Abstract
Pregnancy-associated malaria is a major health problem, which mainly affects primigravidae living in malaria endemic areas. The syndrome is precipitated by accumulation of infected erythrocytes in placental tissue through an interaction between chondroitin sulphate A on syncytiotrophoblasts and a parasite-encoded protein on the surface of infected erythrocytes, believed to be VAR2CSA. VAR2CSA is a polymorphic protein of approximately 3,000 amino acids forming six Duffy-binding-like (DBL) domains. For vaccine development it is important to define the antigenic targets for protective antibodies and to characterize the consequences of sequence variation. In this study, we used a combination of in silico tools, peptide arrays, and structural modeling to show that sequence variation mainly occurs in regions under strong diversifying selection, predicted to form flexible loops. These regions are the main targets of naturally acquired immunoglobulin gamma and accessible for antibodies reacting with native VAR2CSA on infected erythrocytes. Interestingly, surface reactive anti-VAR2CSA antibodies also target a conserved DBL3X region predicted to form an α-helix. Finally, we could identify DBL3X sequence motifs that were more likely to occur in parasites isolated from primi- and multigravidae, respectively. These findings strengthen the vaccine candidacy of VAR2CSA and will be important for choosing epitopes and variants of DBL3X to be included in a vaccine protecting women against pregnancy-associated malaria.
Synopsis
Pregnancy-associated malaria caused by Plasmodium falciparum is characterized by the accumulation of parasite-infected red blood cells in the placenta and is a major health problem in Africa. VAR2CSA is a parasite protein expressed on the surface of malaria-infected red blood cells and mediates the binding to the placental receptor, chondroitin sulphate A. It is believed that a vaccine based on VAR2CSA will protect pregnant women against the adverse effects of pregnancy-associated malaria. However, due to the size and polymorphism of VAR2CSA it is required to define smaller regions that can be included in a vaccine and to analyze the degree and consequences of sequence variation to ensure a broadly protective immune response. The authors have characterized the chondroitin sulphate A-binding DBL3X domain of VAR2CSA with regard to epitopes targeted by naturally acquired antibodies and the influence of sequence variation by bioinformatics and experimental data based on a VAR2CSA peptide array. They identify both variable and conserved surface-exposed epitopes that are targets of naturally acquired immunoglobulin gamma in pregnant women with placental malaria. These findings will be imperative for choosing epitopes and variants of DBL3X to be included in a vaccine protecting pregnant women against malaria.
Introduction
Individuals living in areas with high Plasmodium falciparum transmission acquire immunity to malaria over time and adults have markedly reduced risk of getting severe disease [1]. Pregnant women constitute an important exception to this rule, and this has severe consequences for both mother and child [2]. Pregnancy-associated malaria (PAM) is characterized by selective accumulation of P. falciparum in the intervillous blood spaces of the placenta [3,4]. The main pathophysiological consequences of PAM are delivery of low birth weight babies and maternal anaemia [5]. In areas of high parasite transmission PAM affects mainly primigravidae as immunity is acquired as a function of parity [2]. Parasite sequestration in the placenta is mediated by an interaction between chondroitin sulphate A (CSA) on the syncytiotrophoblasts and proteins expressed on the surface of infected erythrocytes [6]. VAR2CSA, a single and uniquely structured molecule belonging to the Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) family, is currently believed to be the main parasite ligand for placental binding [7]. Var2csa is markedly up-regulated in P. falciparum selected in vitro to bind to CSA [7] and in parasites isolated from the placenta [8]. Antibodies to the surface-expressed VAR2CSA are acquired by women exposed to malaria during pregnancy [9,10], and high levels of anti-VAR2CSA antibodies at delivery are associated with protection from low birth weight [9]. Furthermore, it has been demonstrated that targeted disruption of var2csa results in the loss of [11], or a marked reduction [12] in the ability of parasites to adhere to CSA. Based on these findings, VAR2CSA is recognized as the leading PAM vaccine candidate; however, var2csa is a polymorphic gene and the sequence variation between genes from different parasites ranges from 10%–30% at the nucleic acid level [7,13]. It is thus a major challenge for vaccine development to characterize the importance of the sequence variation and to define smaller epitopes that can be used in a vaccine to protect women against PAM. This study had two objectives. Firstly, to characterize the epitopes of the CSA-binding Duffy-binding-like (DBL) 3X domain of VAR2CSA, which are recognized by naturally acquired antibodies. Secondly, to analyze the degree and consequences of sequence variation and selection pressure within the var2csa subfamily, using the var2csa cDNA sequences of a large number of fresh placental parasite isolates. These studies would also test the validity of B cell epitope predictions and structural modeling of the DBL3X domain.
Results/Discussion
Recombinant VAR2CSA DBL3X Binds CSA and the Affinity Depends on the Primary Amino Acid Sequence
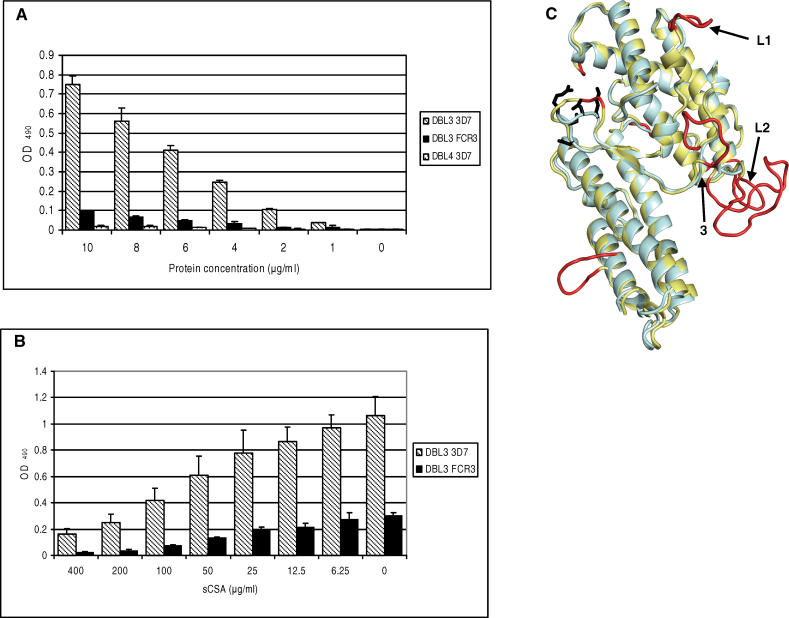
It has previously been shown that DBL3X expressed on the surface of Chinese hamster ovary cells binds CSA in vitro [14]. However, it is important to test the CSA-binding properties of secreted VAR2CSA proteins produced in expression systems that could allow for larger-scale production of a vaccine. For this study, recombinant HIS-tagged proteins were produced in Baculovirus-transfected insect cells and binding to CSA was determined in an enzyme-linked immunosorbent assay (ELISA) system. It has previously been suggested that the FCR3 DBL3X domain binds CSA, whereas the 3D7 DBL3X domain does not [14]. We found that both variants had affinity to CSA and that the 3D7 variant exhibited the strongest binding. Binding of both FCR3 and 3D7 DBL3X was concentration-dependent (Figure 1A) and could be inhibited by soluble CSA in a dose-dependant manner (Figure 1B).
Figure 1. Recombinant DBL3X VAR2CSA Binds CSA and the Structure of the Domain Can Be Modeled on the Basis of the Structure of the DBL Domains of EBA-175.
(A) Binding assay. DBL3X binds to CSA in a protein concentration-specific manner. ELISA plates were coated with soluble CSA and binding of recombinant proteins at different concentrations was determined. DBL3X from 3D7 shows better binding compared to DBL3X from FCR3. The non-CSA binding VAR2CSA DBL4ɛ was included as a control. Results are the mean of three binding assays and the error bars indicate the standard deviation.
(B) Inhibition assay. Recombinant DBL3X proteins (7 μg/ml) were preincubated with increasing amounts of soluble CSA, and binding to CSA-coated plates was determined. Binding of DBL3X is dose-dependently inhibited by soluble CSA. Results are the mean of four inhibition binding assays and error bars indicate the standard deviation.
(C) Model of DBL3X using EBA-175 as template. The figure shows a superposition of the EBA-175 F1 domain in light blue and the DBL3X (PFL0030c amino acids 1,217–1,559) model in yellow with insertions highlighted in red. Arrows indicate insertions, which align with flexible loop regions in Pkα-DBL (L1) and EBA-175 F1 (L2). Side chains of glycan-binding residues in EBA-175 F1 are shown in black. Arrow 3 indicates the corresponding Pkα-DBL Duffy antigen receptor for chemokine-binding site.
