), cry1 (hy4-2.23N) (15), cry2 (fha-1) (17), phot1 (nph1-5) (45), and phot2 (cav1-1) (46). The background ecotype of phot1 and phot2 was Columbia, and that of the others was Landsberg erecta. For crosses of the phot mutants with flu, a flu Columbia line was used that had been obtained by six back-crosses of flu1-3 in Landsberg erecta with wild-type Columbia. The flu, executer1, cry1, phot1, and phot2 mutations were identified phenotypically as described by refs. 21 and 28, respectively. Cry2 was identified by PCR (28). The mutations in individual lines and double and triple mutant lines were confirmed with DNA-based markers (28).
Protoplast Preparation and Determination of Cell Death.
Arabidopsis protoplasts were isolated from 5-day-old seedlings grown under continuous light by incubating leaves for 15 h in the dark in the presence of the digestion medium (Gamborg B5 medium: 0.5 M mannitol/1.2% cellulase/0.8% macerozyme, pH 5.8). During this dark period, cell walls were digested, and, at the same time, Pchlide accumulated in cells of the flu mutant. Protoplasts were then transferred to the light (100 μmol·m−2·s−1) and separated from cellular debris by filtration through a 100-μm mesh filter (Milian, Geneva, Switzerland), followed by flotation on a 0.4 M sucrose solution at 60 × g for 10 min. They were then collected and centrifuged at low speed (60 × g) for 5 min and washed. Finally, protoplasts were resuspended in the culture medium containing MS medium (supplemented with 0.4 M sucrose/0.4 M mannitol). The percentage of dead cells was determined by staining protoplasts with Evans blue dye (Sigma, St. Louis, MO), which was added to the samples to a final concentration of 0.04%. The number of stained cells was determined by using a light microscope.
Real-Time PCR.
Wild-type, flu, cry1, and flu–cry1 plants were grown for five days under continuous light and then darkened for 15 h. Total RNA was extracted and prepared at time 0 and 1, 3, 6, and 12 h after reillumination according to Melzer et al. (47). RNA was treated with RQ1 RNase-Free DNase (Promega, Madison, WI) and reverse-transcribed by using random hexamers and SUPERSCRIPT TM II RNase H reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations. The ABI PRISM 7700 sequence detection system (Applied Biosystems, Foster City, CA) was used to perform quantitative real-time PCR. Relative mRNA abundance was calculated by using the comparative cycle threshold (ΔCt) method and normalized to the corresponding ACTIN2 (At3g18780) gene levels. Equivalent efficiencies between the different probes and our internal standard were observed during the PCRs.
Primers used for the analysis were the following: At4g11280, GTTTGATCCTGACCGGATTG and CCGCAACACTTATGAACTCG; At3g2340, GCAACAAACCTATATTGACTCG and GTCCCACTATTTTCAGAAGACC; At1g61120, CCGATGTTCCAAGGTTGTCT and AATTTCGATGGCAACACCTC; At4g21830, TCCAGCAACTGGATCTTTCC and GGAGGAACCAGCAGAAGAGA; and ACTIN2, GTGTGTCTCACACTGTGC and CAGATCCTTCCTGATATCC.
Microarray Hybridization and Analysis.
Detailed procedures are provided in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Other Methods.
Extraction and analysis of tetrapyrroles were done as described in ref. 13.
Supplementary Material
Acknowledgments
We thank Dr. T. Fitzpatrick for critical reading, Dr. C. Kim for support, Dr. D. Rubli for art work, A. Imboden and M. Nater for technical assistance, M. Geier-Bächtold and U. Baldenweg for editorial work, and Drs. Margaret Ahmad (Université de Paris 6, Paris, France) and Winslow R. Briggs (Carnegie Institution of Washington, Stanford, CA) for seeds of cryptochromes and phototropins mutant lines, respectively. This study was supported by the Swiss Federal Institute of Technology, the Functional Genomic Center Zürich, and the Swiss National Science Foundation.
Abbreviations
- ROS
reactive oxygen species
- PCD
programmed cell death
- PSII
photosystem II
- Pchlide
protochlorophyllide.
Footnotes
The authors declare no conflict of interest.