The Structure of the VAR2CSA DBL3X Domain Can Be Modeled on the Basis of the Structure of the DBL Domains of Erythrocyte-Binding Protein-175
Recently, the structures of the two DBL domains (F1 and F2) of erythrocyte-binding protein (EBA)-175 and a DBL domain in Plasmodium knowlesi (Pk)α-DBL were solved [15,16]. Using the crystal structure of EBA-175 F1 (Protein Data Bank code 1ZRO) as template, a three-dimensional model of DBL3X VAR2CSA was constructed by comparative modeling on the basis of the 3D7 sequence (Figure 1C). The sequence identity between DBL3X and EBA-175 F1 was 28%. From sequence alignments of DBL3X, Pkα-DBL, and EBA-175 (Figure S1) it was apparent that a number of cysteines were conserved, and ten of these from DBL3X aligned with cysteines which form disulfide bridges in the determined structures of EBA-175 [15] and Pkα-DBL [16]. In addition, we identified 34 buried hydrophobic residues in EBA-175 F1, EBA-175 F2, and Pkα-DBL, which corresponded to hydrophobic residues buried in the DBL3X model (Figure S1). Compared to EBA-175 F1, a number of insertions were found in DBL3X, and the majority of these were predicted to have coil secondary structure (Figure 1C). One of the insertions in DBL3X (L1: R56-I63) was found to align structurally next to a region in Pkα-DBL, which is described as a flexible linker with an experimentally determined proteolytic cleavage site [16]. A second insertion (L2: N1417-E1430) was aligned in a loop region where two residues were missing structural information in the structure of EBA-175 F1 [15], which also indicates high flexibility. Thus, the alignment of DBL3X to the solved DBL domain structures seems to match characteristics such as disulfide-bridges, stabilizing hydrophobic interactions, and flexible loop regions. These findings suggest that VAR2CSA DBL3X has the same basic structure as the solved DBL structures [15–17] in spite of the extensive sequence variation. The ability of proteins to form similar structures despite marked sequence variation has also been described for the VSG molecules covering the surface of Trypanosomes [18,19].
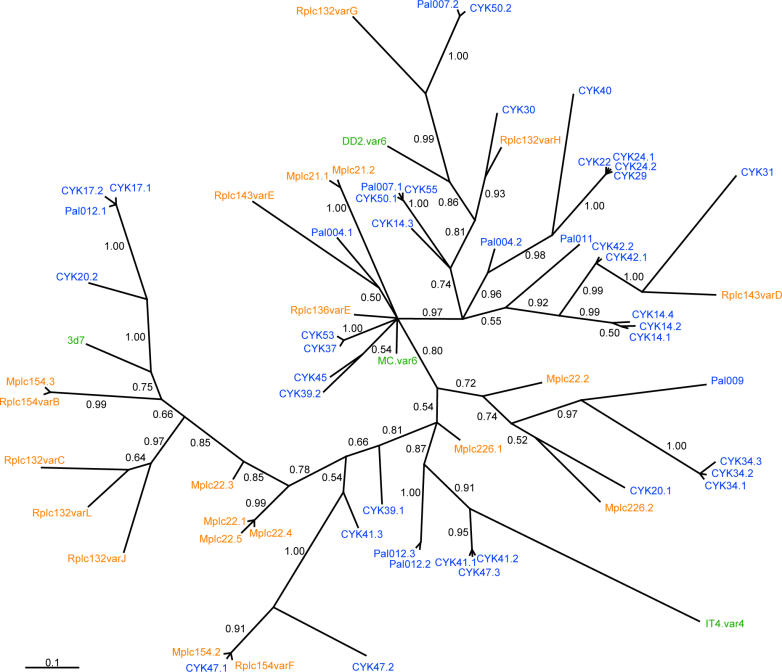
Sequence Variation within the var2csa Family Is Present as Small Hypervariable Blocks and Parasite Diversity Is Similar on a Local and Global Scale
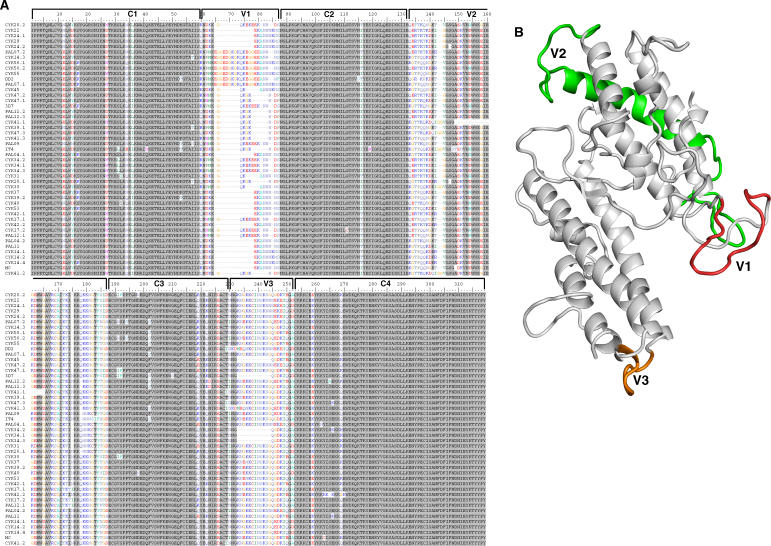
Var2csa is a relatively conserved var gene carried by all P. falciparum genomes; however, sequence polymorphisms are present in the gene. In a previous study, it was established that var2csa is transcribed at high levels by placental parasites isolated at delivery from Senegalese women [8,20]. Using cDNA from 24 placentas from that study, the region encoding DBL3X of var2csa was cloned and sequenced. A multiple alignment of 43 sequences showed that the average nucleotide diversity was low (π = 8.48% ± 0.37%, see Materials and Methods for details) reflecting a limited inter-parasite diversity in these isolates collected from a geographically small and well-defined area of West Africa. To test whether the Senegalese placental DBL3X sequences represented a monophyletic group compared to non-Senegalese DBL3X sequences, a phylogenetic tree, which included VAR2CSA DBL3X sequences available in GenBank, was constructed (Figure 2). These non-Senegalese sequences included four lab-strain parasites with different geographic origins (DD2 from Laos, MC from Thailand, IT4 from Brazil, and 3D7 with unknown origin) and 21 sequences from a sequencing study from Malawi [21]. The four database sequences and the Malawi sequences were scattered evenly in the tree indicating that the Senegalese sequences were representative for the var2csa repertoire in general and that parasite nucleotide diversity is similar on a local and global scale. It is also apparent that there is no clear subgrouping within the DBL3X sequences. The protein alignment of the DBL3X sequences (Figure 3A) suggested that DBL3X could be divided into four relatively conserved regions (C1–4) separated by three shorter variable regions (V1–3). When the variable regions were mapped to the 3-D model (Figure 3B), V1 and V3 were part of flexible loops, whereas V2 included another flexible loop but also extended into a helical region. The length of V1 and V3 varied between sequences and the 3D7 sequence was relatively short in both regions.
Figure 2. Phylogenetic Relationship between Different VAR2CSA DBL3X Sequences.
Bayesian inference tree of 43 placental cDNA sequences from Senegal (blue), 21 Malawian sequences (orange), and database sequences from four isolates (green) of different geographic origin. The posterior probability of the clades is shown at the branches.
Figure 3. Multiple Alignment of VAR2CSA DBL3X Sequences.
(A) cDNA from 43 placental parasite samples were amplified with conserved DBL3X primers and sequenced. Sequences were curated for primer sequence, translated, and aligned. The text to the left indicates the identity of the samples. The DBL3X domain can be divided into four regions, which are highly conserved, C1–C4; and three regions, which are polymorphic and harbor deletions V1–V3.
(B) Model of DBL3X showing the position of V1 (red), V2 (green), and V3 (orange).
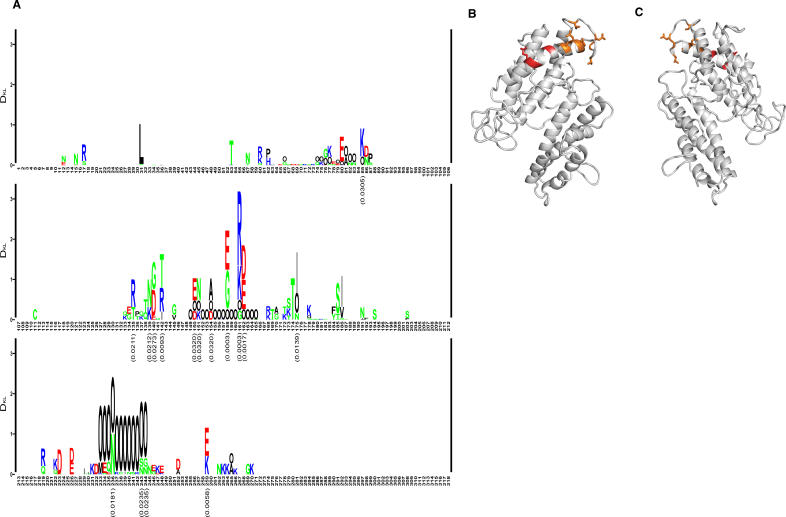
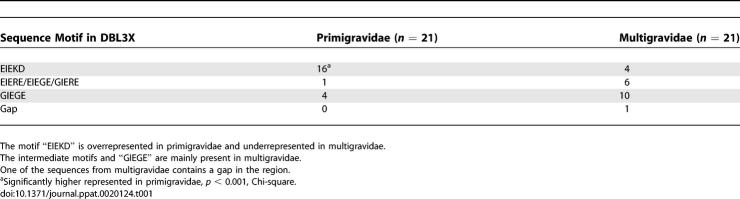
VAR2CSA Sequence Motifs Can Be Linked to the Parity of the Infected Women
We have previously shown that the expression of certain var genes is associated with severe malaria in young children [22] and suggested that var gene expression is hierarchically structured. This could occur because the progeny of parasites expressing var gene products that mediate the most efficient sequestration outgrow the progeny of parasites expressing a molecule mediating less efficient binding [22–24]. A similar process could shape the expression of molecules mediating binding in the placenta. If that was the case, it would be predicted that a systematic difference between molecules mediating binding in primigravidae and multigravidae women could be detected in areas of high P. falciparum transmission where women are exposed to several parasite clones during pregnancy [25]. This was addressed by calculating the Kullback-Leibler distance between 21 VAR2CSA sequences originating from primigravidae and 21 sequences from multigravidae. The overall cumulated Kullback-Leibler distance (DKL) between sequences from the primigravidae and the multigravidae was higher than for randomly chosen groupings of the sequences (p = 0.0075). The DKL was also calculated for each position in the alignment to determine which polymorphisms contributed to the difference between the sequence of parasites from primi- and multigravidae women and visualized in a Kullback-Leibler sequence logo (Figure 4A). This implicated two stretches of amino acids in positions 135–175 and 233–245 located in the V2 and V3 regions, respectively. The region from position 158 to 162 was of special interest since the motif “EIEKD” was mainly found in primigravidae and the motif “GIEGE” mainly in the multigravidae (Table 1). Intermediate motifs like “(G/E)IERE” where the fourth position had changed from lysine to arginine were also found, implying that the following evolutionary pathway may have been operating: lysine (AAA or AAG) ↔ arginine (AGG) ↔ glycine (GGG). The change from glutamate and lysine/arginine, which have large charged side chains, into glycine without a side chain, could result in marked functional and antigenic changes. Thus, it was interesting that positions 1, 4, and 5 in the motif (position 158, 161, and 162) were predicted to be surface-exposed in the structural model, which was also the case for all of the other amino acid positions in V2 that differed significantly in the Kullback-Leibler sequence logo (Figure 4B and 4C). The finding that parasites expressing the EIEKD motif were more prevalent in primigravidae than in multigravidae women (Table 1, Chi-square test: p < 0.001) could indicate that these parasites have a biological advantage in women experiencing their first pregnancy and that parasites expressing the other motifs (G/EIERE or GIEGE) have an advantage in women who have been pregnant before. This could arise because parasites carrying the EIEKD motif are the most efficient mediators of binding and therefore dominate in women with limited immunity against PAM. As immunity develops against these parasite forms, parasites expressing other motifs that are less efficient in binding but not serologically cross-reactive take over. Interestingly, a monoclonal Ghanaian field isolate undergoing full genome sequencing at the Sanger Institute has two copies of var2csa, one with the EIEKD motif and one with the GIEGE. Although the functional background for the observed phenomenon is presently not clear, the systematic sequence variation at positions predicted to be surface-exposed between parasites from primi- and multigravidae women strengthen the concept that VAR2CSA is the main parasite ligand for sequestration of malaria parasites in the placenta.
Figure 4. Sequence Differences in Infections with Different Parity.
(A) Kullback-Leibler sequence logo based on a multiple alignment of the DBL3X region (Figure 3). The sequences in the alignment were split into two groups of equal size: 21 sequences obtained from primigravidae and 21 sequences from multigravidae women. The x-axis shows amino acid positions in the alignment. The height of a position indicates the amount of difference between the two groups according to the symmetric Kullback-Leibler distance, and the letters indicate the amino acids contributing to this difference. The letter “O” has been chosen to signify gaps in the alignment. Polar amino acids are green, basic are blue, acidic are red, and hydrophobic amino acids are black. P-values indicating the significance of DKL based on the parity grouping compared to random groupings are shown below positions where p < 0.05.
(B) DBL3X model showing the position of amino acids that were differentially found in primi-gravidae and multigravidae women. Residues in the region 135–175 of moderately significant difference are shown in orange, and highly significant residues (positions 158, 161, and 162) are shown in red.
(C) The model (B) rotated 180 degrees so it is positioned similar to Figure 1C.
Table 1.
Sequence Signature in DBL3X Position 158–162 of VAR2CSA Expressed by 42 Parasites Infecting either Primigravidae or Multigravidae
VAR2CSA Is Under both Positive and Purifying Selection
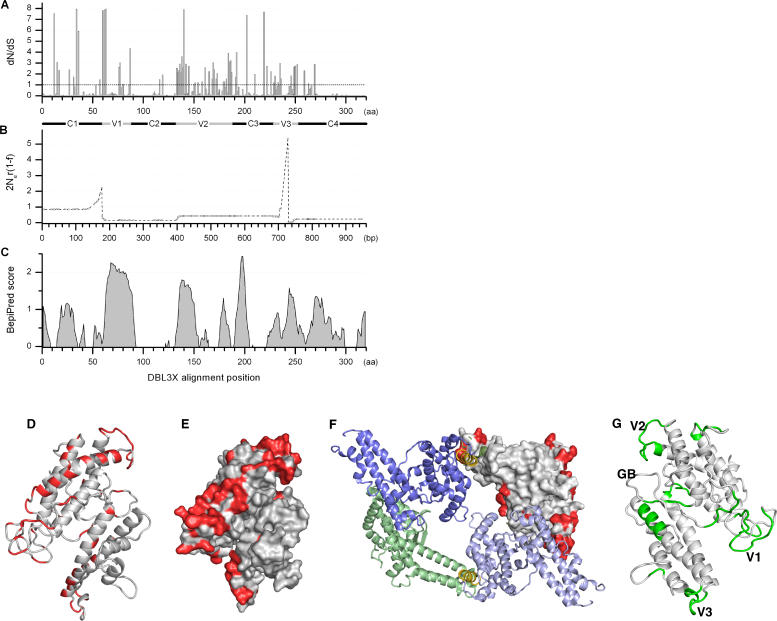
A recent study has suggested that sequence polymorphisms in a region of VAR2CSA upstream to DBL3X largely are due to positive natural selection pressure [13]. To further investigate the nature of sequence diversity in var2csa, the dN/dS ratios (dN, rate of non-synonymous mutations per non-synonymous site and dS, the rate of synonymous mutations per synonymous site) for DBL3X were calculated on the basis of the sequenced DBL3X domains. A dN/dS <1 indicates that the position is under negative or purifying selection pressure leading to conservation of the residue, while dN/dS >1 suggests positive or diversifying selection pressure, and suggests that amino acid changes are evolutionarily advantageous at the position. Purifying selection was mainly found in the conserved regions C1–4 and it was especially pronounced in the C2 and C4 regions, although single sites were observed to be under diversifying selection (Figure 5A). Several blocks appeared to be under strong diversifying selection and these appeared most prominently in regions V1, V2, and V3. It is interesting to note that residues under diversifying selection mainly were situated in regions predicted to be surface-exposed and concentrated on one side of the molecule (Figure 5D and 5E). The two DBL domains of EBA-175 are predicted to form a reverse handshake dimer with the F1 domain of each molecule interacting with F2 of the other [15]. In this four-domain DBL structure, we replaced one of the EBA-175 F1 domains with our VAR2CSA DBL3X model (Figure 5F). It was noticeable that the largely conserved C2 and C4 regions of DBL3X were predicted to take part in the lining of the central cavity of the four-domain structure and form the region next to the cavity facing the membrane in native configuration. The model presented here is unlikely to fatefully reflect the structure of the native molecule, but the positioning of conserved DBL3X regions in the model makes biological sense, and the finding underscores the need to obtain knowledge about possible interactions between VAR2CSA DBL domains. The lack of amino acid positions under diversifying selection in the regions adjacent to the cavity might be due to the possibility that these sites are involved in ligand binding and thus functional constraint. Another explanation could be that the regions forming the predicted cavity and the area predicted to face the membrane are not accessible for antibodies in natively folded molecules.
Figure 5. Sequence Variation and B Cell Epitopes in DBL3.
(A) dN/dS ratio per amino acid based on an alignment of 47 DBL3X sequences. dN/dS ratio higher than the dashed line indicates that the position is under positive selection. V1–V3 and C1–C4 are indicated with solid bars. It should be noted that the inference of selection pressure is less reliable in positions with gaps, which are mainly found in the variable regions (see Figure 3).
(B) The population recombination rate ρ = 2Ner(1 − f) estimated per bp across DBL3X. Circles indicate the SNPs for which ρ is estimated, linear interpolation is shown between SNPs. Two hotspots corresponding to a local raise in r are observed in association with the variable regions V1 and V3.
(C) B cell epitope predictions of the 3D7 sequence performed by the BepiPred server. DD2 and IT-4 were used to fill 3D7 sequence gaps.
(D) Cartoon representation of the DBL3X model showing amino acid positions under diversifying selection in red (dN/dS ratios >1).
(E) DBL3X model with surface topography prediction showing amino acid positions under diversifying selection in red (dN/dS ratios >1).
(F) The homo-dimer of the two domains of RII of EBA-175 has been shown to form a handshake structure with F1 of one molecule dimerizing with F2 of another molecule and vice-versa. Here VAR2CSA DBL3X has been superimposed on the F1 domain of one of the EBA-175 molecules in the dimer. The F1 domain is shown in green, F2 domains are blue, and the linker regions are gold. Amino acids under diversifying selection in DBL3X are shown in red (dN/dS ratios >1). It is clear that most residues inward of the dimer are not under positive selection.
(G) DBL3X model showing residues with BepiPred scores higher than 0.9 in green. Locations of V1–V3 are indicated on the model. GB indicates the glycan-binding site of EBA-175.