References
- 1.Hoeberichts FA, Woltering EJ. BioEssays. 2003;25:47–57. doi: 10.1002/bies.10175. [DOI] [PubMed] [Google Scholar]
- 2.Franklin KA, Whitelam GC. J Exp Bot. 2004;55:271–276. doi: 10.1093/jxb/erh026. [DOI] [PubMed] [Google Scholar]
- 3.Rusterucci C, Aviv DH, Holt BF III, Dangl JL, Parker JE. Plant Cell. 2001;13:2211–2224. doi: 10.1105/tpc.010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gechev TS, Hille J. J Cell Biol. 2005;168:17–20. doi: 10.1083/jcb.200409170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foyer CH, Noctor G. New Phytol. 2000;146:359–388. [Google Scholar]
- 6.Asai T, Stone JM, Heard JE, Kovtun Y, Yorgey P, Sheen J, Ausubel FM. Plant Cell. 2000;12:1823–1836. doi: 10.1105/tpc.12.10.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodersen P, Petersen M, Pike HM, Olszak B, Skov S, Odum N, Jorgensen LB, Brown RE, Mundy J. Genes Dev. 2002;16:490–502. doi: 10.1101/gad.218202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danon A, Rotari VI, Gordon A, Mailhac N, Gallois P. J Biol Chem. 2004;279:779–787. doi: 10.1074/jbc.M304468200. [DOI] [PubMed] [Google Scholar]
- 9.Gray J, Janick-Buckner D, Buckner B, Close PS, Johal GS. Plant Physiol. 2002;130:1894–1907. doi: 10.1104/pp.008441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorrain S, Lin B, Auriac MC, Kroj T, Saindrenan P, Nicole M, Balague C, Roby D. Plant Cell. 2004;16:2217–2232. doi: 10.1105/tpc.104.022038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mateo A, Muhlenbock P, Rusterucci C, Chang CC, Miszalski Z, Karpinska B, Parker JE, Mullineaux PM, Karpinski S. Plant Physiol. 2004;136:2818–2830. doi: 10.1104/pp.104.043646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willekens H, Chamnongpol S, Davey M, Schraudner M, Langebartels C, Van Montagu M, Inze D, Van Camp W. EMBO J. 1997;16:4806–4816. doi: 10.1093/emboj/16.16.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.op den Camp RG, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Gobel C, Feussner I, Nater M, Apel K. Plant Cell. 2003;15:2320–2332. doi: 10.1105/tpc.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briggs WR, Christie JM. Trends Plants Sci. 2002;7:204–210. doi: 10.1016/s1360-1385(02)02245-8. [DOI] [PubMed] [Google Scholar]
- 15.Ahmad M, Cashmore AR. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- 16.Chen M, Chory J, Fankhauser C. Annu Rev Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- 17.Guo H, Yang H, Mockler TC, Lin C. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- 18.Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K. Proc Natl Acad Sci USA. 2001;98:12826–12831. doi: 10.1073/pnas.221252798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hideg E, Kalai T, Hideg K, Vass I. Biochemistry. 1998;37:11405–11411. doi: 10.1021/bi972890+. [DOI] [PubMed] [Google Scholar]
- 20.Flors C, Fryer MJ, Waring J, Reeder B, Bechtold U, Mullineaux PM, Nonell S, Wilson MT, Baker NR. J Exp Bot. 2006;57:1725–1734. doi: 10.1093/jxb/erj181. [DOI] [PubMed] [Google Scholar]
- 21.Wagner D, Przybyla D, op den Camp R, Kim C, Landgraf F, Lee KP, Wursch M, Laloi C, Nater M, Hideg E, Apel K. Science. 2004;306:1183–1185. doi: 10.1126/science.1103178. [DOI] [PubMed] [Google Scholar]
- 22.Danon A, Miersch O, Felix G, op den Camp RG, Apel K. Plant J. 2005;41:68–80. doi: 10.1111/j.1365-313X.2004.02276.x. [DOI] [PubMed] [Google Scholar]