Recombination Is a Factor in the Generation of var2csa Sequence Variation
Previous studies have reported that frequent recombination events generate sequence diversity in the PfEMP1 family [26–28]. To determine the role of recombination for DBL3X sequence variation, we estimated the population recombination rate ρ defined for partially inbreeding haploid species by the compound parameter 2Ner(1− f), where Ne represents the effective population size, r is the per-generation cross-over recombination rate per base pair (bp), and f is the inbreeding coefficient. Variations in ρ across DBL3X correspond to variations in the recombination rate r, as Ne and f are constant for the dataset. Two recombinational hotspots were observed at bp positions 138–178 and 704–730 (Figure 5B). The two best defined recombination breakpoints were present in the C1/V1 borderline at bp 177–179 and within V3 at bp 728–730. Both the V1 and V3 region harbored major deletions/insertions in several of the sequences, which may have arisen as a consequence of unequal cross-over during recombination at the hotspots. Unequal cross-over results in either deletions or insertions of variable number of tandem repeats (VNTR), and the DBL3X V1 region did indeed contain a high amount of VNTR. The loop region of V2 contained a small VNTR insert in some sequences, while VNTR were less obvious in the V3 region. In a recent study of the P. falciparum Chromosome 3, high recombination rates were found in sub-telomeric regions, and African P. falciparum strains showed a much higher population recombination rate than strains from other regions of the world [28]. The overall population recombination rate for the DBL3X region was estimated to ρ = 0.54 per bp (95% CI: 0.41–0.90). This is in accordance with a recent report for a VAR2CSA region upstream to DBL3X around interdomain 1, where the rate was estimated to 0.71 per bp [13], and in agreement with recombination rates in Chromosome 3 of African parasite lines summarized as ρ > 0.1 per bp [28]. The high population recombination rate in DBL3X combined with the observed sequence variation adjacent to the detected recombination hotspots, suggests that recombination is an important factor in generating sequence variation. The V1 region that seems to be most strongly affected by recombination is predicted to form a structurally unrestricted flexible loop allowing for sequence variation, and it is possible that the whole V1 region is under diversifying selection pressure exerted by the immune system, even though this could not be predicted by the dN/dS method due to gaps. This notion is supported by the findings discussed below, showing that this region is part of a major B cell epitope.
VAR2CSA DBL3X B Cell Epitope Prediction
VAR2CSA epitopes exposed on the surface of the protein and accessible for immunoglobulin gamma (IgG) binding could be under diversifying selection resulting in escape mutations and high dN/dS ratios, whereas residues involved in protein folding, stability, and anchoring could be under purifying selection with low dN/dS ratios. To predict linear B cell epitopes, the 3D7 sequence was submitted to the BepiPred server [29] and seven epitopes were predicted within the DBL3X sequence (Figure 5C). Some of these epitopes were located in areas with high dN/dS ratios and there was a weak but statistically significant association between the BepiPred score and the dN/dS ratio (Pearson's r = 0.18 and p = 2.5*10−9). The reason for this weak association could be that antibody epitopes are situated in regions that are functionally constrained or that positive selection pressure is also driven by other forces like MHC-2 binding and T helper cell activation as found for HIV-1 [30]. Furthermore, the BepiPred algorithm predicts linear epitopes, and some of these could be located in parts of the molecule that are not accessible to antibodies in the native folded molecule. Nevertheless, most of the predicted epitopes (Figure 5G) were situated in surface-exposed loop regions of the model, and one of the highest scoring epitopes was in the V1 region. Residues that align directly to the glycan-binding residues of EBA-175 F1 and to the Duffy antigen receptor for chemokines (DARC) binding site in Pkα-DBL were not predicted to be part of epitopes. However, a part of the V2 region in proximity to the putative glycan-binding loop was predicted to be targeted by antibodies and had high dN/dS values.
Fine Epitope Mapping of VAR2CSA Antibodies Acquired during Pregnancy
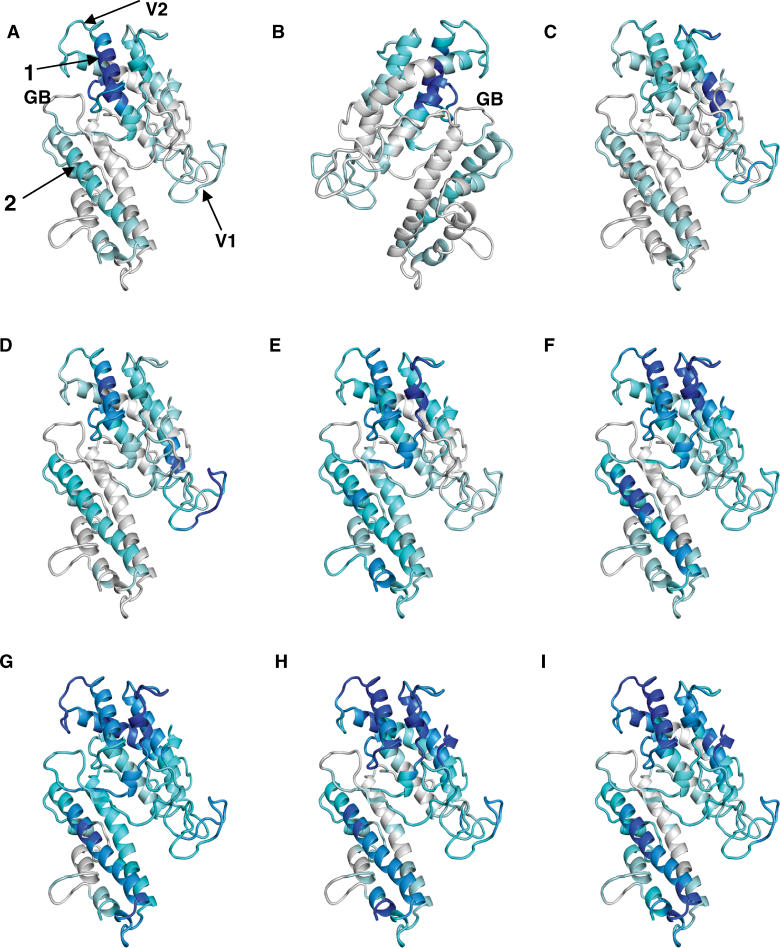
To verify the above bioinformatical predictions, we evaluated the fine specificity of naturally acquired human antibodies to VAR2CSA DBL3X in a peptide array consisting of 442 overlapping 31mer peptides covering exon 1 of VAR2CSA of the 3D7 sequence. Antibody reactivity to individual amino acids was assigned on the basis of an algorithm based on the observation that a major part of antibody binding motifs in a set of conformational epitopes are from two to seven amino acids long, containing either two or three defined residues spaced by undefined amino acids [31]. The VAR2CSA of 3D7 contains 31,149 such motifs and each was assigned an average PepScan value by adding the measured reactivity from the 31mers in which the motif was present, and dividing with the number of times the motif occurred. The method, described in Materials and Methods, was validated by testing serum from rabbits immunized with a VAR2CSA construct, which showed that both the measurements based on individual peptide readings and the algorithm described above, mapped antibody responses to regions present in the antigen used for immunization (Figure S2). Furthermore, there was a high concurrence between antibody peaks defined by the reactivity of the individual peptides and the peaks defined by the algorithm defining single amino acid scores (Figure S2). Plasma from individuals not exposed to malaria did not react with any of the peptides in the array (unpublished data). The IgG reactivity in the plasma of eight Ghanaian women with a known history of a placental malaria infection was analyzed and the peptide array data was visualized on the DBL3X model (Figure 6). The regions with the highest reactivity were on the side of the domain where glycan-binding is found in EBA-175 F1 (Figure 6A and 6B) and therefore IgG reactivity was visualized on a model positioned with this side in the front (Figure 6A and 6C–I). It was clear that the majority of the individuals had specific IgG against the variable regions, V1 or V2. V3 is partly deleted in 3D7, and IgG reactivity could thus not be measured in the peptide array based on the 3D7 sequence. Remarkably, a short α-helix (Figure 6A, arrow 1) in proximity to the loop for glycan-binding in EBA-175 F1 showed the highest antibody reactivity in all serum samples, despite the fact that this region had very low dN/dS ratios (Figure 5A, positions 120–132). Another α-helix was also often recognized by antibodies (Figure 6A, arrow 2). This helix was predicted to contain a B cell epitope by the BepiPred algorithm and had polymorphic residues (Figure 5A and 5C, positions 251–281). The region corresponding to the loops containing the EBA-175 F1 glycan-binding sites was not recognized by any of the serum samples. Conserved regions (Figure 6A, arrow 1) targeted by naturally acquired IgG are of considerable interest in the search for vaccine constructs which could elicit a broad protective immune response.
Figure 6. Defining Targets of Antibodies on DBL3X.
Models of DBL3X in which the intensity of the gray and blue indicates reactivity of plasma tested on a peptide array (PepScan scores). Gray indicates lowest scores and dark blue indicates highest scores. (A and C–I) represent reactivity in eight individual plasma samples. (B) shows the reactivity in the same plasma as tested in (A), but the model has been rotated 180 degrees. Arrow 1 indicates a highly recognized conserved α-helix region, which was not predicted to be an epitope by BepiPred and was characterized by low dN/dS ratios and low sequence variation. Arrow 2 indicates another α-helix with a predicted B cell epitope containing some residues with high sequence variation. Variable regions V1 and V2 are fairly well recognized by most of the serum samples. GB indicates the glycan-binding site of EBA-175.
PepScan Analysis of Affinity-Purified Antibodies
During infection, VAR2CSA will be degraded and antibodies will be acquired against epitopes that are not accessible for antibodies when the protein is in its natural conformation. It is therefore possible that some of the antibody reactivities measured in the peptide array were directed against such epitopes. To address this question, analysis was performed on plasma, which had been affinity purified on recombinant DBL3X or antibody-depleted by incubation with erythrocytes infected with VAR2CSA-expressing parasites.
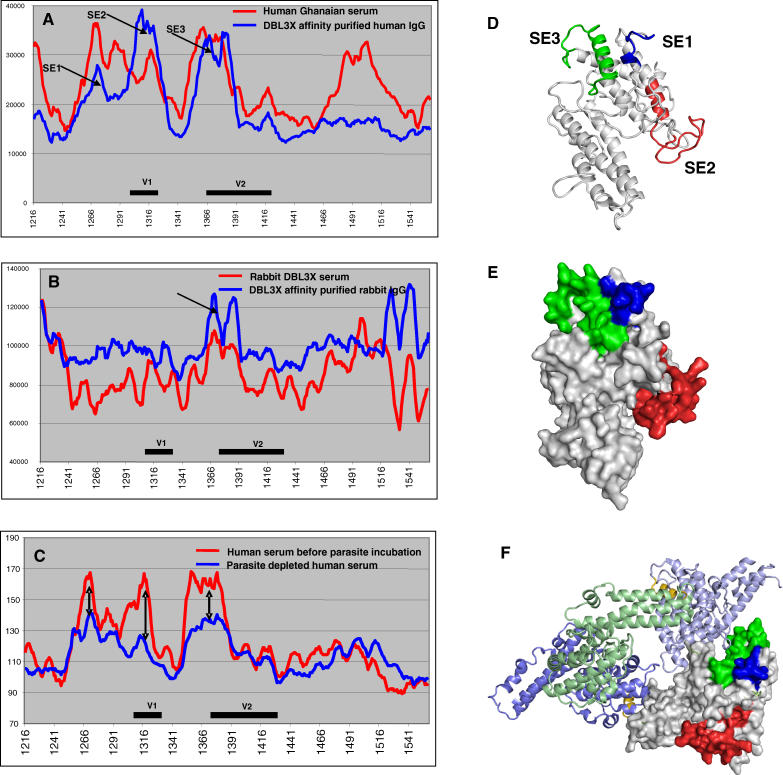
Plasma from a rabbit immunized with recombinant DBL3X and plasma from women who had suffered a placental infection were affinity purified on the recombinant CSA-binding DBL3X protein and analyzed by the peptide array. Before affinity purification, the plasma pool from Ghanaian women showed reactivity corresponding to eight peaks distributed throughout the domain (Figure 7A). In contrast, the reactivity of the affinity-purified IgG was concentrated to three peaks (SE1–SE3) in the C1, V1, and C2/V2 regions. The immunized rabbit had strong IgG reactivity against the epitope in the C2/V2, which was also affinity purified from the female plasma pool (Figure 7B), but the rabbit had not raised an IgG response against the epitopes in C1 and V1. A plasma pool from Tanzanian women was also analyzed and in this case surface reactive antibodies were depleted from the pool by incubation with VAR2CSA-expressing infected erythrocytes. In this pool the main reactivity was against the surface-exposed epitopes (SE1–SE3) defined in the Ghanaian plasma, and the depletion experiment indicated that absorption with infected erythrocytes caused a marked reduction in the reactivity against these epitopes (Figure 7C). These findings indicate that the three identified regions were accessible to antibodies on the native protein and that the folding of the Baculovirus-produced recombinant protein was close to the natural configuration. SE2 and SE3 corresponded to loop regions V1 and V2, which are both protruding from the structure of the DBL3X model (Figure 7D). Interestingly, SE1 and SE3, which are located in separate parts of the primary structure of the domain, form a continuous region on the surface of the predicted DBL3X protein structure (Figure 7E). The model also predicts that all surface-exposed sites are located on one part of the domain indicating that the other parts are buried or engaged in the intact PfEMP1 molecule expressed on the surface of the infected erythrocyte (Figure 7E). Unexpectedly, the highly conserved part of C2, which was well recognized in the peptide array by all women, corresponded to a part of the surface-exposed epitope SE3.
Figure 7. Reactivity of Plasma Affinity Purified on Recombinant DBL3X and Human IgG Depleted for Surface Reactivity by Incubation with Infected Erythrocytes Expressing VAR2CSA.
(A) PepScan IgG reactivity in a Ghanaian pool of pregnancy plasma before (red) and after (blue) affinity purification on recombinant DBL3X protein. Variable regions V1 and V2 are indicated with a black line. Arrows indicate three surface-exposed regions (SE). The x-axis is amino acid position in 3D7 VAR2CSA.
(B) PepScan IgG reactivity in plasma from a rabbit immunized with DBL3X before (red) and after (blue) affinity purification on recombinant DBL3X protein. Arrow indicates the surface-exposed region. N- and C-terminal parts of the recombinant proteins did also appear to be surface-exposed; this could be due to improper folding of the N- and C-terminal parts of the protein.
(C) PepScan IgG reactivity in a Tanzanian pool of pregnancy plasma before (red) and after (blue) depletion of antibodies directed against surfaced-exposed VAR2CSA regions by incubation with infected erythrocytes expressing VAR2CSA on the surface. Arrows indicate three regions where the reactivity was markedly reduced after depletion largely corresponding to SE1, SE2, and SE3 on (A).
(D) DBL3X model showing the location of surface-exposed epitopes, SE1 (blue), SE2 (red), and SE3 (green).
(E) DBL3X surface topography showing SE1–SE3. From the model it is apparent that the SE1 and SE3 form a continuous epitope.
(F) VAR2CSA DBL3X superimposed on the F1 domain of one of the molecules in the EBA-175 dimer. The F1 domain is shown in green, F2 domains are blue, and the linker regions are gold. On the DBL3X domain the three experimentally verified surface-exposed regions are shown in similar coloring as (D and E) The three regions are mainly predicted to be on the opposite side of the central cavity of the dimer, and parts of SE3 correspond to amino acid positions directly involved in the dimerization of EBA-175.
When exchanging the F1 domain of one of the EBA-175 molecules in the EBA-175 dimer with VAR2CSA DBL3X and mapping SE1–SE3 to the model, it is apparent that the surface-exposed regions are on the opposite side of the central cavity of the dimer (Figure 7F). However, the part of DBL3X that may be directly involved in the dimerization extends into the SE3 region (shown in green), indicating that in this model the potential dimerization motifs are accessible for antibodies. EBA-175 is suggested to dimerize upon ligand interaction and it could be that SE3 was “unengaged” due to the lack of CSA during antibody depletion. If several domains need to interact to form a buried CSA-binding site, antibodies targeting conserved residues, like the region in C2 that forms parts of SE3, could function by inhibiting the dimerization of domains.
Our results demonstrate that both conserved and polymorphic surface-exposed regions are targets for VAR2CSA DBL3X antibodies acquired during pregnancy by malaria-infected women. This opens for vaccine, strategies similar to those being pursued for the polymorphic merozoite surface protein (MSP) 1 [32]. The proteolytic processing of MSP1 is a prerequisite for successful parasite invasion of erythrocytes, and one vaccine strategy is based on the induction of antibodies against the conserved C-terminal part of the molecule that inhibits processing [33]. Another MSP1 vaccine strategy employs chimeric vaccine constructs designed to induce antibodies targeting the polymorphic types present in the N-terminal part of the molecule [34]. In a similar fashion, VAR2CSA vaccine constructs could target conserved epitopes like those identified in SE3, or alternatively, constructs should induce antibodies targeting different serological variants like those predicted to be generated by the sequence polymorphisms present in regions SE1 and SE2. The human antibody pools used in this study to identify surface-exposed antigenic targets inhibit parasite binding to CSA in vitro (unpublished data). However, the molecular targets of the inhibitory antibodies have not been identified and knowledge is required about the antigenic targets for antibodies on the other VAR2CSA DBL domains. The high similarity between the DBL structures of EBA-175 [15], Pkα-DBL [16], and the VAR2CSA DBL3X model suggests that the DBL structures in Plasmodium are relatively conserved and that the antigenic characteristics of the DBL3X might be comparable to those of the remaining VAR2CSA DBL domains. However, a more comprehensive analysis of sequence variation, antibody epitopes, and structure of the VAR2CSA DBL domains not belonging to DBL3X is needed to establish the extent of the structural conservation between the domains.
Development of PAM vaccines requires a much better understanding about the molecular interaction between placental parasites and the ligand on the syncytiotrophoblasts, as well as knowledge about the fine specificity of the targets for antibodies inhibiting binding. Native PfEMP1 molecules are difficult to isolate and with current technologies it is difficult to produce correctly folded recombinant material in quantities allowing structure elucidation by crystallography. In this paper, we have combined in silico methods such as model building and sequence analysis and the analysis of antibody reactivity to obtain new information and generate hypotheses about the structure and functional relationship of VAR2CSA.
Materials and Methods
Cloning and expression of VAR2CSA domains.
DBL3X and DBL4ɛ of VAR2CSA were amplified from FCR3 and 3D7 genomic DNA with the following primers: FCR3 DBL3X – 5′ CG GAA TTC ACC AAT ATT AAT AAA AGT GAA and 3′ ATT TGC GGC CGC CAG CAT TAT TAT ATT TGT A, 3D7 DBL3X – 5′CG GAA TTC AAG ATG AAG TCC TCC GAG and 3′ATT TGC GGC CGC CAA AAC AGC CAA GCT GGA, 3D7 DBL4ɛ - 5′CG GAA TTC CAG GTG AAG TAC TAC GAA and 3′CTG TTC CTC CAC GTG CTC CAG. PCR products were digested with EcoRI and NotI for cloning into the Baculovirus vector, pAcGP67-A (BD Biosciences, http://www.bdbiosciences.com), which was modified to contain the V5 epitope upstream of a histidine tag in the C-terminal end of the constructs. Linearised Bakpak6 Baculovirus DNA (BD Biosciences) was co-transfected with pAcGP67-A into Sf9 insect cells for generation of recombinant virus particles. Recombinant protein was purified on Co2+ metal-chelate agarose columns as secreted histidine-tagged proteins from the supernatant of infected High-Five insect cells.