- 23.Montillet JL, Chamnongpol S, Rusterucci C, Dat J, van de Cotte B, Agnel JP, Battesti C, Inze D, Van Breusegem F, Triantaphylides C. Plant Physiol. 2005;138:1516–1526. doi: 10.1104/pp.105.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Hernandez A, Lopez-Ochoa L, Arguello-Astorga G, Herrera-Estrella L. Plant Physiol. 2002;128:1223–1233. doi: 10.1104/pp.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mochizuki T, Onda Y, Fujiwara E, Wada M, Toyoshima Y. FEBS Lett. 2004;571:26–30. doi: 10.1016/j.febslet.2004.06.052. [DOI] [PubMed] [Google Scholar]
- 26.Gorman AA, Rodgers MA. J Photochem Photobiol B. 1992;14:159–176. doi: 10.1016/1011-1344(92)85095-c. [DOI] [PubMed] [Google Scholar]
- 27.Sies H, Menck CF. Mutat Res. 1992;275:367–375. doi: 10.1016/0921-8734(92)90039-r. [DOI] [PubMed] [Google Scholar]
- 28.Ohgishi M, Saji K, Okada K, Sakai T. Proc Natl Acad Sci USA. 2004;101:2223–2228. doi: 10.1073/pnas.0305984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bechtold U, Murphy DJ, Mullineaux PM. Plant Cell. 2004;16:908–919. doi: 10.1105/tpc.015818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.In O, Berberich T, Romdhane S, Feierabend J. Planta. 2005;220:941–950. doi: 10.1007/s00425-004-1410-7. [DOI] [PubMed] [Google Scholar]
- 32.Vieira Dos Santos C, Cuine S, Rouhier N, Rey P. Plant Physiol. 2005;138:909–922. doi: 10.1104/pp.105.062430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asada K. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:601–639. doi: 10.1146/annurev.arplant.50.1.601. [DOI] [PubMed] [Google Scholar]
- 34.Kozaki A, Takeba G. Nature. 1996;384:557–560. [Google Scholar]
- 35.Rance II, Fournier J, Esquerre-Tugaye MT. Proc Natl Acad Sci USA. 1998;95:6554–6559. doi: 10.1073/pnas.95.11.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rusterucci C, Montillet JL, Agnel JP, Battesti C, Alonso B, Knoll A, Bessoule JJ, Etienne P, Suty L, Blein JP, Triantaphylides C. J Biol Chem. 1999;274:36446–36455. doi: 10.1074/jbc.274.51.36446. [DOI] [PubMed] [Google Scholar]
- 37.Genoud T, Buchala AJ, Chua NH, Metraux JP. Plant J. 2002;31:87–95. doi: 10.1046/j.1365-313x.2002.01338.x. [DOI] [PubMed] [Google Scholar]
- 38.Jenkins GI, Long JC, Wade HK, Shenton MR, Bibikova TN. New Phytol. 2001;151:121–131. doi: 10.1046/j.1469-8137.2001.00151.x. [DOI] [PubMed] [Google Scholar]
- 39.Christopher DA, Mullet JE. Plant Physiol. 1994;104:1119–1129. doi: 10.1104/pp.104.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thum KE, Kim M, Christopher DA, Mullet JE. Plant Cell. 2001;13:2747–2760. doi: 10.1105/tpc.010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma J, Panico M, Barber J, Morris HR. J Biol Chem. 1997;272:3935–3943. doi: 10.1074/jbc.272.7.3935. [DOI] [PubMed] [Google Scholar]
- 42.Trebst A. Z Naturforsch. 2003;58:609–620. doi: 10.1515/znc-2003-9-1001. [DOI] [PubMed] [Google Scholar]
- 43.Allison LA. Biochimie. 2000;82:537–548. doi: 10.1016/s0300-9084(00)00611-8. [DOI] [PubMed] [Google Scholar]
- 44.Tsunoyama Y, Ishizaki Y, Morikawa K, Kobori M, Nakahira Y, Takeba G, Toyoshima Y, Shiina T. Proc Natl Acad Sci USA. 2004;101:3304–3309. doi: 10.1073/pnas.0308362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huala E, Oeller PW, Liscum E, Han I-S, Larsen E, Briggs WR. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 46.Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K, Wada M. Science. 2001;291:2138–2141. doi: 10.1126/science.291.5511.2138. [DOI] [PubMed] [Google Scholar]
- 47.Melzer S, Majewski DM, Apel K. Plant Cell. 1990;2:953–961. doi: 10.1105/tpc.2.10.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.