CSA binding assay.
Binding to CSA (C9819, Sigma-Aldrich, http://www.sigmaaldrich.com) was determined in an ELISA system. ELISA plates (Falcon 351172) were coated overnight with CSA (50 μg/ml) in PBS at 4 °C. Coating with 1% BSA in PBS (blocking buffer) was used as negative control. Plates were incubated with blocking buffer for 1 h at room temperature (RT) to inhibit non-specific adsorption to the plate. The VAR2CSA proteins were diluted in blocking buffer (1–10 μg/ml protein), added to the wells, and incubated for 1 h at RT. For the inhibition assays, proteins (7 μg/ml) were pre-incubated with different concentrations of soluble CSA for 30 min. Plates were washed four times in PBS between the different steps. Specific binding was visualized by adding an HRP-conjugated antibody (R960–25, Invitrogen, http://www.invitrogen.com) targeting the V5 epitope of the constructs. Plates were incubated for 1 h with the anti-V5 antibody diluted 1:3000 in blocking buffer. The color reactions were developed for 15 min by the addition of o-phenylenediamine substrate and stopped by adding 2.5 M H2SO4. The optical density was measured at 490 nm.
Cloning and sequencing of placental var2csa genes.
All DBL3X sequences were obtained from cDNA, whereas DBL2X and the overlapping region of DBL4ɛ and DBL5ɛ of var2csa were cloned from genomic DNA of placental parasites. In brief, parasites were dissolved in Trizol LS (Invitrogen) and RNA was prepared according to the manufacturer's instruction. RNA pellets were dissolved in 10 μl of RNase-free water and treated with DNaseI (Sigma-Aldrich) for 25 min at RT, followed by 10 min heat inactivation at 65 °C. All RNA samples were subsequently tested in real-time PCR for contamination with genomic DNA using a primer set for the housekeeping gene, seryl-tRNA synthetase. DNA-free RNA samples were used for synthesis of cDNA by reverse transcriptase (Superscript II, Invitrogen) and random hexamer primers as described by the manufacturer. Following primer sets were used for cloning DBL3X from cDNA into either the Baculovirus vector, pAcGP67-A (BD Biosciences) or the pCR2.1-TOPO vector (Invitrogen): p509 5′ CG GAA TTC GAT ACA AAT GGT GCC TGT and p510 3′ ATT TGC GGC CGC ATA TAC TGC TAT AAT CTC C, p508 5′ CG GAA TTC ACA CAA AAT TTA TGT GTT and p510, p503 5′ GAG ATA CAA ATG GTG CCT GT and p505 3′ AAA TTT GCT GAT ATA CAT TCA G. PCR products aimed for the Baculovirus vector were digested with EcoRI and NotI before ligation. Three to six colonies of each cloning and corresponding plasmids were sequenced by Macrogen (http://www.macrogen.com).
Genomic DNA was extracted from filter paper using a chelex-based method [35]. Briefly, filter spots were dissolved in 0.5% saponin in PBS using a microtiter plate and incubated overnight on a shaker at RT. After washing the filter spots twice in PBS, a solution of 50 μl of H2O and 100 μl of 10% chelex mixture was added to each well. The plate was boiled for 8 min and subsequently cooled down for 10 min at RT. A PCR reaction was run with primers for the seryl-tRNA synthetase gene to control for the DNA content. Around 1–3 μl of DNA was used for the PCR reactions amplifying the different var2csa regions. All PCR products were cloned into the pCR2.1-TOPO vector and the inserts sequenced on a 3100-Avant Genetic Analyzer (Applied Biosystems, http://www.appliedbiosystems.com). The origin of parasites are described in [20].
Phylogenetic reconstruction.
The alignment of 43 placental and four database VAR2CSA DBL3X sequences, covering the 3D7 amino acid positions 1256–1549, was constructed using the software RevTrans [36], and subsequently manually corrected for errors. To cover more of the DBL3X domain, an alignment of 17 database sequences was constructed in the same way, covering the 3D7 positions 1217–1255. For the phylogenetic tree, 21 Malawian sequences [21] were aligned with the sequences mentioned above, again using RevTrans and manual correction, resulting in a 609-bp alignment. The program MrModeltest version 2.2 [37] was used to find the most appropriate nucleotide substitution model based on the Akaike information criterion [38]. Phylogenetic trees based on the above alignments were constructed by Bayesian inference using the program MrBayes version 3.1.1 [39]. In all cases Markov chain Monte Carlo (MCMC) sampling was performed for 10,000,000 generations with eight chains. Convergence was confirmed by comparing the results of two independent runs. Burn-in was determined using Tracer [40] and 50% majority rule consensus trees were constructed.
Model fitting and Akaike weighted dN/dS average.
The program codeml from the PAML package version 3.14 [41] was used to fit a range of codon-based evolutionary models to the VAR2CSA DBL3X region using the alignments and Bayesian trees mentioned above. The 11 codon-based models were tested using codeml: M0, M1a, M2a, M3 (with either 3, 4, 5, 6, or 7 site categories), M5, M7, and M8 [42–44]. All 11 models were fitted using the F3x4 (different nucleotide frequencies for each codon position) approach for estimating codon frequencies. Convergence was confirmed by comparing the results of several independent runs started with different parameter vectors.
The Akaike information criterion (AIC) was used to assess model fits [38,45,46]. Briefly, AIC estimates the expected relative Kullback-Leibler distance (i.e., AIC is an estimate of the amount of information that is lost when a given model is used to approximate the full truth). AIC is a function of the maximized log-likelihood (lnL) and the number of estimated parameters (K) for a model. Specifically, AIC = −2lnL + 2K where lower AIC values indicate better models. From AIC it was possible to compute Akaike weights, which can be interpreted as the conditional probability of the model given the data and the set of initial models [38,46]. On this basis, dN/dS ratios for codon positions were calculated as an average of the dN/dS ratios estimated from each of the 11 models, weighted by the Akaike weights for the corresponding model.
Recombination and mutation rates, diversity, and sequence logo creation.
The population recombination rate ρ was estimated for the VAR2CSA DBL3X domain in LDhat version 2.0 [47], which based on population genetics uses coalescent methods specially adapted to account for the possibility of recurrent or back mutation and for an AT-rich genome such as that of P. falciparum [48]. As argued by Mu et al. [28], the coalescent recombination estimate can, for partially inbreeding haploid species such as P. falciparum, be interpreted as the compound parameter ρ = 2Ner(1 − f), where Ne represents the effective population size of the DBL3X population, r denotes the rate of recombination cross-over events per generation per bp, and f is the inbreeding coefficient. The effective population size should be thought of as the size of an ideal population [48,49] with the same magnitude of random genetic drift as the actual population with size N [50].
To test if the placental DBL3X sequence data showed evidence for deviation from the neutral model of evolution assumed by the coalescent method, we calculated Tajima's D statistic [51] and Fu and Li's D* and F* statistic [52]. All three statistics were insignificantly different from zero (p > 0.1 in all cases), indicating that the coalescent approach could be applied. The hypothesis of no recombination was rejected using the likelihood permutation test [48] with 1,000 permutations of segregating sites, of which none produced a higher maximum composite likelihood than for the DBL3X data. The hypothesis of a constant recombination rate across the analyzed region was also rejected (p = 0.048 with 10,000 simulations) using the method described in [47], indicating significance in the recombination rate variations over DBL3X.
For calculation of the population recombination rate ρ, we used the Bayesian reversible-jump Markov chain Monte Carlo (RJMCMC) method with a block penalty of 10, running for 10,000,000 iterations with 2,000 iterations per sample and a burn in of 50 samples. The overall region estimate was converted to bp units using the average length of the analyzed sequences. Recombination hotspots were defined as intervals containing SNPs where the population recombination rate mean was above the upper limit of the 95% confidence interval for the overall region estimate of 2Ner(1 − f). Fu and Li's D* and F* and Tajima's D were calculated using DnaSP version 4.10.6 [53]. The average nucleotide diversity and its variance were calculated according to Nei [54] equation 10.5 and 10.7, respectively (gapped columns were included).
The Kullback-Leibler sequence logo was created by calculating the distance between the two groups of sequences for each amino acid position in the alignment using the symmetric Kullback-Leibler distance:
where p and p′ are the frequencies of an amino acid type in each of the two groups, and AA indicates that the sum is over all the amino acid types. The cumulated Kullback-Leibler distance was calculated as the sum of Dkl for all positions in the alignment. Gaps in the alignment were in this analysis assigned the letter “O” and treated as an amino acid class. To test if the mentioned grouping according to parity gave two significantly different sequence groups, we created 10,000 random groupings and for each of these summed the DKL over all amino acid positions. Similarly for the individual positions in the logo, the distribution of DKL for 10,000 random shuffles of sequence grouping was noted specifically for each position, and the p-values were based on these distributions.
PepScan motif analysis.
442 overlapping 31mer peptides covering the exon 1 of 3D7 VAR2CSA were synthesized as solid phase peptide synthesis (SPPS) with a stepwise addition of the different amino acids attached to a solid resin. The long peptides were synthesized with a cysteine at amino acid (aa) position 15 allowing some secondary structure. This approach allows identification of antigenic sites that cannot be mapped using short, linear peptides (PepScan Systems, the Netherlands). The raw data from a PepScan experiment consists of figures measuring the amount of IgG bound to each of the overlapping 31mer peptides. We used the overlap in primary sequence to determine more specifically what the antibodies have affinity for.
The motif analysis is based on the concept that polyclonal IgG response consists of subpopulations of monoclonal antibodies each binding a certain set of amino acid sequence motifs. We then made a list of all possible binding motifs and transferred the information from the peptide array to these, giving each motif a score indicating the IgG affinity for the motif. The PepScan assay is designed primarily to determine linear epitopes, and thus we are mainly interested in short binding motifs and gaps. On this basis we performed the PepScan motif analysis, using motifs containing either two or three defined residues spaced by undefined amino acids up to a certain maximal length of 5, 7, 10, 15, 20, or 25 residues. We found that the different maximal motif lengths gave similar results, but with different detail resolutions (long motifs had a smoothing effect), and a binding motif with a maximal length of seven residues was selected as being most informative. Thus, the presented results are based on the assumption that motifs are two to seven amino acids long, and contain either two or three defined residues spaced by undefined amino acids, e.g., a possible motif could be “WXXXDXE” or simply “KN.” The method was validated by comparison to the raw data, where rabbit serum from a rabbit immunized with a DBL5 construct was used in the PepScan assay (Figure S2). The figure shows that the motif analysis produces peaks approximately at the same positions as in the raw data, and that the analysis does not introduce bias in the other regions of the protein. As control for the human IgG used in Figure 6, we used non-immune Dutch serum as well as a malaria-exposed nulliparous female individual.
Affinity purification of antibodies and depletion of serum on parasites.
Affinity purification of antibodies was done according to manufacturer's instructions. In brief, 1 mg of recombinant protein was dialyzed against 0.2 M NaHCO3, 0.5 M NaCl (pH 8.3), and applied to a NHS-activated HiTrap 1-ml column (GE Healthcare, http://www.gehealthcare.com) that had been equilibrated with 3 × 2 ml 4 °C 1 mM HCl. After coupling, the columns were washed with 0.5 M ethanolamine, 0.5 M NaCl (pH 8), 0.1 M acetate, 0.5 M NaCl (pH 4), and a final wash with PBS (pH 7.4). 1 ml of a plasma pool (28 women from Ghana) was then applied to the column. After washing in 10 ml PBS, affinity-bound antibodies were eluted by CH3COONH4 (pH 3) and neutralized in 1 M Tris (pH 7.5). The specificity of the purified antibodies was tested in ELISA against (1) the domain used for affinity purification, (2) other VAR2CSA domains, and (3) glutamine rich protein (GLURP) [55]. Affinity-purified antibodies used for PepScan analysis were all negative in ELISA against control proteins and positive against the homologous domain (unpublished data).
Surface reactive antibodies in a pool of pregnancy plasma (from 15 pregnant women from Korogwe, Tanzania) were depleted using a parasite line selected for VAR2CSA expression using VAR2CSA specific antibodies [9]. In brief, 40 μl of the plasma pool were incubated with 2.0 × 108 MACS purified intact late stage trophozoite- and schizont-infected erythrocytes for 20 min at 4 °C. Hereafter, the cells were centrifuged at 800 g for 8 min, and the supernatant used to suspend a new pellet of 2.0 × 108 infected erythrocytes. This procedure was repeated four times. The depletion of surface reactive antibodies from the plasma pool was confirmed using a flow cytometry assay [9] after the final depletion.
3-D modeling of the 3D7 DBL3X domain.
The 3-D structure of the 3D7 sequence (PFL0030c aa 1217–1559) was modeled using the HHpred server with default settings [56]. Briefly, the HHpred method is specialized in remote homology detection using hidden Markow models (HMMs) built from PSI-BLAST profiles and secondary structure. The crystal structure of EBA-175 F1 (Protein Data Bank code 1ZRO chain A, [15]) was used as template and had the highest sequence and secondary structure alignment scores. The HHpred alignment was corrected in a short template loop sequence (Figure S1, positions 215–219) positioned next to a gap. The correction shifted the position of the gap and allowed for the modeling of a disulfide bridge in DBL3X, which was conserved in the EBA-175 F1, F2, and Pkα-DBL domains. HHpred HMMs for DBL3X and the template continued to match. Finally the corrected alignment was used to generate a 3-D model using MODELLER [57] with the protocol setup in the HHpred server toolkit. A superimposition of the EBA-175 F1 structure and the DBL3X model was obtained by the HHpred toolkit. Naccess version 2.1.1 [58] was used to calculate relative surface-exposed areas (RSAs) in single chains of EBA-175 F1, F2, and the Pkα-DBL domain [16]. The MAMMOTH-mult alignment server [59] was used to make a multiple structure superimposition of DBL3X model on the EBA-175 F1 and F2 DBL domains [15] and the Pkα-DBL domain [16]. The resulting alignment was inspected to identify conserved positions of cysteines and buried hydrophobic residues (RSA < 30%). Structural visualizations were made using PyMol [60].
Supporting Information
Cysteines in conserved disulfide bonds are highlighted in blue, conserved tryptophans buried in the structures are shown in red, and other conserved, buried, and hydrophobic positions are shown in green.
(51 KB PTT)
(A) Plasma from a rabbit immunized with a DBL5 construct was tested in 1:1000 on the VAR2CSA PepScan array. The upper diagram is the raw PepScan values with the PepScan score (y-axis) for each of the 420 peptides (x-axis).
(B) The same PepScan scores as (A) but calculated using a motif algorithm assigning a value to each amino acid.
(160 KB PPT)
Accession Numbers
All sequence data are available at GenBank (http://www.ncbi.nlm.nih.gov/Genbank) under the accession numbers DQ995590–DQ995632.
Acknowledgments
We acknowledge the women who donated serum and parasites for this study. We thank Gilean McVean for advice regarding recombination analysis, Kristoffer Rapacki for help with logo generation, and Anne Corfitz for excellent technical assistance. PepScan systems (the Netherlands) are thanked for technical assistance on the data work on the serum PepScan data.
Abbreviations
- bp
base pair
- CSA
chondroitin sulphate A
- DBL
Duffy-binding-like
- DKL
Kullback-Leibler distance
- dN
non-synonymous
- dS
synonymous
- EBA
erythrocyte-binding antigen
- ELISA
enzyme-linked immunosorbent assay
- IgG
immunoglobulin gamma
- PAM
pregnancy-associated malaria
- PfEMP1
Plasmodium falciparum erythrocyte membrane protein 1
- Pkα-DBL
Plasmodium knowlesi α-Duffy-binding-like
- VNTR
variable number of tandem repeats
Footnotes
Competing interests. The authors have declared that no competing interests exist.
Author contributions. MD, TSR, AGP, TGT, and AS conceived and designed the experiments. MD, TSR, MAN, MR, and AS performed the experiments. MD, TSR, PHA, OL, TGT, and AS analyzed the data. MD, PHA, NTN, LT, PD, and LH contributed reagents/materials/analysis tools. MD, TSR, PHA, and AS wrote the paper.
Funding. This study was primarily funded by the European Malaria Vaccine Initiative grant 0012–2004. This study also received financial support from the Danish Medical Research Council (SSVF) (grants 22-02-0220 and 22-03-0333), the Danish Research Council for Development Research (RUF) (grant 104.Dan.8.L.306 and 8.L.306), and the Danish National Research Foundation (Danish Platform for Integrative Biology). AS is supported by a postdoctoral grant from SSVF. MD is supported by a PhD studentship from RUF. MAN is supported by a postdoctoral grant from Hovedstadens Sygehusfaellesskab. NTN is supported by a postdoctoral grant from the Fondation Recherche Médicale.
References
- Christophers SR. The mechanism of immunity against malaria in communities living under hyper-endemic conditions. Ind J Med Res. 1924;12:273–294. [Google Scholar]
- Brabin BJ. An analysis of malaria in pregnancy in Africa. Bull World Health Organ. 1983;61:1005–1016. [PMC free article] [PubMed] [Google Scholar]
- Walter PR, Garin Y, Blot P. Placental pathologic changes in malaria. A histologic and ultrastructural study. Am J Pathol. 1982;109:330–342. [PMC free article] [PubMed] [Google Scholar]
- Bray RS, Anderson MJ. Falciparum malaria and pregnancy. Trans R Soc Trop Med Hyg. 1979;73:427–431. doi: 10.1016/0035-9203(79)90170-6. [DOI] [PubMed] [Google Scholar]
- McGregor IA, Wilson ME, Billewicz WZ. Malaria infection of the placenta in Gambia, West Africa: Its incidence and relationship to stillbirth, birth-weight, and placental weight. Trans R Soc Trop Med Hyg. 1983;77:232–244. doi: 10.1016/0035-9203(83)90081-0. [DOI] [PubMed] [Google Scholar]
- Fried M, Duffy PE. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science. 1996;272:1502–1504. doi: 10.1126/science.272.5267.1502. [DOI] [PubMed] [Google Scholar]
- Salanti A, Staalsoe T, Lavstsen T, Jensen AT, Sowa MP, et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- Tuikue Ndam NG, Salanti A, Bertin G, Dahlback M, Fievet N, et al. High level of var2csa transcription by Plasmodium falciparum isolated from the placenta. J Infect Dis. 2005;192:331–335. doi: 10.1086/430933. [DOI] [PubMed] [Google Scholar]
- Salanti A, Dahlback M, Turner L, Nielsen MA, Barfod L, et al. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J Exp Med. 2004;200:1197–1203. doi: 10.1084/jem.20041579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuikue Ndam NG, Salanti A, Le-Hesran JY, Cottrell G, Fievet N, et al. Dynamics of anti-VAR2CSA immunoglobulin G response in a cohort of Senegalese pregnant women. J Infect Dis. 2006;193:713–720. doi: 10.1086/500146. [DOI] [PubMed] [Google Scholar]
- Viebig NK, Gamain B, Scheidig C, Lepolard C, Przyborski J, et al. A single member of the Plasmodium falciparum var multigene family determines cytoadhesion to the placental receptor chondroitin sulphate A. EMBO J. 2005;6:775–781. doi: 10.1038/sj.embor.7400466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy MF, Maier AG, Byrne TJ, Marty AJ, Elliott SR, et al. VAR2CSA is the principal ligand for chondroitin sulfate A in two allogeneic isolates of Plasmodium falciparum. Mol Biochem Parasitol. 2006;148:117–124. doi: 10.1016/j.molbiopara.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Trimnell AR, Kraemer SM, Mukherjee S, Phippard DJ, Janes JH, et al. Global genetic diversity and evolution of var genes associated with placental and severe childhood malaria. Mol Biochem Parasitol. 2006;148:169–180. doi: 10.1016/j.molbiopara.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Gamain B, Trimnell AR, Scheidig C, Scherf A, Miller LH, et al. Identification of multiple chondroitin sulfate A (CSA)-binding domains in the var2CSA gene transcribed in CSA-binding parasites. J Infect Dis. 2005;191:1010–1013. doi: 10.1086/428137. [DOI] [PubMed] [Google Scholar]
- Tolia NH, Enemark EJ, Sim BK, Joshua-Tor L. Structural basis for the EBA-175 erythrocyte invasion pathway of the malaria parasite Plasmodium falciparum. Cell. 2005;122:183–193. doi: 10.1016/j.cell.2005.05.033. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hora R, Belrhali H, Chitnis CE, Sharma A. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature. 2006;439:741–744. doi: 10.1038/nature04443. [DOI] [PubMed] [Google Scholar]
- Howell DP, Samudrala R, Smith JD. Disguising itself: Insights into Plasmodium falciparum binding and immune evasion from the DBL crystal structure. Mol Biochem Parasitol. 2006;148:1–9. doi: 10.1016/j.molbiopara.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Blum ML, Down JA, Gurnett AM, Carrington M, Turner MJ, et al. A structural motif in the variant surface glycoproteins of Trypanosoma brucei. Nature. 1993;362:603–609. doi: 10.1038/362603a0. [DOI] [PubMed] [Google Scholar]
- Carrington M, Boothroyd J. Implications of conserved structural motifs in disparate trypanosome surface proteins. Mol Biochem Parasitol. 1996;81:119–126. doi: 10.1016/0166-6851(96)02706-5. [DOI] [PubMed] [Google Scholar]
- Tuikue Ndam NG, Fievet N, Bertin G, Cottrell G, Gaye A, et al. Variable adhesion abilities and overlapping antigenic properties in placental Plasmodium falciparum isolates. J Infect Dis. 2004;190:2001–2009. doi: 10.1086/425521. [DOI] [PubMed] [Google Scholar]
- Duffy MF, Caragounis A, Noviyanti R, Kyriacou HM, Choong EK, et al. Transcribed var genes associated with placental malaria in Malawian women. Infect Immun. 2006;74:4875–4883. doi: 10.1128/IAI.01978-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen AT, Magistrado P, Sharp S, Joergensen L, Lavstsen T, et al. Plasmodium falciparum associated with severe childhood malaria preferentially expresses PfEMP1 encoded by group A var genes. J Exp Med. 2004;199:1179–1190. doi: 10.1084/jem.20040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hviid L, Staalsoe T. Malaria immunity in infants: A special case of a general phenomenon? Trends Parasitol. 2004;20:66–72. doi: 10.1016/j.pt.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Lavstsen T, Magistrado P, Hermsen CC, Salanti A, Jensen AT, et al. Expression of Plasmodium falciparum erythrocyte membrane protein 1 in experimentally infected humans. Malar J. 2005;4:21. doi: 10.1186/1475-2875-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari-Guemouri S, Boudin C, Fievet N, Ndiaye P, Deloron P. Plasmodium falciparum genotype population dynamics in asymptomatic children from Senegal. Microbes Infect. 2006;8:1773–1670. doi: 10.1016/j.micinf.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Taylor HM, Kyes SA, Newbold CI. Var gene diversity in Plasmodium falciparum is generated by frequent recombination events. Mol Biochem Parasitol. 2000;110:391–397. doi: 10.1016/s0166-6851(00)00286-3. [DOI] [PubMed] [Google Scholar]
- Freitas-Junior LH, Bottius E, Pirrit LA, Deitsch KW, Scheidig C, et al. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- Mu J, Awadalla P, Duan J, McGee KM, Joy DA, et al. Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biol. 2005. e335. doi: 10.371/journal.pbio.0030335. [DOI] [PMC free article] [PubMed]
- Larsen JE, Lund O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2:2. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Bielawski JP, Yang Z. Widespread adaptive evolution in the human immunodeficiency virus type 1 genome. J Mol Evol. 2003;57:212–221. doi: 10.1007/s00239-003-2467-9. [DOI] [PubMed] [Google Scholar]
- Andersen PH, Nielsen M, Lund O. Prediction of residues in discontinuous B cell epitopes using protein 3-D structures. Protein Sci. 2006. In press. [DOI] [PMC free article] [PubMed]
- Holder AA, Guevara Patino JA, Uthaipibull C, Syed SE, Ling IT, et al. Merozoite surface protein 1, immune evasion, and vaccines against asexual blood stage malaria. Parassitologia. 1999;41:409–414. [PubMed] [Google Scholar]
- Blackman MJ, Scott-Finnigan TJ, Shai S, Holder AA. Antibodies inhibit the protease-mediated processing of a malaria merozoite surface protein. J Exp Med. 1994;180:389–393. doi: 10.1084/jem.180.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetteh KK, Cavanagh DR, Corran P, Musonda R, McBride JS, et al. Extensive antigenic polymorphism within the repeat sequence of the Plasmodium falciparum merozoite surface protein 1 block 2 is incorporated in a minimal polyvalent immunogen. Infect Immun. 2005;73:5928–5935. doi: 10.1128/IAI.73.9.5928-5935.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce RJ, Drakeley C, Chandramohan D, Mosha F, Roper C. Molecular determination of point mutation haplotypes in the dihydrofolate reductase and dihydropteroate synthase of Plasmodium falciparum in three districts of northern Tanzania. Antimicrob Agents Chemother. 2003;47:1347–1354. doi: 10.1128/AAC.47.4.1347-1354.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernersson R, Pedersen AG. RevTrans: Multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA. MrModeltest, version 2.2. Department of Systematic Zoology, Uppsala University; 2006. Available: http://www.ebc.uu.se/systzoo/staff/nylander.html. Accessed April 2006. [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: Advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond A. Tracer. Evolutionary Biology Group, University of Oxford. 2006. Available: http://evolve.zoo.ox.ac.uk/software.html?id=tracer. Accessed April 2006.
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Hasegawa M. Models of amino acid substitution and applications to mitochondrial protein evolution. Mol Biol Evol. 1998;15:1600–1611. doi: 10.1093/oxfordjournals.molbev.a025888. [DOI] [PubMed] [Google Scholar]
- Akaike H. Information theory and an extension of the maximum likelihood principle. In: Petrov BN, Csaki F, editors. Second international symposium on information theory. Budapest: Akademiai Kiado; 1973. pp. 267–281. [Google Scholar]
- Burnham KP, Anderson DR. Model selection and multimodel inference: A practical information-theoretic approach. New York: Springer-Verlag; 2002. 488 [Google Scholar]
- McVean GA, Myers SR, Hunt S, Deloukas P, Bentley DR, et al. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Two-locus sampling distributions and their application. Genetics. 2001;159:1805–1817. doi: 10.1093/genetics/159.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Clark AG. Principles of population genetics. 3rd edition. Sunderland (Massachusetts): Sinauer; 1997. 542 [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. 512 [Google Scholar]
- Dodoo D, Theisen M, Kurtzhals JA, Akanmori BD, Koram KA, et al. Naturally acquired antibodies to the glutamate-rich protein are associated with protection against Plasmodium falciparum malaria. J Infect Dis. 2000;181:1202–1205. doi: 10.1086/315341. [DOI] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Potterton L, Yuan F, van VH, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- Hubbard Simon J, Janet M. T. NACCESS [computer program]. University College of London: Department of Biochemistry and Molecular Biology; 1993. Available: http://wolf.bms.umist.ac.uk/naccess. Accessed July 2006. [Google Scholar]
- Lupyan D, Leo-Macias A, Ortiz AR. A new progressive-iterative algorithm for multiple structure alignment. Bioinformatics. 2005;21:3255–3263. doi: 10.1093/bioinformatics/bti527. [DOI] [PubMed] [Google Scholar]
- DeLano Warren. The PyMOL Molecular Graphics System. San Carlos (California): DeLano Scientific; 2002. Available: http://www.pymol.org. Accessed July 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cysteines in conserved disulfide bonds are highlighted in blue, conserved tryptophans buried in the structures are shown in red, and other conserved, buried, and hydrophobic positions are shown in green.
(51 KB PTT)
(A) Plasma from a rabbit immunized with a DBL5 construct was tested in 1:1000 on the VAR2CSA PepScan array. The upper diagram is the raw PepScan values with the PepScan score (y-axis) for each of the 420 peptides (x-axis).
(B) The same PepScan scores as (A) but calculated using a motif algorithm assigning a value to each amino acid.
(160 KB PPT)